Abstract
Summary: Oligonucleotide microarray probes are designed to match specific transcripts present in databases that are regularly updated. As a consequence probes should be checked every new database release. We thus developed an informatics tool allowing the semi-automatic update of probe collections of long oligonucleotides and applied it to the mouse RefSeq database.
Availability: http://www.bio.espci.fr/sol/
Contact: marie-claude.potier@espci.fr
Supplementary information: Supplementary data are available at http://www.bio.espci.fr/sol/
1 INTRODUCTION
The National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) database provides a curated non-redundant collection of sequences representing genomic data, transcripts and proteins for different species (Pruitt et al., 2007). Since the first release in July 2003, there has been 28 new releases (release 29, September 5, 2008). With the promise of whole-genome analysis methods, such as microarrays, the challenge now is to design specific probes for each exon of every single gene. These whole-genome probe collections will not be exhaustive and totally accurate unless gene databases are stable (Perez-Iratxeta and Andrade, 2005). In order to address the question of reliability of a probe collection every new database release, we have developed an informatics tool that allows the update of a probe collection. This tool was applied to the mouse RefSeq database. The starting probe set was designed on the sixth release of RefSeq using a new algorithm of Selection of OLigonucleotides (SOL), and RefSeq update has been followed until version 29.
2 RESULTS
2.1 SOL algorithm
The goal of SOL was to generate a database of all possible long oligonucleotide probes of a given length L that have the specificity fitted to the microarray hybridization conditions. For each identifier of mouse RefSeq, all the possible oligonucleotides of length L that did not contain any tetra-nucleotide repeat using a sliding window of L/2 were BLASTed against the mouse RefSeq database (−W=7 −F=F –E=100). Thermodynamic analysis of BLAST sequence alignment outputs allowed the selection of gene-specific probes by eliminating the ones that gave at least one cross-hybridization with a melting temperature Tm below the hybridization temperature (Thyb). Tms were computed using the nearest neighbour thermodynamic modelimplemented in the MELTING software (Le Novére, 2001) slightly modified to include the effect of formamide concentration, a widely used denaturing agent, in the hybridization solution. Formamide-corrected Tm (Tmf)=Tm−(0.75−GC%*0.0025) × %formamide. Finally, the Tm associated to the more stable non-target probe duplex will define the MAXTm parameter that will be used to characterize the probe specificity. In addition, probe Tm, GC content, probe position on the original RefSeq sequence and ΔG of the most stable probe secondary structure calculated using MFOLD-software (Zuker et al., 2003). Among all softwares for DNA microarray probe design, SOL resembles most to OLIGOARRAY2 (Rouillard et al., 2003): the oligonucleotide specificity is computed by considering the thermodynamic properties of its hybridization to non-specific targets. However, OLIGOARRAY2 provides the optimal probe for each transcript based on Tm, length and GC content in a database for distant users only (Le Brigand et al., 2006), while SOL and its interface for probe collection and update gives the list of all specific probes that are fully accessible on site after installation. SOL has been used to design a specific human chromosome 21 oligonucleotide microarray (Ait Yahya-Graison et al., 2007).
2.2 Interface to update oligonucleotide collections
One of the main drawbacks with oligonucleotide design is the difficulty to update oligonucleotide collections during time. We thus developed an interface based on SOL algorithm to follow each new release of the mouse RefSeq library. This interface is split into two parts: the first part is dedicated to administration tasks, such as library modifications; the second part is dedicated to user queries to update local databases.
