Abstract
Epidermal growth factor receptor (EGFR) is an activated oncogene in many cancers. It can be transactivated by ligands of G protein-coupled receptors (GPCRs). We show here that a novel gene, human rhomboid family-1 (RHBDF1), which was recently reported to have a pivotal role in epithelial cancer cell growth in culture and in xenograft tumors, participates in the modulation of GPCR-mediated EGFR transactivation. The RHBDF1 protein localizes mainly in the endoplasmic reticulum. Silencing the RHBDF1 gene in head and neck squamous cancer cell line 1483 cells with siRNA causes an inhibition of gastrin-releasing peptide (GRP) -induced phosphorylation of EGFR and EGFR-dependent signaling proteins p44/42 MAPK and AKT, accompanied by an inhibition of GRP-induced survival, proliferation, and invasion of the cells. The EGFR signaling pathway itself remains intact, however, as the cells remain responsive to exogenous EGF. In addition, RHBDF1 gene silencing disrupts GRP-stimulated secretion of EGFR ligand TGF-α, but not the production of latent TGF-α, whereas engineered overexpression of RHBDF1 markedly accelerates the secretion of TGF-α. These findings are consistent with the view that RHBDF1 is critically involved in a GPCR ligand-stimulated process leading to the activation of latent EGFR ligands.—Zou, H., Thomas, S. M., Yan, Z.-W., Grandis, J. R., Vogt, A., Li, L.-Y. Human rhomboid family-1 gene RHBDF1 participates in GPCR-mediated transactivation of EGFR growth signals in head and neck squamous cancer cells.
Keywords: siRNA, mitogenesis, oncogenesis, secretion, therapeutic
The epidermal growth factor receptor (EGFR) plays a major role in cancers as an activated oncogene (1,2,3,4,5). Elevated expression and activation of EGFR are frequently detected in a wide variety of carcinomas, including breast, lung, head and neck, and cervical cancers, and have been correlated with poor prognosis (6,7,8,9,10,11,12,13,14). The family of EGFR ligands includes EGF, transforming growth factor-α (TGF-α), amphiregulin (AR), heparin-binding EGF-like growth factor (HB-EGF), betacellulin (BTC), epiregulin (EPR), and epigen (15). These proteins are synthesized as latent ligands that need to be proteolytically processed to yield active growth factors. The EGFR growth signals can also be activated via stimulation of the activation of another family of cell surface receptors, namely, the G protein-coupled receptors (GPCRs) (16). Gastrin-releasing peptide (GRP) is the ligand for gastrin-releasing peptide receptor (GRPR), which belongs to the GPCR family (17). GRP promotes cancer cell survival, proliferation, and invasion in a number of cancer cell lines (18,19,20,21). GRP-induced signals have been linked to significantly enhanced proteolytic release of EGFR ligands and, consequently, activation of EGFR and its downstream signals p44/42 MAPK and AKT (19, 21,22,23). The metalloprotease-disintegrin tumor necrosis factor α-converting enzyme (TACE) and the Src family kinases have been implicated in GRP-induced EGFR transactivation (20, 21). However, the key steps in the transactivation of EGFR by GPCR activation remain incompletely understood.
We recently reported that a novel gene, the human rhomboid family-1 gene (RHBDF1), is essential to epithelial cancer cell growth (24). The rhomboid family of genes carry out a wide range of important functions in a variety of organisms (25), although little is known about the function of human RHBDF1. We found that RHBDF1 mRNA levels are significantly elevated in clinical specimens of invasive ductal carcinoma of the breast, and the protein is readily detected in human breast cancer or head and neck cancer cell lines. Silencing the RHBDF1 gene with siRNA results in apoptosis in breast cancer MDA-MB-435 cells and autophagy in head and neck squamous cell cancer (HNSCC) 1483 cells. The treatment also leads to significant down-regulation of activated AKT and p44/42 MAPK in the cells, suggesting that critically diminished strength of these growth signals may be the key attributes of the induction of the cell death. Furthermore, silencing the RHBDF1 gene in MDA-MB-435 or 1483 xenograft tumors on athymic nude mice by using intravenously administered histidine-lysine polymer nanoparticle encapsulated siRNA results in marked inhibition of tumor growth. These findings indicate that RHBDF1 plays a pivotal role in sustaining growth signals in epithelial cancer cells, and thus may serve as a therapeutic target for treating epithelial cancers.
Human RHBDF1 has a high degree of sequence homology to the Drosophila rhomboid 1 gene. Fruit flies possess a single EGFR, which is highly homologous to all four mammalian EGFR/ErbB receptors (26,27,28). Similar to its mammalian counterpart, the Drosophila EGFR exhibits multiple functions, including control of differentiation, cell survival, and proliferation. The principal ligand of the Drosophila EGFR is Spitz, which is homologous to mammalian TGF-α (29). Spitz must be proteolytically released as a soluble extracellular fragment to be functional. Rhomboid-1 is directly involved in the proteolytic cleavage of Spitz. This has been demonstrated to represent a highly conserved mechanism for eukaryotes and prokaryotes (29,30,31). It was reported recently that RHBDF1 interacts with the TGF-α family of ligands (31). However, it seems unlikely that RHBDF1 functions as a serine protease to process latent EGFR ligands, as a serine residue catalytically critical to the fruit fly enzyme and other rhomboid family of proteases is absent in the corresponding position in RHBDF1 (29, 31).
