Abstract
Summary: The MiMI molecular interaction repository integrates data from multiple sources, resolves interactions to standard gene names and symbols, links to annotation data from GO, MeSH and PubMed and normalizes the descriptions of interaction type. Here, we describe a Cytoscape plugin that retrieves interaction and annotation data from MiMI and links out to multiple data sources and tools. Community annotation of the interactome is supported.
Availability: MiMI plugin v3.0.1 can be installed from within Cytoscape 2.6 using the Cytoscape plugin manager in ‘Network and Attribute I/0’ category. The plugin is also preloaded when Cytoscape is launched using Java WebStart at http://mimi.ncibi.org by querying a gene and clicking ‘View in MiMI Plugin for Cytoscape’ link.
Contact: dstates@umich.edu
1 INTRODUCTION
There has been an explosion of molecular interaction data in recent years, and relevant information is available from many sources. Unfortunately, these data sources are inconsistent in the syntax, nomenclature and semantics that they use. Further, the annotations provided by different sources vary in scope ranging from experimental technique to molecular context. The MiMI molecular interaction repository integrates and normalizes data from multiple sources using an intelligent merging approach based on alignment of hierarchical data structures (Jayapandian et al., 2007). In addition, MiMI annotates interactions using MeSH, OMIM and the Gene Ontology.
Cytoscape is a widely used open source software tool for the analysis of biomolecular interaction networks (Shannon et al., 2003). Cytoscape began as a standalone tool, but the power of Cytoscape is most apparent when it is coupled to databases of protein–protein, protein–DNA and genetic interactions. With plugins to query specific interaction databases (Cerami et al., 2006; Salwinski and Eisenberg, 2007), pathway repositories (Ferro et al., 2007; Le Fevre et al., 2007) and model organism repositories (Avila-Campillo et al., 2007; Shannon et al., 2006), Cytoscape can access a great deal of data, but integrating data between sources remains a major challenge. Here, we describe the MiMI plugin for Cytoscape to visualize, annotate and disseminate molecular interaction information retrieved from the MiMI integrated molecular interaction repository. MiMI normalizes data across many of the most widely used repositories for molecular interaction data and integrates interactions with multiple sources of annotation. The MiMI Plugin for Cytoscape is distributed using the Cytoscape Plugin Manager and is now one of the most popular downloads from the Cytoscape site.
2 OVERVIEW
2.1 MiMI plugin and MiMI database
The MiMI repository gathers data from multiple data sources (Supplementary Table S1) and uses an intelligent integration strategy that aligns hierarchical data structures on shared elements (‘deep merging’). Important enhancements to the MiMI resource include an RDBMS implementation to facilitate multi-user and arbitrary query functionality, the inclusion of KEGG and Reactome pathway data, resolution of many more molecular identifiers from source databases to standard gene names and the addition of MeSH as well as GO annotations on interactions. Annotation of interaction type varies between different source databases. Where possible, the MiMI database normalizes interaction types to the PSI standard Molecular Interaction Ontology (Orchard et al., 2007). Where source repositories specified interaction types not represented in the PSI ontology, extensions to the standard have been proposed.
The MiMI plugin communications with the MiMI database using a PHP server that acts as a database interface. The interface is implemented using stored procedures to modularize the code and avoid embedding of SQL queries in PHP scripts. In this way, the database query is isolated from the Cytoscape MiMI plugin. This allows changes and upgrades to be made to the database without affecting the MiMI plugin.
In order to improve plugin performance, we generate pre-computed tables for MiMI. Query processing and data retrieval based on precomputed tables takes ∼21 s for a network with 1004 nodes and 6234 edges with 15 types of node attributes, and 7 types of edge attributes. Queries were performed using Cytoscape v2.6.0 on a 3.4 GHz, Pentium 4 desktop with 2.0 GB of RAM running on Windows XP.
2.2 The MiMI plugin user interface
Users can begin a session by uploading interactions from the MiMI server using the MiMI plugin to specify a seed list of genes, optionally filtered by species, molecule type and data source. Alternatively, users can load a file containing gene name into the MiMI plugin. After the user has loaded a set of genes, they can use the MiMI plugin to retrieve different interaction views. The MiMI plugin allows users to query interactions by finding nearest neighbors. In addition, the plugin can optionally retrieve interactions only amongst the query set of genes, or between neighbors shared by more than one gene in the query set.