2.2.1 Update of oligonucleotide specificity against RefSeq library
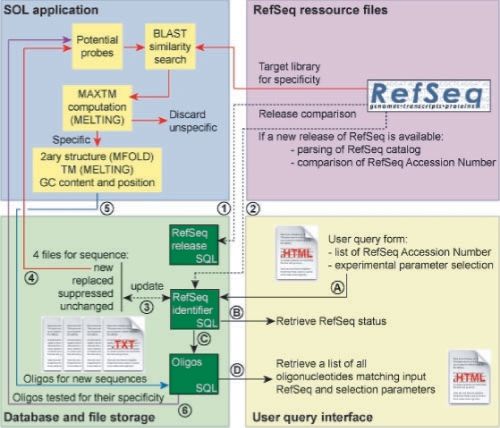
When a new release of the RefSeq library is detected (Fig. 1, arrow 1) as compared with our local database, RefSeq files are parsed and accession numbers are queried against the local database (Fig. 1, arrow 2). Four files containing new, unchanged, suppressed (permanently or temporarily) and replaced sequences are created (Fig. 1, arrow 3). In the case of replaced sequences, RefSeq FTP site gives the reference of the new sequence that replaces the old one. The RefSeq version of unchanged sequences is updated and the oligonucleotide status of the suppressed and replaced sequences is modified. For new identifiers, their corresponding sequences are used as an input to SOL algorithm to design oligonucleotide probes using the latest RefSeq target library for specificity (Fig. 1, arrow 4). Specific oligonucleotides are stored in the database (Fig. 1, arrow 5). Finally, SOL algorithm is launched using the older oligonucleotide collection (without the new ones) against the new RefSeq library (Fig. 1, arrow 6). Following these steps, we confirm that all the oligonucleotides found in the local database are specific against the latest RefSeq release. In addition, all RefSeq identifier modifications are followed to keep track of all changes concerning oligonucleotide design. An example is given for the mouse RefSeq database (Supplementary Material). When a user queries the database interface using a list of RefSeq accession numbers (A), RefSeq status for each identifier is retrieved (B). For each identifier available in the database, an SQL query retrieves all corresponding oligonucleotides (C). Next, a list of oligonucleotides that match the experimental parameters entered by the user is sent back to the browser (D).
Fig. 1.
SOL update functions in action. This figure shows how the oligonucleotide database is updated when a new release of RefSeq has been made available and how user can query the local database to follow the evolution of customized oligonucleotide collections. See text for details on how the SOL algorithm and the update interface are working. Dashed lines correspond to the update of RefSeq database and solid lines to the probe design and specificity calculations.
2.2.2 User interface to follow updates of a custom oligoset of oligonucleotide specificity against RefSeq library
When a user queries the database using a list of RefSeq accession numbers (Fig. 1, arrow A), RefSeq status is retrieved from the database for each submitted identifier (Fig. 1, arrow B). For replaced sequences, the new accession number is displayed. For suppressed sequences, the information is available to the user. For each identifier still present in the database, a SQL query retrieves all corresponding oligonucleotides (Fig. 1, arrow C). Finally, a list of oligonucleotides that match the experimental parameters entered by the user is sent back to the browser (Fig. 1, arrow D). This interface can be used to retrieve an updated collection of mouse-specific oligonucleotides and to follow-up oligonucleotide collections during time. When testing the latest mouse oligonucleotide probe collections of Illumina, Agilent and the RNG/MRC (Le Brigand et al., 2006), we found that, respectively, 33.4%, 50.3% and 79.2% of oligonucleotides were specific against Refseq 28 database, demonstrating the need to regularly update the specificity of probes.
Funding: EEC grant AnEUploidy; Fondation Jérôme Lejeune.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Ait Yahya-Graison E, et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am. J. Hum. Genet. 2007;81:475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Brigand K, et al. An open-access long oligonucleotide microarray resource for analysis of the human and mouse transcriptomes. Nucleic Acids Res. 2006;34:e87. doi: 10.1093/nar/gkl485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novère N. MELTING, computing the melting temperature of nucleic acid duplex. Bioinformatics. 2001;17:1226–1227. doi: 10.1093/bioinformatics/17.12.1226. [DOI] [PubMed] [Google Scholar]
- Perez-Iratxeta C, Andrade MA. Inconsistencies over time in 5% of NetAffx probe-to-gene annotations. BMC Bioinformatics. 2005;6:183. doi: 10.1186/1471-2105-6-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, et al. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard JM, et al. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Bioinformatics. 2002;18:486–487. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.