We studied the role of RHBDF1 in EGFR signaling in the 1483 cells and found that RHBDF1 gene function is essential to the cancer cells. We show here that silencing the RHBDF1 gene with siRNA leads to diminished GRP-induced EGFR activation, decreased TGF-α secretion, and decreased abilities of the cancer cells to survive, proliferate, and invade. These findings indicated that RHBDF1 has a key role in GPCR-mediated EGFR transactivation.
MATERIALS AND METHODS
Materials
Human HNSCC line 1483 was described previously (32). HeLa cells were purchased from American Type Culture Collection (Manassas, VA, USA). The siRNA molecules were synthesized (Dharmacon, Lafayette, CO, USA). The sequences of the RHBDF1 siRNA molecules are as described previously (24). ELISA kit for TGF-α was purchased from R&D Systems (Minneapolis, MN, USA). Human GRP and EGF were purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA). Antibodies against p44/42 MAPK, phosphorylated p44/42 MAPK, Flag, and green fluorescent protein (GFP) were purchased from Cell Signaling (Danvers, MA, USA). Antibodies for Golgin 97 and endoplasmic reticulum (ER) tracker were purchased from Invitrogen (Carlsbad, CA, USA). Rabbit polyclonal antibody against RHBDF1 was generated by immunizing a rabbit with a synthetic RHBDF1 peptide MSEARRDSTS SLQRKKPC, purified by affinity chromatography, and verified by the loss of the antibody’s ability to detect the target protein in Western blot analysis in the presence of the peptide.
siRNA-mediated RHBDF1 gene silencing
The RHBDF1 siRNA or a scrambled control siRNA (Ambion, Austin, TX) was transfected into the HNSCC 1483 cells using a HiperFect transfection protocol (Qiagen, Valencia, CA). Briefly, cells were seeded (2×104/well) in a 24-well plate. One microgram of the siRNA was mixed with 3 μl of HiperFect transfection reagent and incubated for 10 min at room temperature. The mixture was then added dropwise onto the cells. The cells were incubated for 6 h before changing to fresh medium.
Western blot analysis
Approximately 50 μg total protein was separated using a 7.5% SDS-PAGE gel and transferred onto a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with Tris-buffered saline Tween containing 10% nonfat dry milk for 1 h at room temperature. After being probed with the primary antibody overnight at 4°C, the membrane was incubated with the second antibody for 1 h at room temperature. The signal was developed with enhanced chemiluminescence reagent (Amersham Biosciences, Piscataway, NJ). The band intensity was quantitatively determined with ImageJ software (U.S. National Institutes of Health, Bethesda, MD, USA).
p44/42 MAPK ELISA
The 1483 cells were seeded in 96-well plates (1×104/well) and then were treated with either the control siRNA or the RHBDF1 siRNA and incubated overnight. The culture medium was replaced with FBS-free medium, and cells were incubated for 48 h. p44/42 MAPK phosphorylation was then measured by using an ELISA assay method (SuperArray Bioscience, Frederick, MD, USA), which uses specific primary antibodies against either phospho-p44/42 MAPK or total p44/42 MAPK and horseradish peroxidase-conjugated secondary antibodies, according to the manufacturer’s instructions.
TGF-α ELISA
The 1483 cells were seeded at a density of 2 × 104 cells/ml in a 100-mm dish overnight. Cells were transfected with Lipofectamine 2000 (Invitrogen; 10 μl/plate) and siRNA (5 μg/ml) against either RHBDF1 or control siRNA for 16 h before changing to serum-free Dulbecco modified Eagle medium (DMEM) and incubated for another 48 h. The cells were then treated with either vehicle control (water), GRP (400 nM), or EGF (10 ng/ml) for 10 min, basically as described (33, 34). Cell supernatants were collected for ELISA analyses of TGF-α (R&D Systems, Minneapolis, MN, USA) following manufacturer’s instructions. Cells were harvested and lysates were analyzed by Western blot analysis, as described previously (34).
RHBDF1-facilitated TGF-α release
Human cervical cancer cell line HeLa cells (American Type Culture Collection) were seeded in collagen-coated 384-well plates (BD Biosciences, San Jose, CA, USA) and cotransfected with GFP-TGF-α and pCI/RHBDF1 or an empty vector using Lipofectamine 2000 (Invitrogen). Cells were fixed with 4% formaldehyde and stained with Hoechst 33342 to visualize nuclei. The plates were then analyzed by multiparametric single-cell analysis on an ArrayScan II solid-phase cytometer (Cellomics, Pittsburgh, PA, USA), as described previously (35). A threshold for green fluorescence intensity was defined as the average green fluorescence intensity ± sd from 4 wells transfected with RHBDF1 alone. Cells were classified as positive for GFP-TGF-α expression if their average GFP intensity exceeded this threshold. At least 1000 cells per well were analyzed.
Cell survival
The 1483 cells were seeded at a density of 5000 cells/well in 96-well plates and incubated at 37°C with 5% CO2 overnight. The cells were treated with either control siRNA or RHBDF1 siRNA for 6 h. The medium was changed with serum-free DMEM and incubated for 48 h. The cells then were treated with vehicle control (water), GRP, or EGF for another 24 h and MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay was carried out to determine the effect of siRNA treatment. Fold of cell survival was calculated by normalizing the values of experiment groups to the control groups.
Cell proliferation
The 1483 cells seeded in 24-well plates were treated with siRNA as mentioned above. Subsequently, cells were incubated with GRP or EGF in FBS-free DMEM. The control cultures were untreated. After 72 h, cells were harvested and cell numbers were counted under a microscope.
