Abstract
Motivation: MHC:peptide binding plays a central role in activating the immune surveillance. Computational approaches to determine T-cell epitopes restricted to any given major histocompatibility complex (MHC) molecule are of special practical value in the development of for instance vaccines with broad population coverage against emerging pathogens. Methods have recently been published that are able to predict peptide binding to any human MHC class I molecule. In contrast to conventional allele-specific methods, these methods do allow for extrapolation to uncharacterized MHC molecules. These pan-specific human lymphocyte antigen (HLA) predictors have not previously been compared using independent evaluation sets.
Result: A diverse set of quantitative peptide binding affinity measurements was collected from Immune Epitope database (IEDB), together with a large set of HLA class I ligands from the SYFPEITHI database. Based on these datasets, three different pan-specific HLA web-accessible predictors NetMHCpan, adaptive double threading (ADT) and kernel-based inter-allele peptide binding prediction system (KISS) were evaluated. The performance of the pan-specific predictors was also compared with a well performing allele-specific MHC class I predictor, NetMHC, as well as a consensus approach integrating the predictions from the NetMHC and NetMHCpan methods.
Conclusions: The benchmark demonstrated that pan-specific methods do provide accurate predictions also for previously uncharacterized MHC molecules. The NetMHCpan method trained to predict actual binding affinities was consistently top ranking both on quantitative (affinity) and binary (ligand) data. However, the KISS method trained to predict binary data was one of the best performing methods when benchmarked on binary data. Finally, a consensus method integrating predictions from the two best performing methods was shown to improve the prediction accuracy.
Contact: mniel@cbs.dtu.dk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Cytotoxic T lymphocytes (CTL) play a central role in defeating intracellular infections with pathogens, such as viruses and certain bacteria. The CTL T-cell receptor (TCR) recognizes foreign peptides in complex with major histocompatibility complex (MHC) class I molecules on the surface of the infected cells. MHC class I molecules preferably bind and present nine amino acid long peptides, which mainly originates from proteins expressed in the cytosol of the presenting cell. In most mammals, MHCs exist in a number of different allelic variants each of which binds to a specific and very limited set of peptides.
During the last decades, a number of prediction methods have been developed to identify which peptides will bind a given MHC molecule (see review Lundegaard et al., 2007). Today, the best methods are able to predict MHC class I binding with very high accuracy (Lin et al., 2008; Peters et al., 2006). The human class I version of MHC, the human lymphocyte antigen (HLA), is expressed by genes at three loci (HLA-A, -B, and -C) at chromosome 6. Several hundred alleles expressing MHCs with slightly different amino acid sequence are hosted at each locus, and each product potentially binds a different set of peptides. Most of the available data of MHC:peptide binding has been originating from a limited number of alleles and thus prediction systems have been biased towards these alleles. Lately, the number of available binding data has increased significantly both in terms of amount of different peptides and the number of different MHC alleles. This fact has in itself influenced the MHC prediction systems that now cover a large number of different HLA alleles (Peters et al., 2006). More interestingly, however, it has enabled the development of new so called pan-specific algorithms that can predict peptide binding to alleles for which limited or even no experimental data are available (Jacob and Vert, 2008; Jojic et al., 2006; Nielsen et al., 2007; Zhang et al., 2005). A common feature of these pan-specific methods is that they go beyond the conventional single allele approach, and take both the peptide sequence and the MHC contact environment into account. A large set of publicly available allele-specific MHC class I predictors that predict peptide binding only to the alleles on which they have been trained have been benchmarked latest by Peters et al. (2006) and Lin et al. (2008). The pan-specific predictors, on the other hand, have not to date been evaluated on a large independent dataset. The Immune Epitope database (IEDB) (Sette et al., 2005) is growing rapidly, and a large number of MHC class I binding data has become available since the training of the publicly available online versions of the pan-specific predictors. In this work, 6533 such novel quantitative peptide binding affinity measurements covering 33 different human MHC class I alleles were collected. As the resources necessary for biochemical determination of MHC:peptide affinity are significant, many of the data-points are presumably selected either based on other biological evidence or by use of in silico predictions like the ones tested in this work. It is known for a fact that NetMHC has been used to predict a number of data subsequently measured and deposited in the IEDB database. To test if this bias will give any difference in evaluation performance, a second evaluation set of 5137 data points covering 27 HLA-I alleles was created by removal of data-points identified by the authors of the NetMHC and NetMHCpan methods. For evaluation of the ability to distinguish between ligands and non-ligands, a set of 566 HLA ligands covering 34 HLA class I alleles was created by download from the SYFPEITHI database. For each evaluation dataset, peptide data used in the training of the NetMHC-3.0 (Buus et al., 2003; Nielsen et al., 2003) and NetMHCpan-1.0 (Nielsen et al., 2007) methods was removed. The benchmark was run against adaptive double threading (ADT) (Jojic et al., 2006), the artificial neural network (ANN)-based NetMHCpan-1.0 (Nielsen et al., 2007) and the SVM-based: kernel-based inter-allele peptide binding prediction system (KISS) (Jacob and Vert, 2008) methods. These methods are all publicly available via web interfaces. In addition, the quality of the pan-specific predictors was compared with one of the best available conventional allele-specific MHC predictor: NetMHC-3.0 (Buus et al., 2003; Lundegaard et al., 2008; Nielsen et al., 2003).
