Abstract
Summary: TOPALi v2 simplifies and automates the use of several methods for the evolutionary analysis of multiple sequence alignments. Jobs are submitted from a Java graphical user interface as TOPALi web services to either run remotely on high-performance computing clusters or locally (with multiple cores supported). Methods available include model selection and phylogenetic tree estimation using the Bayesian inference and maximum likelihood (ML) approaches, in addition to recombination detection methods. The optimal substitution model can be selected for protein or nucleic acid (standard, or protein-coding using a codon position model) data using accurate statistical criteria derived from ML co-estimation of the tree and the substitution model. Phylogenetic software available includes PhyML, RAxML and MrBayes.
Availability: Freely downloadable from http://www.topali.org for Windows, Mac OS X, Linux and Solaris.
Contact: iain.milne@scri.ac.uk
1 INTRODUCTION
The statistical revolution in molecular phylogenetics (Felsenstein, 2001) continues to gather pace with the development of faster and/or more sophisticated maximum likelihood (ML) and Bayesian inference methods, with recent software implementations including PhyML (Guindon and Gascuel, 2003), RAxML (Stamatakis, 2006) and MrBayes (Ronquist and Huelsenbeck, 2003). To fully utilize these methods, it is also important to select a model of evolution with an appropriate level of complexity for the dataset by making use of model selection procedures, e.g. as in the ModelTest software (Posada and Crandall, 1998). However, there are opportunities for improved practice in model testing, particularly for protein-coding DNA where appropriate models for each codon position (CP) are usually required (Bofkin and Goldman, 2007) and are often underused (Shapiro et al., 2005). While biologists are being encouraged to adopt these improved methods (Whelan et al., 2000), there are still obstacles, including the lack of a single easy-to-use interface for analyzing a multiple sequence alignment (MSA) with a range of methods (e.g. model selection and phylogenetic tree estimation) and sophisticated access to computer power. While computational resources are becoming available for phylogenetic analysis (e.g. Dereeper et al., 2008), the interfaces available are basic and analysis options are often not stored for amendment.
TOPALi (tree TOPology-related analysis of ALignments Interface) version 1 (Milne et al., 2004) specialized in the detection of recombination in an MSA and ran only on desktops. We have extended and redesigned TOPALi to carry out additional analyses, particularly automated model selection linked to phylogenetic analyses, resulting in rich graphical output, and to utilize, via a sophisticated interface with access to previous option choices, the increased power of high performance computing (HPC) clusters and multi-core desktops. In particular, TOPALi v2 reduces the learning curve for biologists to carry out high quality phylogenetic analyses, by hiding the complexities of setting up and running many analyses.
2 FEATURES
Alignment handling: TOPALi can import/export DNA, RNA and protein MSAs in many formats and create DNA alignments from a protein MSA and corresponding unaligned DNA. Several MSAs can be stored within a TOPALi project allowing working with a group of related MSAs. TOPALi can quickly render alignments facilitating quality checks: the alignment overview shows the relative position of the zoomed region to the full alignment. The user can semi-automatically or manually select a reduced number of sequences for analysis and can also restrict the columns, e.g. exons could be extracted from a genomic alignment and saved as a new alignment.
Model selection: the menu launches models available in MrBayes (24 nucleotide models, 36 amino acid models) or in PhyML (56 and 40, respectively) or in RAxML (no nucleotide model choice, 40 amino acid models). The optimal model is automatically selected (and passed to the phylogenetic analysis launch menus) based on calculations involving either hierarchical likelihood ratio tests (hLRTs), Akaike information criterion (AIC), or Bayesian information criterion (BIC), generally following the ModelTest approach, except that (i) the model parameters (substitution, rate heterogeneity) and the phylogenetic tree are estimated by running a separate PhyML job for each model resulting in more accurate estimates of the log likelihood and derived quantities (AIC, BIC) and (ii) the hLRT tests among the 56 PhyML nucleotide models are based on single pairwise LRT tests. CP model selection treats the coding region as three separate alignments. Model components that are similar across CPs can be linked to share parameter estimation in subsequent MrBayes analysis.