Cell invasion
Biocoat tumor invasion system (BD Biosciences) was used. RHBDF1 or control siRNA-treated 1483 cells were plated in the 24-well Matrigel-coated chambers at a density of 10,000 cells/well in the presence of control, GRP, or EGF. Cells were then incubated at 37°C with 5% CO2 for 24 h. After the noninvading cells were removed from filter inserts with a cotton-tipped swab, the filter inserts were fixed and stained with Fisher Protocol Hema 3 kit (Fisher Scientific, Pittsburgh, PA, USA), according to the manufacturer’s instructions. The number of the invading cells was determined at ×200.
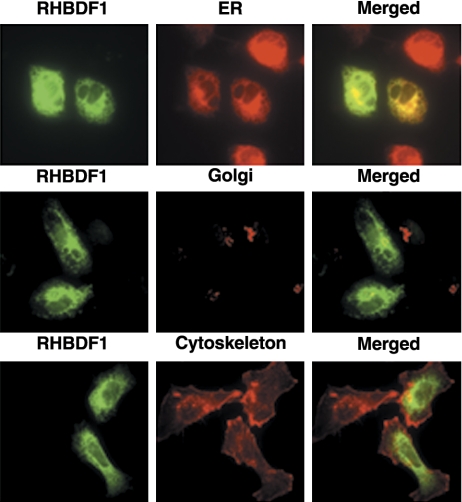
Intracellular localization
The 1483 cells were seeded in 4-well chambered slides and transfected with FLAG-RHBDF1. The cells were fixed after 24 h with 4% PFA, permeabilized with 0.1% Triton, blocked with 5% BSA, incubated with an anti-FLAG antibody alone or together with Golgin 97, then incubated with fluorescence-conjugated secondary antibodies for 1 h at RT. To determine colocalization of RHBDF1 and ER or F-actin, FLAG-antibody-treated cells were incubated with the secondary antibody and either fluorescence-conjugated ER tracker, or fluorescence-conjugated F-actin antibody, for 1 h at RT. The slides were then treated with antifade agent, covered, and observed with a confocal microscope (Fluoview 1000; Olympus, Tokyo, Japan).
RESULTS
RHBDF1 siRNA inhibits GRP-stimulated activation of EGFR signaling pathway
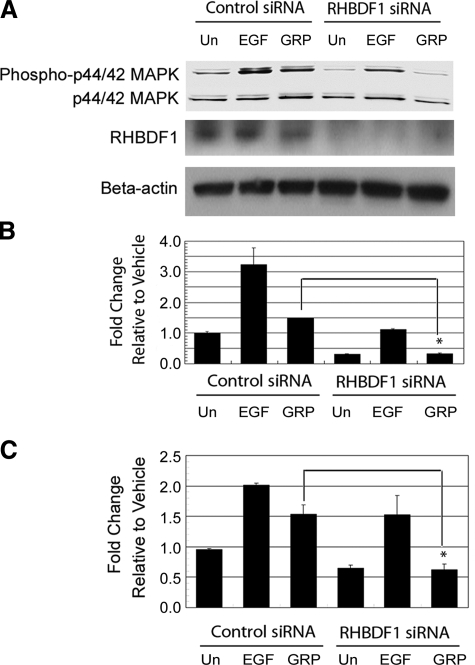
We found previously that silencing the RHBDF1 gene in HNSCC 1483 cells caused a down-regulation of p44/p42 MAPK (24), which is an EGFR-derived growth signal. We were, therefore, interested in whether RHBDF1 is involved in the modulation of EGFR growth signaling pathway, using the HNSCC line 1483 that expresses high levels of EGFR and exhibits an EGFR-dependent phosphorylation of p44/p42 MAPK in response to GRP stimulation. First, we determined the effect of RHBDF1 gene silencing on EGFR tyrosine phosphorylation in response to GRP stimulation. The cells were treated with either the control siRNA or the RHBDF1 siRNA prior to GRP treatment. The RHBDF1 siRNA treatment resulted in a nearly complete depletion of the RHBDF1 protein in the cells within 48 h (Fig. 1A). Phosphorylated EGFR was isolated by immunoprecipitation with an antibody against EGFR, then identified with an anti-phosphotyrosine antibody by Western blot analysis. We found that GRP-stimulated EGFR phosphorylation significantly diminished in RHBDF1 siRNA-treated cells to the background level seen in vehicle-treated cells (Fig. 1). This suggests that RHBDF1 is a critical component in the signaling pathway mediating EGFR transactivation by GRP. We then analyzed the effect on EGFR-derived signals. The addition of EGF or GRP to the control siRNA-treated 1483 cells typically gives rise to a 3- or 1.5-fold increase, respectively, of the p44/p42 MAPK phosphorylation under the experimental conditions (Fig. 2A, B). However, treatment of the cells with the RHBDF1 siRNA not only abolished the GRP-stimulated p44/42 MAPK phosphorylation but also caused a further down-regulation of the baseline MAPK phosphorylation. The addition of exogenous EGF to the culture medium gave rise to a partial recovery of the effect of the RHBDF1 siRNA on the p44/p42 MAPK phosphorylation, suggesting that the EGFR signaling pathway itself remains functional. These findings were corroborated using an ELISA assay as an alternative method to detect p44/42 MAPK phosphorylation (Fig. 2C), which confirmed that GRP-induced p44/42 MAPK phosphorylation was inhibited when the 1483 cells were pretreated with the RHBDF1 siRNA, that this effect was significantly lessened if EGF instead of GRP was used to stimulate the cells, and that there was a further decline of the baseline level of p44/p42 MAPK phosphorylation compared with that in control siRNA-treated cells. These results indicate that silencing the RHBDF1 gene causes a decrease of the signals from GRP/GRPR leading to the activation of EGFR, but has no effect on the signaling pathways downstream of the activated EGFR.