In many areas where different predictors have been developed, it has been demonstrated how consensus methods defined by averaging over several different predictions can improve the prediction accuracy beyond that of each individual predictors (Bujnicki et al., 2001). This is often due to the fact that the methods have been trained on different data and thus have learned potentially different features of the sequence space. However, also methods trained on the exact same data can contribute with non-redundant information and thus gain superiority by combination, as shown for example in Nielsen et al. (2007), Nielsen et al. (2003) and Petersen et al. (2000). Although such so-called meta predictors have been tried several times for MHC class II prediction (Huang et al., 2006; Karpenko et al., 2008; Mallios, 2001), no such scheme has to our knowledge been published using the wide diversity of MHC class I predictors. To investigate the possible benefits of such an approach, the predictions of the individual methods were compared with a consensus method defined by averaging the allele-specific MHC NetMHC-3.0 and pan-specific NetMHCpan-1.0 predictors.
2 MATERIALS AND METHODS
2.1 Datasets
The 9mer peptides associated with quantitative binding data were retrieved from the Immune Epitope Database and Analysis Resource (IEDB).
All peptides used in the training of NetMHC and NetMHCpan methods were discarded. This resulted in a dataset of 6533 experimental measurements covering 33 HLA-I alleles. These data were released after a point of time when the methods NetMHC-3.0, NetMHCpan-1.0 and ADT were trained and published. By doing so, it can be assured that the data had not been used in training of the above methods. This dataset is named EvaluationSet-1. Table 1 (panel A) gives an overview of dataset EvaluationSet-1.
Table 1.
Overview of dataset EvaluationSet-1, dataset EvaluationSet-2 and the SYFPEITHI dataset EvaluationSet-3
| Allele | # | Allele | # | Allele | # |
|---|---|---|---|---|---|
| Panel A: EvaluationSet-1 | |||||
| A0101 | 446 | A2902 | 329 | B3501 | 77 |
| A0201 | 442 | A3002 | 329 | B3901 | 106 |
| A0202 | 194 | A3101 | 224 | B4001 | 230 |
| A0203 | 193 | A3301 | 224 | B4002 | 92 |
| A0206 | 198 | A6801 | 224 | B4402 | 92 |
| A0301 | 329 | A6802 | 202 | B4403 | 92 |
| A1101 | 217 | B0702 | 231 | B4501 | 92 |
| A2301 | 329 | B0801 | 119 | B5101 | 77 |
| A2402 | 367 | B1501 | 114 | B5301 | 67 |
| A2403 | 111 | B1801 | 92 | B5401 | 66 |
| A2601 | 428 | B2705 | 98 | B5801 | 102 |
| Panel B: EvaluationSet-2 | |||||
| A0101 | 354 | A2601 | 305 | B3501 | 75 |
| A0201 | 347 | A2902 | 328 | B4001 | 85 |
| A0202 | 192 | A3002 | 328 | B4002 | 91 |
| A0203 | 191 | A3101 | 222 | B4402 | 91 |
| A0206 | 196 | A3301 | 222 | B4403 | 91 |
| A0301 | 213 | A6801 | 222 | B4501 | 91 |
| A1101 | 214 | A6802 | 200 | B5101 | 75 |
| A2301 | 328 | B0702 | 88 | B5301 | 66 |
| A2402 | 366 | B1801 | 91 | B5401 | 65 |
| Panel C: SYFPEITHI dataset EvaluationSet-3 | |||||
| A0101 | 1157 | A3001 | 669 | B3501 | 736 |
| A0201 | 3089 | A3002 | 92 | B4001 | 1078 |
| A0202 | 1447 | A3101 | 1869 | B4002 | 118 |
| A0203 | 1443 | A3301 | 1140 | B4402 | 119 |
| A0206 | 1437 | A6801 | 1141 | B4403 | 119 |
| A0301 | 2094 | A6802 | 1434 | B4501 | 114 |
| A1101 | 1985 | A6901 | 833 | B5101 | 244 |
| A2301 | 104 | B0702 | 1262 | B5301 | 254 |
| A2402 | 197 | B0801 | 708 | B5401 | 255 |
| A2403 | 254 | B1501 | 978 | B5701 | 59 |
| A2601 | 672 | B1801 | 118 | B5801 | 988 |
| A2902 | 160 | B2705 | 969 | ||
Allele, name of the alleles; #, number of peptides.
There was a concern that a significant part of the data deposited into IEDB dataset was selected for measurement by the NetMHC prediction method. In this way the dataset could be biased and in favor of NetMHC and possibly NetMHCpan as well. To test this, all data submitted to the IEDB database by the authors of the NetMHC and NetMHCpan methods, were removed and this resulted in a dataset containing 5137 data points covering 27 HLA-I alleles. This dataset is called EvaluationSet-2. Table 1 (panel B) summarizes the data in dataset EvaluationSet-2. Alleles in both datasets containing less than 10 data points were not included in the evaluation. Note, that both EvaluationSet-1 and EvaluationSet-2 have a very high ratio of binding versus non-binding peptides (50% on average). This highly non-biological fraction of binders (due to the procedure used to select peptides for experimental validation) will make the predictive performance values lower than on the biological data, where the ratio of binders to non-binders is of the order 1–2% (Nielsen et al., 2003; Yewdell and Bennink, 1999).