Phylogenetic tree estimation: web services run the MrBayes, PhyML and RAxML programs on either nucleotide or protein MSAs. For nucleotide data, MrBayes analysis can use a model for all positions or a CP model. The user then accepts, or overrules the model selection choices and enters the MrBayes analyses settings (nRuns, nGenerations, Sample Frequency and Burn-in percentage). For ML analysis, PhyML offers only one model for all positions, so the user accepts or overrules the model selection choice and analysis settings (including number of bootstrap runs). RAxML has three rate heterogeneity models (including the Gamma distribution) but only one parameter-rich model (GTR) for nucleotide analysis, although the model parameters can be estimated separately for each CP. Tree manipulation tools include midpoint rooting and editing to simplify the display of support values.
3 IMPLEMENTATION
TOPALi's analysis methods have been designed to run either on remote HPC clusters (the default setting), or on standard desktops. With no underlying code duplication, we have devised a novel approach that runs tasks as independent processes within their own Java Virtual Machine (when executed on separate cluster nodes), or as semi-independent processes within a typical multithreaded application (when running on a desktop), managed by a local process manager that sets the number of CPUs to be used.
In addition to the obvious speed benefits, HPC usage also eliminates any compilation or configuration issues a user may encounter when running jobs locally as some sub-components of the analyses are handled by C or C++ programs from third parties, and must therefore be compiled for local use.
TOPALi is designed to be user-friendly and thus includes functionality that allows the user to work with a project locally (loading or examining alignments for instance) and then to submit one or more analysis jobs for remote processing. The client can be closed and reopened later, and the progress of the jobs will be updated from the server. Previously, completed jobs can also be reselected and a new job submission can be created that mirrors the settings from the original job, with or without further modifications.
Making use of a newly developed web services resource broker (I.Milne et al., manuscript in preparation), TOPALi queries the broker monitoring a pool of remote HPC clusters hosting TOPALi web services (currently at the Scottish Crop Research Institute and University of Dundee) that can then intelligently decide which cluster is most suitable for the job. We can also manage load by rejecting jobs submitted with very high numbers of sequences (on a per analysis basis). Readers who are interested in hosting TOPALi services on their own Sun Grid Engine enabled cluster (either for private or further public use) may contact us for advice on configuration.
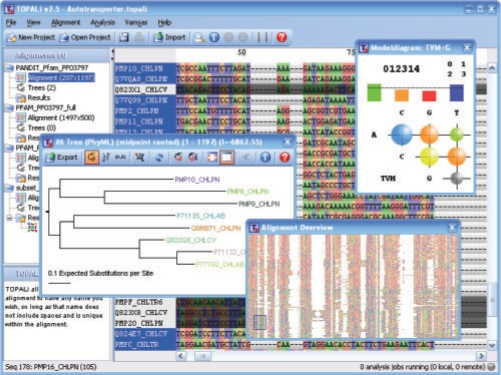
Fig. 1.
TOPALi's main interface showing alignment handling, tree estimation and model selection features.
TOPALi is coded in Java for platforms supporting Java version 1.5.0 and above. We provide installable versions with everything required to run the application, including a suitable Java runtime.
Funding: UK BBSRC/EPSRC Bioinformatics/E-science Initiative (BBSB16615); the Scottish Government; Scottish Funding Council; Scottish Enterprise.
Conflict of Interest: none declared.
REFERENCES
- Bofkin L, Goldman N. Variation in evolutionary processes at different codon positions. Mol. Biol. Evol. 2007;24:513–521. doi: 10.1093/molbev/msl178. [DOI] [PubMed] [Google Scholar]
- Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucl. Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. The troubled growth of statistical phylogenetics. Syst. Biol. 2001;50:465–467. doi: 10.1080/10635150119297. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Milne I, et al. TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics. 2004;20:1806–1807. doi: 10.1093/bioinformatics/bth155. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Shapiro B, et al. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RaxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Whelan S, et al. Molecular phylogenetics: state-of-the-art methods for looking into the past. Trends Genet. 2000;17:262–272. doi: 10.1016/s0168-9525(01)02272-7. [DOI] [PubMed] [Google Scholar]