Figure 1.
RHBDF1 siRNA inhibits GRP-induced EGFR phosphorylation in the 1483 cells. A) Western blotting analysis of EGFR phosphorylation. B) Quantitative representation of the Western blotting results. Con siRNA, control siRNA; Un, untreated. Data are from 3 independent experiments. *P < 0.01, Student’s t test.
Figure 2.
RHBDF1 siRNA inhibits GRP-induced p44/p42 MAPK phosphorylation in the 1483 cells. A) Western blot analysis of p44/p42 MAPK phosphorylation. B) Quantitative representation of the Western blot analysis results. C) ELISA analysis of p44/p42 MAPK phosphorylation. Un, untreated. Data are from 3 independent experiments. *P < 0.01, Student’s t test.
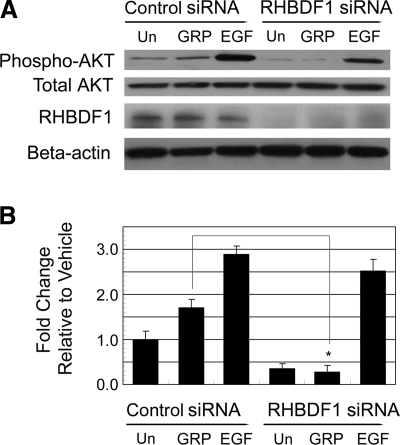
RHBDF1 gene-silencing also resulted in an inhibition of AKT phosphorylation, another EGFR downstream signaling event. Using Western blot analysis, we found that treatment of control siRNA-treated 1483 cells with GRP or EGF gave rise to an ∼1.5- to 3-fold increase in AKT phosphorylation compared to that of vehicle treatment (Fig. 3). Again, GRP-induced AKT phosphorylation was nearly completely abolished when the cells were treated with the RHBDF1 siRNA. Similar to our findings with p44/42 MAPK activation, the RHBDF1 siRNA had no effect on EGF-induced AKT activation. In addition, a further decline of the baseline level of AKT phosphorylation was observed in RHBDF1 siRNA-treated cells. These findings indicate that RHBDF1 gene-silencing causes a disruption of the signaling pathway leading from GRP/GRPR to EGFR, but not of the signaling pathway derived from activated EGFR, since the RHBDF1 siRNA-treated cells retain the ability to respond to exogenous EGF.
Figure 3.
RHBDF1 siRNA inhibits GRP-induced AKT phosphorylation. A) Western blot analysis of AKT phosphorylation. B) Quantitative representation of the Western blot analysis results. Un, untreated. Data are from 3 independent experiments. *P < 0.01, Student’s t test.
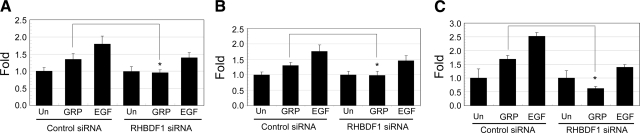
Silencing the RHBDF1 gene inhibits cell survival, proliferation, and invasion
We determined the effect of RHBDF1 siRNA treatment on GRP-simulated 1483 cancer cell survival, growth, and migration, cellular activities known to be enhanced as the result of GRP-stimulated activation of EGFR-dependent signaling pathway. The 1483 cells were treated with either the control siRNA or the RHBDF1 siRNA, serum-starved, then treated with either vehicle, GRP, or EGF (Fig. 4). Analyzed for viability within 24 h, cells survived from starvation 30 and 80% more in the presence of GRP and EGF, respectively, in the control siRNA-treated group (Fig. 4A). The ability of the cells to respond to GRP to survive was abolished due to the prior treatment with the RHBDF1 siRNA. As expected, the cells remained responsive to EGF in survival. The experiments were repeated to determine the number of cells increased within 72 h in the presence of GRP or EGF (Fig. 4B). The addition of GRP or EGF to the culture medium gave rise to a 30 and 80% increase, respectively, of the number of cells in the control siRNA-treated group. Again, GRP but not EGF, failed to promote the proliferation of the cells pretreated with the RHBDF1 siRNA. Furthermore, we examined the ability of the cells to migrate through filters coated with a mixture of extracellular matrix proteins (Matrigel) as a measure of the capacity of cell invasion (Fig. 4C). The addition of GRP or EGF to the culture medium resulted in an 80 and 150% increase of the number of the control siRNA-treated cancer cells to move through the filters, whereas the ability of GRP or EGF to promote cell invasion diminished when the cells were pretreated with the RHBDF1 siRNA. These results indicate that the 1483 cells depend on RHBDF1 to benefit from GRP-stimulated, EGFR-derived signals for cell survival, proliferation, and invasion.
Figure 4.
RHBDF1 siRNA inhibits GRP-stimulated survival, proliferation, and invasion of the 1483 cells. A) Changes in cell viability in 24 h on treatment. B) Changes in cell viability in 72 h on treatment. C) Changes in the ability of the cells to migrate into Matrigel. Un, untreated. Data are from 3 independent experiments. *P < 0.01, Student’s t test.