2.2 Endogenously presented peptides
Both the NetMHCpan and ADT methods were trained only on quantitative peptide MHC binding data. The KISS method on the other hand was trained on qualitative binding versus non-binding data from a broad range of data sources including IEDB (Sette et al., 2005), SYFPEITHI (Rammensee et al., 1999), Los Alamos HIV database (http://www.hiv.lanl.gov) and MHCBN (Bhasin et al., 2003). To test if this inherent difference in the training data would make the NetMHCpan and ADT perform advantageous on quantitative data and the KISS method favor qualitative data, an evaluation set of HLA ligands were downloaded from the SYFPEITHI database (Rammensee et al., 1999). Only data restricted to HLA alleles covered by all methods and not included in the training of any of the three methods NetMHCpan, ADT and KISS were included. This set consists of 566 HLA ligands restricted to 33 different HLA-A and -B alleles. For every peptide, the source protein was found in the UniProt database (The UniProt Consortium, 2008). The source protein was split into overlapping 9mer peptide sequences, and all peptides except the annotated HLA ligand were taken as negative. When using this definition of positive and negative peptides, one has to take into account that some peptides will falsely be classified as negatives because the SYFPEITHI database is incomplete. Since the MHC class I molecules are very specific, binding only a highly limited repertoire of peptides, this misclassified proportion will, however, be very small (Yewdell and Bennink, 1999). Further, the KISS method does not provide prediction values for peptides included in the training. These peptides were removed from the negative set. For each protein–HLA ligand pair, the predictive performance was estimated as the AUC value (Swets, 1988). This benchmark dataset is referred to as EvaluationSet-3, and is summarized in Table 1 (panel C).
All benchmark datasets 1–3 are available online at http://www.cbs.dtu.dk/immunology/pan-eval.php
2.3 Prediction methods
All predictions were made using public web interfaces with default parameters of the method in question. Table 2 gives a summary of the different methods in terms of data resources and release dates.
Table 2.
Prediction methods
| Method | Data source | Date |
|---|---|---|
| NetMHCpan-1.0 | IEDB, in house data | August, 2007 |
| NetMHC-3.0 | IEDB, in house data, SYFPEITHI | August, 2006 |
| ADT | IEDB | July, 2006 |
| KISS | IEDB, SYFPEITHI, LANL, and MHCDB | December, 2007 |
Data source gives the source of data used to train the method. Date gives date of publication.
NetMHC-3.0: http://www.cbs.dtu.dk/services/NetMHC/
NetMHCpan-1.0: http://www.cbs.dtu.dk/services/NetMHCpan-1.0/
Adaptive Double Threading:
http://atom.research.microsoft.com/hlabinding/hlabinding.aspx
KISS: http://cbio.ensmp.fr/kiss/
Due to the late release date of the KISS method, it is likely that some overlap between the present evaluation set and the training set exists. Indeed close to 3% of the data in the EvaluationSet-1 are present in the training data for the KISS method. It is hence likely that the performance of the KISS method was slightly overestimated.
2.4 Criteria for performance
Several performance measures can be used to evaluate the prediction performance of a given method. As a first assumption the methods included in the benchmark give predictions that are linearly proportional to the logarithm of the actual measurement of affinity. The intuitive assessment is, therefore, to evaluate the strength of this correlation. Such a quantifiable relationship can be found with the Pearson correlation coefficient (Pearson CC) (Press et al., 1992). In cases where there is no linear relation between the prediction scores and the binding affinity, it is more appropriate to use the Spearman's Rank-order correlation (Spearman's RC) (Press et al., 1992). For an indication of how well the methods can separate peptide binders from peptide non-binders, the area under the receiving operator characteristics curve (AUC) (Swets, 1988) was calculated using the generally accepted affinity threshold of 500 nM.
2.5 Statistical analysis of results
Binomial tests were used to evaluate the statistical significance of the observed difference in predictive performance between the different methods. P-values <0.05 were taken to define a significant result. For each dataset, the number of alleles where one method outperformed one another was counted. Performing a one-tailed binomial test based on this count, and the number of alleles (excluding ties) in the dataset gives the P-value for accepting the null-hypothesis that the first method does not outperform the other. On the other hand, a significant P-value leads to the rejection of the null-hypothesis and to the acceptance of the alternative hypothesis, which states that the first method does consistently outperform the other. Note that some alleles in the benchmark data share highly similar binding specificities, making the assumption that predictions for different alleles are independently false. This could imply that the quoted P-values in some case should be interpreted with caution.
2.6 Consensus method
A consensus method was defined as the simple average of the raw log-transformed prediction scores from the NetMHC-3.0 and NetMHCpan-1.0 methods.
3 RESULTS
The results are divided into three sections according to the dataset used for testing: EvaluationSet-1, EvaluationSet-2, and the HLA ligand set in EvaluationSet-3. Within the first two sections, tests were conducted with the three different criteria for performance: Pearson's CC, Spearman's RC and AUC. For the HLA ligand data only AUC was applied.
3.1 EvaluationSet-1
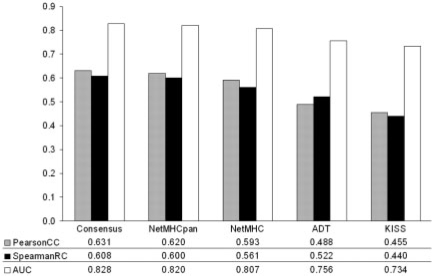
Figure 1 summarizes the predictive performance of the different methods on the data in EvaluationSet-1.
Fig. 1.
Benchmark performance for EvaluationSet-1. The average predictive performance for the different methods included in the benchmark in terms of the Pearson's CC, the Spearman's RC and the AUC.
The figure suggests the ranking of the different methods as follows: Consensus > NetMHCpan-1.0 > NetMHC-3.0 > ADT > KISS.
The ADT method appears to achieve a higher performance value when measured in terms of the Spearman's RC compared with the Pearson's CC. This could be explained by the fact that the prediction made by the method is a kind of unitless energy, which might not be linearly correlated to the logarithm of the binding affinity. For such methods, the Spearman's RC and the AUC values are therefore more appropriate measures of predictive performance.