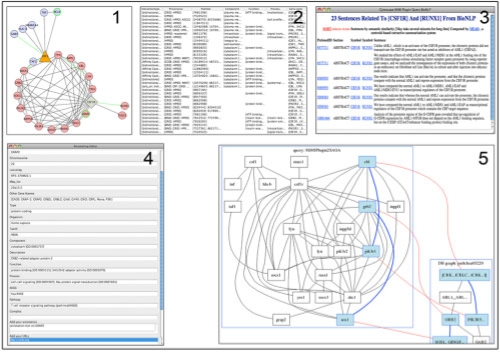
To facilitate navigation of complex networks, the MiMI plugin creates a custom visual style in Cytoscape that distinguishes between the seed nodes and neighboring nodes using different node colors and shapes. Users can iteratively extend a network by double clicking on selecting nodes of interest to identify interactions to these nodes that are not yet in the network and can collapse the extended network by double clicking the selected nodes in reverse order of extending (Fig. 1.1).
Fig. 1.
Panel 1 shows the csf1r interaction network with extended network on gene csf1. Panel 2 shows ‘provenance’ and ‘PubMed’ attributes of interaction. Panel 3 shows literature information retrieved from BioNLP database. Panel 4 is an annotation editor that shows the attributes from MiMI and the annotations added by the users. Panel 5 shows the match of KEGG pathway and selected network using SAGA.
The plugin implements an interactive annotation editor to view attribute information (Fig. 1.2) from the MiMI database, modify attribute values and add annotations and URLs to a node or an edge (Fig. 1.4). Users′ annotations are saved on the MiMI database server for individual users and automatically displayed when the user logs in again. User annotations can also be shared with other users to support collaborative network annotation. The node border color or edge color will be changed to green when users add their annotations to node/edge and make their annotations public.
The MiMI plugin extends the Cytoscape linkout to add links to the MiMI web site, Gene Ontology and MeSH annotations, and the Gene Interaction Network (GIN) service for users to compare and supplement information from MiMI.
Sentence-level PubMed text tagged by genes on the selected edge can be retrieved using the ‘BioNLP’ item from the Pop-Up menu (Fig. 1.3). The PubMed database is generated by a pipeline that processes articles retrieved from public abstract repositories (NCBI Pubmed/Pubmed Central) into a standardized format that identifies gene entities within individual sentences. The database is updated nightly. The sentence results can be ranked using a semantic similarity score computed by MEAD (Radev et al., 2001) by clicking ‘Sort’ in the text window.
Approximate graph match is another way that the MiMI plugin links out from Cytoscape. Users can define a subgraph in a network by the selected nodes and edges, and then apply SAGA (subgraph index for approximate graph alignment) (Tian et al., 2007) to identify similar subgraphs in the KEGG pathway database (Fig. 1.5).
Tutorials and case studies on the use of the MiMI plugin are available at http://mimiplugin.ncibi.org/.
Since the first release of the MiMI plugin in August 2007, there has been more than 2000 downloads and thenumber continues to grow. Feedback from this user community provides important guidance to improve both the plugin and MiMI resource.
Funding
National Institutes of Health (R01 LM008106, P41 RR018627 and U54 DA021519).
Conflict of Interest: none declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jeffery deWet and Marci Brandenburg for data review and their assistance in refining the MiMI plugin user interface, and the SAGA, MEAD and GIN groups in the NCIBI.
References
- Avila-Campillo I, et al. BioNetBuilder: automatic integration of biological networks. Bioinformatics. 2007;23:392–393. doi: 10.1093/bioinformatics/btl604. [DOI] [PubMed] [Google Scholar]
- Cerami EG, et al. cPath: open source software for collecting, storing, and querying biological pathways. BMC Bioinformatics. 2006;7:497. doi: 10.1186/1471-2105-7-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro A, et al. NetMatch: a Cytoscape plugin for searching biological networks. Bioinformatics. 2007;23:910–912. doi: 10.1093/bioinformatics/btm032. [DOI] [PubMed] [Google Scholar]
- Jayapandian M, et al. Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic Acids Res. 2007;35:D566–D571. doi: 10.1093/nar/gkl859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fevre F, et al. Cyclone: java-based querying and computing with pathway/genome databases. Bioinformatics. 2007;23:1299–1300. doi: 10.1093/bioinformatics/btm107. [DOI] [PubMed] [Google Scholar]
- Orchard S, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- Radev DR, et al. First Document Understanding Conference. New Orleans, LA: 2001. Experiments in single and multi-document summarization using MEAD. [Google Scholar]
- Salwinski L, Eisenberg D. The MiSink Plugin: Cytoscape as a graphical interface to the database of interacting proteins. Bioinformatics. 2007;23:2193–2195. doi: 10.1093/bioinformatics/btm304. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon PT, et al. The Gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, et al. SAGA: a subgraph matching tool for biological graphs. Bioinformatics. 2007;23:232–239. doi: 10.1093/bioinformatics/btl571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.