RHBDF1 siRNA inhibits GRP-stimulated secretion of EGFR ligand TGF-α
We determined whether RHBDF1 takes part in the modulation of the processing of latent ligands of EGFR in response to GRP stimulation. Because enhanced secretion of TGF-α has been described in GPCR-mediated EGFR transactivation in head and neck cancer, we analyzed the effect of RHBDF1 siRNA treatment on the release of TGF-α by the 1483 cells into the culture medium. Using an ELISA assay to measure the amount of soluble TGF-α in the culture medium, we found that, while the addition of GRP to the control siRNA-treated 1483 cells resulted an ∼2-fold increase of the amount of TGF-α in the culture medium, the GRP-induced release of TGF-α diminished to a background level in the conditioned medium of the cells pretreated with the RHBDF1 siRNA (Fig. 5A). To determine whether the RHBDF1 siRNA had any effect on the production of latent TGF-α, we determined the amount of cell-associated TGF-α. We found that an ∼70% higher level of the TGF-α protein was present in RHBDF1 siRNA-treated cells as compared with that in the control siRNA-treated cells, suggesting an accumulation of TGF-α in RHBDF1 siRNA-treated cells (Fig. 5B). These results indicate that silencing the RHBDF1 gene leads to a disruption of TGF-α secretion, but not TGF-α production.
Figure 5.
RHBDF1 siRNA inhibits the release of TGF-α by the 1483 cells determined by ELISA. A) TGF-α concentrations in conditioned medium of GRP-stimulated cells (solid bars) relative to control cells without GRP treatment (open bars). B) Levels of TGF-α protein associated with GRP-stimulated cells treated with either the control siRNA (Con siRNA) or the RHBDF1 siRNA. Data are from 3 independent experiments. *P < 0.01, Student’s t test.
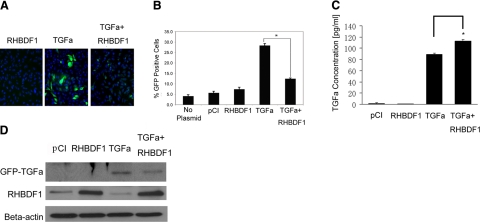
Overexpression of RHBDF1 facilitates TGF-α secretion
The above findings prompted us to determine whether elevated RHBDF1 levels will facilitate TGF-α release into the culture medium. The TGF-α protein was conjugated with the GFP at the N terminus and transfected into HeLa cells, a human cervical epitheloid carcinoma cell line. Unlike other cancer cells that form multilayered clusters in cultures, HeLa cells form a monolayer suitable for high-throughput fluorescence microscope analysis, a method we used to simultaneously determine the fluorescence intensity of several thousand cells (Fig. 6A). We cotransfected HeLa cells with the GFP-TGF-α and a plasmid encoding RHBDF1. Empty plasmids were used in control experiments. We found that 28% of the cells cotransfected with GFP-TGF-α and the control plasmid were GFP-positive, compared to a baseline of 5–7% of fluorescent cells transfected with either the control plasmid or the RHBDF1 alone (Fig. 6B). When the cells were cotransfected with the plasmids encoding RHBDF1 and GFP-TGF-α, the percentage of green fluorescent cells decreased to 12% (Fig. 6B). These findings indicate that an increased presence of RHBDF1 leads to a lowered level of GFP-TGF-α in the cells, likely due to an enhanced export of GFP-TGF-α.
Figure 6.
RHBDF1 overexpression facilitates TGF-α release. A) Fluorescent images of HeLa cell cultures. Blue denotes cell nuclei; green denotes GFP-TGF-α. B) Percentage of GFP-positive cells in the cultures. C) ELISA analysis of TGF-α concentrations in the conditioned medium from the cultures. D) Western blot analysis of TGF-α protein levels in cells isolated from the cultures. RHBDF1, cells transfected with pCI/RHBDF1; TGF-α, cells cotransfected with GFP-TGF-α and empty pCI; TGF-α+RHBDF1, cells cotransfected with GFP-TGF-α and pCI/RHBDF1; pCI, cells transfected with empty vector. Data are from 2 independent experiments. *P < 0.01, Student’s t test.
In addition, we measured the concentration of TGF-α in the conditioned medium of the cells transfected with TGF-α, either alone or cotransfected with RHBDF1, and compared with that of the cells transfected with the empty vectors, using an ELISA assay. We found that the TGF-α-transfected cells secreted a marked amount of TGF-α in the culture medium and that cotransfection of the cells with TGF-α and RHBDF1 gave rise to an ∼25% increase of TGF-α secretion in comparison (Fig. 6C). Consistently, the amount of cell-associated TGF-α decreased markedly in cells cotransfected with TGF-α and RHBDF1, compared with that transfected with TGF-α alone (Fig. 6D). These results indicate that engineered overexpression of RHBDF1 coincides with facilitated secretion of TGF-α.
Intracellular localization of RHBDF1 on ER
In order to establish the intracellular localization where RHBDF1 functions, we transfected the 1483 cells with a plasmid encoding RHBDF1 conjugated with the FLAG-tag at the N terminus. We determined whether the protein colocalizes with the ER, the Golgi apparatus, or the plasma membrane, which were labeled, respectively, with the Golgi-specific marker Golgin 97, the ER-specific marker ER tracker, or an antibody against F-actin (Fig. 7). We found that RHBDF1 mainly colocalizes in the ER and to a much lesser degree also in the Golgi, but not in the plasma membrane. These results indicate that the intracellular site of RHBDF1 function is primarily the ER.