The details of the evaluation as estimated in terms of the AUC values is shown in Table 3 (data for the Pearson's CC and Spearman's RC is shown in Supplementary Table S1a and c, respectively).
Table 3.
The performance for EvaluationSet-1 measured in terms of the AUC
| Allele | # | MHC | Pan | ADT | KISS | Cons | #b |
|---|---|---|---|---|---|---|---|
| A0101 | 446 | 0.931 | 0.913 | 0.848 | 0.880 | 0.927 | 148 |
| A0201 | 442 | 0.921 | 0.923 | 0.871 | 0.858 | 0.927 | 1331 |
| A0202 | 194 | 0.738 | 0.768 | 0.699 | 0.658 | 0.755 | 649 |
| A0203 | 193 | 0.896 | 0.890 | 0.750 | 0.730 | 0.897 | 761 |
| A0206 | 198 | 0.776 | 0.768 | 0.727 | 0.495 | 0.780 | 741 |
| A0301 | 329 | 0.781 | 0.803 | 0.777 | 0.753 | 0.798 | 615 |
| A1101 | 217 | 0.878 | 0.887 | 0.848 | 0.822 | 0.889 | 762 |
| A2301 | 329 | 0.803 | 0.825 | 0.776 | 0.603 | 0.820 | 96 |
| A2402 | 367 | 0.831 | 0.825 | 0.775 | 0.811 | 0.832 | 131 |
| A2403 | 111 | 0.862 | 0.916 | 0.905 | 0.825 | 0.919 | 85 |
| A2601 | 428 | 0.908 | 0.921 | 0.789 | 0.843 | 0.922 | 93 |
| A2902 | 329 | 0.746 | 0.755 | 0.736 | 0.685 | 0.766 | 68 |
| A3002 | 329 | 0.650 | 0.741 | 0.675 | 0.591 | 0.726 | 29 |
| A3101 | 224 | 0.865 | 0.877 | 0.866 | 0.817 | 0.873 | 489 |
| A3301 | 224 | 0.926 | 0.923 | 0.886 | 0.834 | 0.931 | 184 |
| A6801 | 224 | 0.880 | 0.891 | 0.890 | 0.829 | 0.890 | 498 |
| A6802 | 202 | 0.865 | 0.885 | 0.841 | 0.707 | 0.884 | 397 |
| B0702 | 231 | 0.812 | 0.818 | 0.677 | 0.715 | 0.822 | 268 |
| B0801 | 119 | 0.777 | 0.790 | 0.661 | 0.642 | 0.814 | 31 |
| B1501 | 114 | 0.769 | 0.726 | 0.677 | 0.757 | 0.749 | 272 |
| B1801 | 92 | 0.782 | 0.787 | 0.813 | 0.713 | 0.798 | 81 |
| B2705 | 98 | 0.884 | 0.856 | 0.717 | 0.764 | 0.882 | 88 |
| B3501 | 77 | 0.795 | 0.773 | 0.733 | 0.672 | 0.787 | 274 |
| B3901 | 106 | 0.779 | 0.759 | 0.535 | 0.632 | 0.810 | 36 |
| B4001 | 230 | 0.883 | 0.907 | 0.789 | 0.764 | 0.912 | 72 |
| B4002 | 92 | 0.623 | 0.766 | 0.751 | 0.662 | 0.717 | 39 |
| B4402 | 92 | 0.784 | 0.784 | 0.604 | 0.738 | 0.818 | 44 |
| B4403 | 92 | 0.736 | 0.772 | 0.582 | 0.683 | 0.798 | 34 |
| B4501 | 92 | 0.627 | 0.619 | 0.671 | 0.729 | 0.661 | 49 |
| B5101 | 77 | 0.698 | 0.731 | 0.676 | 0.630 | 0.717 | 85 |
| B5301 | 67 | 0.711 | 0.803 | 0.693 | 0.734 | 0.778 | 106 |
| B5401 | 0.893 | 0.878 | 0.898 | 0.844 | 0.898 | 81 | |
| B5801 | 102 | 0.832 | 0.777 | 0.748 | 0.789 | 0.817 | 162 |
| Ave | 6533 | 0.807 | 0.820 | 0.754 | 0.734 | 0.828 | 8799 |
Allele, name of alleles; #, number of peptides in the evaluation set; MHC, NetMHC-3.0; Pan, NetMHCpan-1.0; Cons, the consensus method; #b, number of binders in the data used for NetMHC-3.0 training. The best performing method is highlighted in bold for each allele.
From the data in Table 3, it is clear that not one single method was consistently superior in predictive performance to all other methods. By way of example, the NetMHCpan-1.0 method outperformed NetMHC-3.0 for 20 of the 33 alleles, but had a lower or equal performance for the remaining 13 cases.
By performing one-tailed binomial tests on the difference in performance scores, the significance of such observed differences in predictive performance could be assessed. In doing so, the following ranking of the different methods as measured in terms of the AUC values was found. (Similar results were found for the Pearson's and Spearman's Rank correlation).
Consensus > NetMHCpan-1.0 ≥ NetMHC-3.0 > ADT ≥ KISS Here ‘>’ indicates a significant difference (P < 0.05) and ‘≥’ indicates a non-significant difference. The methods can based on this be grouped into (Consensus), (NetMHCpan-1.0, NetMHC-3.0) and (ADT, KISS). The grouping indicates that the Consensus method performed significantly better than both NetMHCpan-1.0 and NetMHC-3.0, and both of the latter performed significantly better than any of the rest; ADT and KISS performed at the same level. Details of the statistical analysis are found in Supplementary Table S1b, d and f.