Figure 7.
Confocal microscopic images depicting intracellular localization of RHBDF1. FLAG-RHBDF1-transfected 1483 cells are immunostained with fluorescence-conjugated antibodies against FLAG (green), ER-tracker (red, top row), Glogin97 (red, middle row), or F-actin (red, bottom row). Original view ×2000.
DISCUSSION
Our findings indicate that RHBDF1, a human rhomboid family gene known very little thus far, may have a critical role in GPCR-mediated transactivation of EGFR. Two pathways, one EGFR ligand dependent and the other independent, have been proposed to mediate the crosstalk between GPCR and EGFR (16). The EGFR ligand-dependent pathway implies that GPCR-derived signals can induce the cleavage of membrane-bound EGFR latent ligands to yield mature, soluble ligands, which are released from the cells and then activate EGFR through an autocrine mechanism. This pathway involves GPCR ligand-induced activation of PI3K, Src, and PDK1, which then activate metalloproteinases such as TACE, to carry out proteolytic processing of EGFR proligands (20). We found that elimination of RHBDF1 from the 1483 cells leads to greatly reduced GRP-stimulated secretion of EGFR ligand TGF-α and that engineered overexpression of RHBDF1 and latent TGF-α results in facilitated export of TGF-α. These data strongly suggest that RHBDF1 is a critical component of the mechanism responsible for the production of activated EGFR ligands. However, RHBDF1 has not yet been shown to possess a protease activity or exhibit a discernable effect on TACE activity when RHBDF1 gene is silenced (data not shown). It is thus unlikely that RHBDF1 is directly involved in the proteolytic processing of latent TGF-α.
Alternatively, it is plausible that RHBDF1 is critically involved in the production or subcellular transportation of latent TGF-α. Other investigators reported previously that the RHBDF1 protein interacts with TGF-α physically, as determined by coimmunoprecipitation, and that RHBDF1 localizes primarily in the ER when transfected into HeLa cells (31). We were able to reproduce the coimmunoprecipitation of RHBDF1 and TGF-α (data not shown) and found that RHBDF1 mainly localizes in the ER when transfected into the 1483 cells. These findings suggest that RHBDF1 may function as an intracellular transporter for protein secretion, or as a chaperone to assist the folding or intracellular translocation of latent EGFR ligands. Similar functions of other gene products that interact with proteins containing EGF-like domains may serve as examples in this regard. One example, based on the genetic analyses of Drosophila during development, is cornichon, a putative 3-transmembrane protein that may be involved in polarized localization of the TGF-α-like protein Gurken to the cell surface, presumably by guiding its transport through the endoplasmic reticulum (37). The genes for the type II transmembrane protein Star (38) and the 7-transmembrane spanning Rhomboid (39) were identified as required for the signaling activity of the TGF-α-like protein Spitz. Star is needed for trafficking of Spitz from the ER to the Golgi (36, 40), where rhomboid-1 resides and functions as a serine protease to cleave the transmembrane segment of Spitz, thus allowing the release of Spitz as a soluble protein (29, 36). Alternatively, RHBDF1 could function as a chaperone of membrane proteins enriched with EGF-like domains, similar to the role of Boca, a protein located in the ER that ensures correct folding of EGF modules of low-density lipoprotein receptors (41). That the 1483 cells undergo autophagy when the RHBDF1 gene is silenced (24) is consistent also with this view, for autophagy could be the result of ER stress, a consequence of loss of protein functions critical for protein folding, or transportation (42).
Another possibility is that RHBDF1 is an essential component of the ligand-independent EGFR activation pathway that involves signals derived from GPCR activation. This model implies that the GPCR ligand-stimulated signals may lead to direct phosphorylation of the juxtamembrane tyrosine kinase domain of EGFR. Candidates of this route of signals include the Src family of kinases (43) or PKC (44). This type of receptor transactivation is attributable to the modest clinical responses to EGFR-targeted therapies to date, given the highly heterogeneous expression of GPCR in tumors. Such GPCR-EGFR “crosstalk” has been implicated in the progression of colon, lung, breast, head and neck, prostate, and ovarian cancers (19, 45,46,47,48,49). We found that elimination of RHBDF1 from the 1483 cells leads to disruption of GRP-induced MAPK and AKT phosphorylation and that the presence of exogenous EGF largely restores MAPK and AKT phosphorylation in RHBDF1-depleted cells. These results indicate that EGFR itself and the signaling pathway derived from EGFR are intact in these cells since the cells remain responsive to EGF stimulation. It is, therefore, plausible to place the position of RHBDF1 action in the chain of GRP-induced signals before they reach EGFR. In other words, RHBDF1 activity could be needed for GRP-induced activation of EGFR ligand-independent signals. Thus, it will be interesting to determine the effect of RHBDF1 gene-silencing on these signals.
In summary, our findings strongly suggest that the RHBDF1 gene plays a critical role in the signaling pathways involved in GPCR-mediated transactivation of EGFR, which is implicated in a wide variety of cancers as a crucial factor in chronic inflammation, angiogenesis, cancer development, and tumor growth and metastasis. Further research on the function and mechanism of action of RHBDF1 is likely to provide important insights into the role of GPCR-EGFR crosstalk in cancers. Our data also demonstrate that disruption of RHBDF1 results in inhibition of GRP-induced head and neck cancer cell survival, proliferation, and invasion, indicating that RHBDF1 may serve as a new target for the development of anticancer therapeutic agents.