3.2 EvaluationSet-2
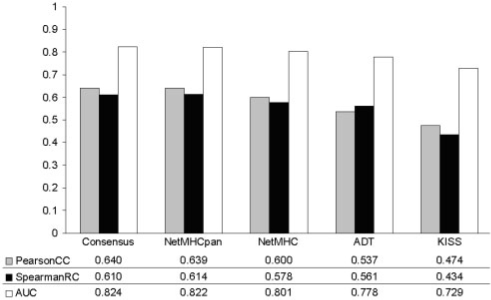
Figure 2 summarizes the predictive performance of the different methods on the data in EvaluationSet-2.
Fig. 2.
Benchmark performance for EvaluationSet-2. The average predictive performance for the different methods included in the benchmark in terms of the Pearson's CC, the Spearman's RC and the AUC.
The figure gives the average predictive performance for the different methods included in the benchmark in terms of the Pearson's CC, the Spearman's RC and the AUC. Details of the evaluation in terms of the AUC values are shown in Table 4 (further evaluation performance values are shown in Supplementary Tables S2a, c and e, respectively).
Table 4.
The performance for EvaluationSet-2 measured in terms of the AUC
| Allele | # | MHC | Pan | ADT | KISS | Cons | #b |
|---|---|---|---|---|---|---|---|
| A0101 | 354 | 0.925 | 0.910 | 0.859 | 0.872 | 0.921 | 148 |
| A0201 | 347 | 0.916 | 0.918 | 0.866 | 0.847 | 0.921 | 1331 |
| A0202 | 192 | 0.742 | 0.773 | 0.702 | 0.659 | 0.760 | 649 |
| A0203 | 191 | 0.892 | 0.885 | 0.746 | 0.733 | 0.892 | 761 |
| A0206 | 196 | 0.779 | 0.773 | 0.730 | 0.497 | 0.784 | 741 |
| A0301 | 213 | 0.834 | 0.858 | 0.840 | 0.754 | 0.852 | 615 |
| A1101 | 214 | 0.885 | 0.887 | 0.848 | 0.828 | 0.892 | 762 |
| A2301 | 328 | 0.802 | 0.824 | 0.775 | 0.601 | 0.819 | 96 |
| A2402 | 366 | 0.831 | 0.825 | 0.775 | 0.810 | 0.831 | 131 |
| A2601 | 305 | 0.828 | 0.849 | 0.793 | 0.721 | 0.844 | 93 |
| A2902 | 328 | 0.746 | 0.754 | 0.736 | 0.688 | 0.765 | 68 |
| A3002 | 328 | 0.649 | 0.740 | 0.674 | 0.589 | 0.724 | 29 |
| A3101 | 222 | 0.863 | 0.875 | 0.865 | 0.818 | 0.871 | 489 |
| A3301 | 222 | 0.926 | 0.922 | 0.887 | 0.835 | 0.931 | 184 |
| A6801 | 222 | 0.879 | 0.890 | 0.889 | 0.829 | 0.889 | 498 |
| A6802 | 200 | 0.865 | 0.884 | 0.842 | 0.707 | 0.883 | 397 |
| B0702 | 88 | 0.790 | 0.795 | 0.723 | 0.701 | 0.798 | 268 |
| B1801 | 91 | 0.783 | 0.788 | 0.815 | 0.715 | 0.800 | 81 |
| B3501 | 75 | 0.793 | 0.769 | 0.729 | 0.672 | 0.783 | 274 |
| B4001 | 85 | 0.838 | 0.927 | 0.820 | 0.781 | 0.904 | 72 |
| B4002 | 91 | 0.629 | 0.767 | 0.748 | 0.672 | 0.720 | 39 |
| B4402 | 91 | 0.787 | 0.784 | 0.597 | 0.736 | 0.818 | 44 |
| B4403 | 91 | 0.742 | 0.772 | 0.575 | 0.680 | 0.800 | 34 |
| B4501 | 91 | 0.621 | 0.624 | 0.683 | 0.729 | 0.657 | 49 |
| B5101 | 75 | 0.692 | 0.725 | 0.674 | 0.635 | 0.710 | 85 |
| B5301 | 66 | 0.708 | 0.803 | 0.690 | 0.730 | 0.777 | 106 |
| B5401 | 65 | 0.890 | 0.874 | 0.898 | 0.840 | 0.895 | 81 |
| Ave | 190 | 0.801 | 0.822 | 0.770 | 0.729 | 0.824 | 8125 |
Allele, name of alleles; #, number of evaluation set; MHC, NetMHC-3.0; Pan, NetMHCpan-1.0; Cons, the consensus method; #b, number of binders in the data used for NetMHC-3.0 training. The best performing method is highlighted in bold for each allele.
For the results shown in Figure 2, the following rank of the different prediction methods was found
Consensus > NetMHCpan-1.0 > NetMHC0-3.0 > ADT > KISS
The significance of this ranking was assessed using a one-tailed binomial test. In terms of the AUC values, the following results were obtained;
Consensus ≥ NetMHCpan-1.0 > NetMHC-3.0 > ADT > KISS
where ‘>’ indicates a significant difference (P < 0.05) and ‘≥’ indicates a non-significant difference. The methods could therefore be grouped as (Consensus, NetMHCpan-1.0), (NetMHC-3.0), (ADT) and (KISS). For the EvaluationSet-2, the analysis thus did not show a significant difference between the Consensus and NetMHCpan-1.0 methods. However, all neural network methods (Consensus, NetMHCpan-1.0 and NetMHC-3.0) were, also for this benchmark, shown to significantly outperform both ADT and KISS methods. Details of the analysis are found in Supplementary Table S2b, d and f.
3.3 Training performance
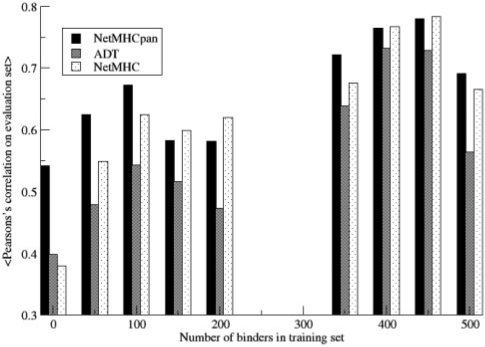
It is clear from the performance values on the two evaluation sets, that the overall ranking of the different pan-specific methods included in the benchmark for quantitative predictions is Consensus,NetMHCpan-1.0, ADT and KISS. In order to investigate if the higher performance of the NetMHCpan-1.0 method is due to the quality and size of the training data, the predictive performance of the NetMHCpan-1.0 and ADT methods were compared when trained on an identical dataset. The dataset and data partitioning were taken from the work by Peters et al. (2006). The dataset consists of more than 29 000 quantitative HLA peptide-binding data covering 35 HLA-A and HLA-B alleles. The performance was estimated in terms of the AUC value for each allele. The performance values for the ADT method were taken from the ADT server web site. The average performance values were 0.92 and 0.87 for the NetMHCpan-1.0 and ADT methods, respectively. The NetMHCpan-1.0 achieved the highest predictive performance for 34 of the 35 alleles, making this difference highly statistically significant (P < 10−5, binomial test). The details of the analysis are given in Supplementary Table S3. This result strongly indicates that the performance gain of the NetMHCpan-1.0 method is not solely due to a difference in the training data, but also is caused by the algorithmic differences between the two methods. This analysis can further be extended to demonstrate the power of the pan-specific methods to interpolate binding motif information from neighboring HLA molecules, enabling prediction of binding motifs for HLA molecules for which no or only limited binding data exist. Such an analysis has been carried out in detail in the original NetMHCpan-1.0 publication (Nielsen et al., 2007), where it was demonstrated how a pan-specific prediction method could accurately be used to describe the binding motif for hereto uncharacterized HLA molecules. To give further support for the power of the pan-specific approach, Figure 3 shows the predictive performance of the NetMHCpan-1.0, ADT and NetMHC-3.0 methods when all methods are trained on the Peters et al. (2006) dataset and evaluated on the EvaluationSet-1. This figure displays a histogram of the predictive performance of the three methods for the different alleles in the evaluation set as a function of the number of peptide binders in the training data for each allele.
Fig. 3.
Histogram of the average predictive performance of the alleles in the EvaluationSet-1 as a function of the numner of binding pepdite in the training data for same allele. Each methods is trained on the data in the Peters et al. (2006) dataset.
The figure clearly demonstrates that for alleles characterized by a large set of peptide binders (>100) both single allele (NetMHC-3.0) and pan-specific (NetMHCpan-1.0) methods achieved similar predictive accuracy. For alleles characterized by no or limited binding data in the training, the figure on the other hand show the power of the pan-specific NetMHCpan-1.0 method. For these alleles, the single allele method, NetMHC-3.0 failed to produce accurate predictions whereas the pan-specific method, NetMHCpan-1.0, maintained a high accuracy. One extreme allele is the HLA-B *3901 that is absent from the training data. This allele is naturally unpredictable by the NetMHC-3.0 method, but is predicted with an accuracy of 0.26 by the NetMHCpan-1.0 method (data not shown). These results thus clearly demonstrate the power of the pan-specific methods to go beyond the data in the training set and accurately predict binding also for uncharacterized HLA molecules. Intriguingly, it seems that the lower predictive performance of the ADT method is consistent and independent of the number of peptide binding examples in the training data, suggesting that the linear algorithm underlying the ADT method has poorer predictive power compared with the higher order neural network algorithm underlying the NetMHCpan-1.0 method.
To allow for a comparison of the predictive performance of the different methods, a Table S4 was included in Supplementary Material comparing the predictive performance as stated in the original publication, and the corresponding evaluation performance for each allele in EvaluationSet-1. The analysis of the data demonstrates that all methods have around 10% drop in predictive performance when comparing the evaluation set to the training set.
3.4 HLA ligand data
Next, the performance on the HLA ligand EvaluationSet-3 was analyzed. The results of this analysis are shown in Table 5.
Table 5.
Predictive performance for the HLA ligand dataset EvaluationSet-3 dataset as measured in terms of the AUC.