Acknowledgments
The study has been supported in part by grants from Flight Attendant Medical Research Institute (L.Y.L.), the Henry L. Hillman Foundation (L.Y.L.), Pennsylvania Department of Health (L.Y.L.), DOD BC073645 (L.Y.L.), P01CA78039 (A.V.), P50CA097190 (J.R.G.) and R01CA098372 (J.R.G.).
References
- Akiyama T, Kadooka T, Ogawara H. Purification of the epidermal growth factor receptor by tyrosine-Sepharose affinity chromatography. Biochem Biophys Res Commun. 1985;131:442–448. doi: 10.1016/0006-291x(85)91822-4. [DOI] [PubMed] [Google Scholar]
- Besmer P, Murphy J E, George P C, Qiu F H, Bergold P J, Lederman L, Snyder H W, Jr, Brodeur D, Zuckerman E E, Hardy W D. A new acute transforming feline retrovirus and relationship of its oncogene v-kit with the protein kinase gene family. Nature. 1986;320:415–421. doi: 10.1038/320415a0. [DOI] [PubMed] [Google Scholar]
- Burgess A W, Cho H S, Eigenbrot C, Ferguson K M, Garrett T P, Leahy D J, Lemmon M A, Sliwkowski M X, Ward C W, Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- King C R, Borrello I, Bellot F, Comoglio P, Schlessinger J. Egf binding to its receptor triggers a rapid tyrosine phosphorylation of the erbB-2 protein in the mammary tumor cell line SK-BR-3. EMBO J. 1988;7:1647–1651. doi: 10.1002/j.1460-2075.1988.tb02991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff-Jamison S, Chen K, Cohen S. Epidermal growth factor induces the tyrosine phosphorylation and nuclear translocation of Stat 5 in mouse liver. Proc Natl Acad Sci U S A. 1995;92:4215–4218. doi: 10.1073/pnas.92.10.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullick W J. Prevalence of aberrant expression of the epidermal growth factor receptor in human cancers. Br Med Bull. 1991;47:87–98. doi: 10.1093/oxfordjournals.bmb.a072464. [DOI] [PubMed] [Google Scholar]
- Harris A L, Nicholson S, Sainsbury R, Wright C, Farndon J. Epidermal growth factor receptor and other oncogenes as prognostic markers. J Natl Cancer Inst Monogr. 1992;11:181–187. [PubMed] [Google Scholar]
- Libermann T A, Nusbaum H R, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield M D, Ullrich A, Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- Berger M S, Greenfield C, Gullick W J, Haley J, Downward J, Neal D E, Harris A L, Waterfield M D. Evaluation of epidermal growth factor receptors in bladder tumours. Br J Cancer. 1987;56:533–537. doi: 10.1038/bjc.1987.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanne B, Richards C S, Hendler F, Burns D, Gusterson B. Over-expression of the EGF receptor is a hallmark of squamous cell carcinomas. J Pathol. 1986;149:9–14. doi: 10.1002/path.1711490104. [DOI] [PubMed] [Google Scholar]
- Kersemaekers A M, Fleuren G J, Kenter G G, Van den Broek L J, Uljee S M, Hermans J, Van de Vijver M J. Oncogene alterations in carcinomas of the uterine cervix: overexpression of the epidermal growth factor receptor is associated with poor prognosis. Clin Cancer Res. 1999;5:577–586. [PubMed] [Google Scholar]
- Emens L A. Trastuzumab: targeted therapy for the management of HER-2/neu-overexpressing metastatic breast cancer. Am J Ther. 2005;12:243–253. [PubMed] [Google Scholar]
- Rubin Grandis J, Zeng Q, Tweardy D J. Retinoic acid normalizes the increased gene transcription rate of TGF-alpha and EGFR in head and neck cancer cell lines. Nat Med. 1996;2:237–240. doi: 10.1038/nm0296-237. [DOI] [PubMed] [Google Scholar]
- Todd R, Donoff B R, Gertz R, Chang A L, Chow P, Matossian K, McBride J, Chiang T, Gallagher G T, Wong D T. TGF-alpha and EGF-receptor mRNAs in human oral cancers. Carcinogenesis. 1989;10:1553–1556. doi: 10.1093/carcin/10.8.1553. [DOI] [PubMed] [Google Scholar]
- Harris R C, Chung E, Coffey R J. EGF receptor ligands. Exp Cell Res. 2003;284:2–13. doi: 10.1016/s0014-4827(02)00105-2. [DOI] [PubMed] [Google Scholar]
- Bhola N E, Grandis J R. Crosstalk between G-protein-coupled receptors and epidermal growth factor receptor in cancer. Front Biosci. 2008;13:1857–1865. doi: 10.2741/2805. [DOI] [PubMed] [Google Scholar]
- DeMichele M A, Davis A L, Hunt J D, Landreneau R J, Siegfried J M. Expression of mRNA for three bombesin receptor subtypes in human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1994;11:66–74. doi: 10.1165/ajrcmb.11.1.8018339. [DOI] [PubMed] [Google Scholar]
- Terashi H, Itami S, Tadokoro T, Takeyama M, Katagiri K, Takayasu S. Growth stimulation of normal melanocytes and nevocellular nevus cells by gastrin releasing peptide (GRP) J Dermatol Sci. 1998;17:93–100. doi: 10.1016/s0923-1811(97)00079-0. [DOI] [PubMed] [Google Scholar]