| Allele | N | pan | ADT | KISS |
|---|---|---|---|---|
| A0201 | 104 | 0.955 | 0.925 | 0.948 |
| A0207 | 4 | 0.961 | 0.867 | 0.933 |
| A0301 | 5 | 0.986 | 0.977 | 0.987 |
| A1101 | 3 | 0.959 | 0.966 | 0.889 |
| A2402 | 2 | 0.998 | 0.988 | 0.998 |
| A2602 | 2 | 0.903 | 0.809 | 0.989 |
| A2603 | 1 | 0.807 | 0.898 | 0.977 |
| A3101 | 3 | 0.981 | 0.978 | 0.982 |
| A3301 | 2 | 0.918 | 0.798 | 0.632 |
| A6602 | 7 | 0.956 | 0.989 | 0.978 |
| A6603 | 2 | 0.999 | 0.999 | 0.996 |
| A6801 | 7 | 0.992 | 0.987 | 0.993 |
| A6802 | 1 | 0.999 | 0.999 | 0.994 |
| B0702 | 14 | 0.990 | 0.982 | 0.986 |
| B0801 | 20 | 0.986 | 0.829 | 0.962 |
| B0802 | 3 | 0.997 | 0.924 | 1.000 |
| B1509 | 1 | 0.866 | 0.842 | 1.000 |
| B1801 | 35 | 0.993 | 0.990 | 0.992 |
| B2702 | 2 | 0.989 | 0.987 | 0.996 |
| B2703 | 9 | 0.992 | 0.964 | 0.987 |
| B2704 | 23 | 0.985 | 0.951 | 0.984 |
| B2705 | 58 | 0.985 | 0.966 | 0.987 |
| B2706 | 25 | 0.980 | 0.945 | 0.980 |
| B2709 | 26 | 0.982 | 0.940 | 0.984 |
| B3901 | 61 | 0.967 | 0.563 | 0.884 |
| B4001 | 4 | 0.998 | 0.987 | 0.983 |
| B4101 | 12 | 0.978 | 0.915 | 0.689 |
| B4402 | 27 | 0.988 | 0.927 | 0.983 |
| B4403 | 1 | 0.970 | 0.967 | 0.997 |
| B4501 | 4 | 0.975 | 0.937 | 0.990 |
| B4701 | 18 | 0.909 | 0.604 | 0.789 |
| B4901 | 101 | 0.992 | 0.903 | 0.861 |
| B5001 | 8 | 0.990 | 0.955 | 0.801 |
| B5101 | 1 | 1.000 | 1.000 | 1.000 |
| Ave per allele | 34 | 0.968 | 0.919 | 0.945 |
| Ave per protein | 566 | 0.976 | 0.886 | 0.931 |
Allele gives the allele name. N gives the number of HLA ligands included in the benchmark for each allele. Pan (NetMHCpan-1.0), ADT and KISS give the AUC values averaged over all ligand:protein pairs for a given HLA allele for each of the three prediction methods included in the benchmark. Ave per allele gives the average of the per-allele performance values. Ave per protein gives the average over all 566 ligand:protein pairs included in the benchmark. For each allele, the best performing method is highlighted in bold.
From this table, it is apparent that the KISS method did perform significantly better when it came to HLA ligand identification compared with the prediction of quantitative peptide binding affinities. Taking the average performance per allele, the rank of the three methods is
NetMHCpan-1.0 > (≥) KISS > ADT
Where ‘>’ as before indicates a significant difference (P < 0.05), and ‘≥’ indicates a non-significant difference. The KISS method thus significantly outperformed the ADT method when it came to HLA ligand identification. When looking at the performance per protein, the NetMHCpan-1.0 method significantly outperformed both the other methods (>). However, when grouping the data for the different alleles did the KISS and NetMHCpan-1.0 methods achieve similar predictive performances (≥). There are many possible reasons for this difference in relative rank of the different methods depending on the type of evaluation data. The most important reason is probably the source of training data. The KISS method was trained on HLA ligand data, and thus implicitly incorporates both the potential bias in the data imposed by the experimental method used to detect the ligands as well as signals from other players in the MHC class I presentation pathway like TAP and proteasome. This additional information is not incorporated into the NetMHCpan-1.0 and ADT method, since these are trained only on in vitro derived quantitative peptide:MHC binding data.
4 DISCUSSION
The large amount of peptide binding data made available to the scientific community has recently made the development of so-called pan-specific MHC-binding methods possible. Such methods are capable of providing accurate prediction of the peptide binding strength to any MHC molecule of known protein sequence. Large-scale benchmark evaluations of prediction algorithms for peptide:MHC binding have been performed in several studies for allele-specific methods (method trained on single allele data) (Bui et al., 2005; Lin et al., 2008; Nielsen et al., 2003; Peters et al., 2006; Yu et al., 2002). These studies have provided a ranking of the vast list of methods publicly available for peptide:MHC binding prediction and thereby aiding the non-expert user in selecting which prediction method to use for a given task.
For pan-specific methods, however, no such benchmark has been performed, and it remains unclear to what extent the publicly available methods differ in accuracy. Also, it has not, in an independent evaluation, been evaluated to what extent pan-specific methods can compete with allele-specific methods for alleles already represented in the peptide binding databases.
Here, such a large-scale benchmark has been carried out trying to answer these questions. The predictive performance of three publicly available pan-specific peptide:MHC binding prediction methods NetMHCpan-1.0 (Nielsen et al., 2007), ADT (Jojic et al., 2006) and KISS (Jacob and Vert, 2008) was compared on a large set of quantitative peptide MHC binding data downloaded from the IEDB database. The data were released on the IEDB database post-training of the NetMHCpan-1.0, and ADT methods. Further, to test if the data in the IEDB database had a potential bias due to large data submissions from the authors involved in the NetMHC-3.0 and NetMHCpan-1.0 methods, a second benchmark set was designed excluding data submitted to the IEDB by these authors. In the benchmark, we further included one of the best available allele-specific MHC binding prediction methods, NetMHC-3.0 (Buus et al., 2003; Nielsen et al., 2003) as well as a consensus method defined by integrating the prediction scores from the NetMHCpan-1.0 and NetMHC-3.0 methods. The KISS method was trained on qualitative (categorized) data and in order to investigate to what extent this has implications for which type of the data this method best predicts a qualitative benchmark dataset consisting of HLA ligands downloaded from the SYFPEITHI database was designed.
In both quantitative benchmark calculations it was found consistently that the Consensus and NetMHCpan-1.0 methods perform better than any of the other pan-specific methods. Further, the NetMHCpan-1.0 method performed noticeably better than NetMHC-3.0. Interestingly, the top ranking quantitatively trained method did perform better or equal to the KISS method trained on binary data when tested on this type of data (EvaluationSet-3). The strong predictive power of the pan-specific prediction methods was clearly illustrated in the benchmark calculation on HLA ligand data. In this benchmark, a large fraction of the HLA alleles (42%) were unknown to the NetMHCpan-1.0 method. For these alleles the NetMHCpan-1.0 method achieved an average predictive performance of 0.96 in terms of the AUC value. This AUC value translates into a false positive ratio of 0.04, meaning that in a protein of length 200 amino acids less than 10 peptides will have to be tested in order to identify the ligand.
It is clear when comparing the NetMHC-3.0 and NetMHCpan-1.0 performances on a single allele basis (see Table 2 and Supplementary Material) that the latter had a superior performance especially in the cases where the training data for that particular allele was scarce. For the data in the EvaluationSet-2, it was for instance found that the five alleles with maximal difference in predictive performance between theNetMHC-3.0 and the NetMHCpan-1.0 methods (HLA-B*4002, HLA-B*5301, HLA-A*3002, HLA-B*4001 and HLA-B*5101) all had a number of peptide binders in the training dataset that was consistently less than 100. The NetMHC-3.0 method thus achieved its poorest predictions for alleles covered by limited data (an average of 66 in the examples shown here). Similar observations were found using the data in EvaluationSet-1. Earlier work has shown a similar result, namely that MHC binding prediction algorithms rely on a sufficient number (in the order of 100) of peptide binders to be available in order to achieve high predictive value (Yu et al., 2002). This illustrates very nicely the limits faced when deriving allele-specific predictive methods and strength of the pan-specific approaches like NetMHCpan-1.0, to benefit from data of related alleles.
Funding
National Institute of Health (contracts HHSN2662004 00025C, HHSN266200400083C, and HHSN26620040006C); sixth Framework program of the European commission (grant LSHB-CT-2003-503231).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Bhasin M, et al. MHCBN: a comprehensive database of MHC binding and non-binding peptides. Bioinformatics. 2003;19:665–666. doi: 10.1093/bioinformatics/btg055. [DOI] [PubMed] [Google Scholar]
- Bui HH, et al. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–314. doi: 10.1007/s00251-005-0798-y. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM, et al. LiveBench-2: large-scale automated evaluation of protein structure prediction servers. PROTEINS: Structure, Function, and Genetics Suppl. 2001;5:184–191. doi: 10.1002/prot.10039. [DOI] [PubMed] [Google Scholar]
- Buus S, et al. Sensitive quantitative predictions of peptide-MHC binding by a ‘Query by Committee’ artificial neural network approach. Tissue antigens. 2003;62:378–384. doi: 10.1034/j.1399-0039.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- Huang L, et al. A meta-predictor for MHC class II binding peptides based on naive Bayesian approach. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006;1:5322–5325. doi: 10.1109/IEMBS.2006.259832. [DOI] [PubMed] [Google Scholar]
- Jacob L, Vert JP. Efficient peptide-MHC-I binding prediction for alleles with few known binders. Bioinformatics. 2008;24:358–366. doi: 10.1093/bioinformatics/btm611. [DOI] [PubMed] [Google Scholar]
- Jojic N, et al. Learning MHC I-peptide binding. Bioinformatics. 2006;22:e227–e235. doi: 10.1093/bioinformatics/btl255. [DOI] [PubMed] [Google Scholar]
- Karpenko O, et al. A probabilistic meta-predictor for the MHC class II binding peptides. Immunogenetics. 2008;60:25–36. doi: 10.1007/s00251-007-0266-y. [DOI] [PubMed] [Google Scholar]
- Lin HH, et al. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunology. 2008;9:8. doi: 10.1186/1471-2172-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C, et al. Modeling the adaptive immune system: predictions and simulations. Bioinformatics. 2007;23:3265–3275. doi: 10.1093/bioinformatics/btm471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C, et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36(Web Server issue):W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallios RR. Predicting class II MHC/peptide multi-level binding with an iterative stepwise discriminant analysis meta-algorithm. Bioinformatics. 2001;17:942–948. doi: 10.1093/bioinformatics/17.10.942. [DOI] [PubMed] [Google Scholar]
- Nielsen M, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS ONE. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B, et al. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput. Biol. 2006;2:e65. doi: 10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, et al. Prediction of protein secondary structure at 80% accuracy. Proteins. 2000;41:17–20. [PubMed] [Google Scholar]
- Press WH, et al. Numerical Recipies in C: The Art of Scientific Computing. Cambridge: Cambridge University Press; 1992. [Google Scholar]
- Rammensee H, et al. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- Sette A, et al. A roadmap for the immunomics of category A-C pathogens. Immunity. 2005;22:155–161. doi: 10.1016/j.immuni.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium The universal protein resource (UniProt) Nucleic Acids Res. 2008;36(Database issue):D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- Yu K, et al. Methods for prediction of peptide binding to MHC molecules: a comparative study. Mol. Med. 2002;8:137–148. [PMC free article] [PubMed] [Google Scholar]
- Zhang GL, et al. MULTIPRED: a computational system for prediction of promiscuous HLA binding peptides. Nucleic Acids Res. 2005;33:W172–W179. doi: 10.1093/nar/gki452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.