- Thomas S M, Grandis J R, Wentzel A L, Gooding W E, Lui V W, Siegfried J M. Gastrin-releasing peptide receptor mediates activation of the epidermal growth factor receptor in lung cancer cells. Neoplasia. 2005;7:426–431. doi: 10.1593/neo.04454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Thomas S M, Lui V W, Xi S, Siegfried J M, Fan H, Smithgall T E, Mills G B, Grandis J R. Phosphorylation of TNF-alpha converting enzyme by gastrin-releasing peptide induces amphiregulin release and EGF receptor activation. Proc Natl Acad Sci U S A. 2006;103:6901–6906. doi: 10.1073/pnas.0509719103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Thomas S M, Xi S, Smithgall T E, Siegfried J M, Kamens J, Gooding W E, Grandis J R. SRC family kinases mediate epidermal growth factor receptor ligand cleavage, proliferation, and invasion of head and neck cancer cells. Cancer Res. 2004;64:6166–6173. doi: 10.1158/0008-5472.CAN-04-0504. [DOI] [PubMed] [Google Scholar]
- McCole D F, Keely S J, Coffey R J, Barrett K E. Transactivation of the epidermal growth factor receptor in colonic epithelial cells by carbachol requires extracellular release of transforming growth factor-alpha. J Biol Chem. 2002;277:42603–42612. doi: 10.1074/jbc.M206487200. [DOI] [PubMed] [Google Scholar]
- Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- Yan Z W, Zou H F, Tian F, Grandis J R, Mixson A J, Lu P Y, Li L Y. Human rhomboid family-1 (RHBDF1) gene-silencing causes apoptosis or autophagy to epithelial cancer cells and inhibits xenograft tumor growth. Mol Cancer Therap. 2008;7:1355–1364. doi: 10.1158/1535-7163.MCT-08-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S. Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 2006;20:3054–3068. doi: 10.1101/gad.1488606. [DOI] [PubMed] [Google Scholar]
- Freeman M. The spitz gene is required for photoreceptor determination in the Drosophila eye where it interacts with the EGF receptor. Mech Dev. 1994;48:25–33. doi: 10.1016/0925-4773(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Golembo M, Raz E, Shilo B Z. The Drosophila embryonic midline is the site of Spitz processing, and induces activation of the EGF receptor in the ventral ectoderm. Development. 1996;122:3363–3370. doi: 10.1242/dev.122.11.3363. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Howes R, Smith R, Shilo B Z, Freeman M. Inhibition of Drosophila EGF receptor activation by the secreted protein Argos. Nature. 1995;376:699–702. doi: 10.1038/376699a0. [DOI] [PubMed] [Google Scholar]
- Urban S, Lee J R, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- Gallio M, Sturgill G, Rather P, Kylsten P. A conserved mechanism for extracellular signaling in eukaryotes and prokaryotes. Proc Natl Acad Sci U S A. 2002;99:12208–12213. doi: 10.1073/pnas.192138799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Guichard A, Castro C P, Xiao Y, Rizen M, Zhang H Z, Hu D, Bang A, Helms J, Bier E, Derynck R. Characterization of a human rhomboid homolog, p100hRho/RHBDF1, which interacts with TGF-α family ligands. Dev Dyn. 2005;233:1315–1331. doi: 10.1002/dvdy.20450. [DOI] [PubMed] [Google Scholar]
- Sacks P G, Parnes S M, Gallick G E, Mansouri Z, Lichtner R, Satya-Prakash K L, Pathak S, Parsons D F. Establishment and characterization of two new squamous cell carcinoma cell lines derived from tumors of the head and neck. Cancer Res. 1988;48:2858–2866. [PubMed] [Google Scholar]
- Kijima T, Niwa H, Steinman R A, Drenning S D, Gooding W E, Wentzel A L, Xi S, Grandis J R. STAT3 activation abrogates growth factor dependence and contributes to head and neck squamous cell carcinoma tumor growth in vivo. Cell Growth Differ. 2002;13:355–362. [PubMed] [Google Scholar]
- Lui V W, Thomas S M, Zhang Q, Wentzel A L, Siegfried J M, Li J Y, Grandis J R. Mitogenic effects of gastrin-releasing peptide in head and neck squamous cancer cells are mediated by activation of the epidermal growth factor receptor. Oncogene. 2003;22:6183–6193. doi: 10.1038/sj.onc.1206720. [DOI] [PubMed] [Google Scholar]
- Vogt A, Kalb E N, Lazo J S. A scalable high-content cytotoxicity assay insensitive to changes in mitochondrial metabolic activity. Oncol Res. 2004;14:305–314. doi: 10.3727/096504003773994842. [DOI] [PubMed] [Google Scholar]
- Lee J R, Urban S, Garvey C F, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–171. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg F S, Barcelo G, Schupbach T. Cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell. 1995;81:967–978. doi: 10.1016/0092-8674(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Kolodkin A L, Pickup A T, Lin D M, Goodman C S, Banerjee U. Characterization of Star and its interactions with sevenless and EGF receptor during photoreceptor cell development in Drosophila. Development. 1994;120:1731–1745. doi: 10.1242/dev.120.7.1731. [DOI] [PubMed] [Google Scholar]
- Bier E, Jan L Y, Jan Y N. Rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev. 1990;4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- Tsruya R, Schlesinger A, Reich A, Gabay L, Sapir A, Shilo B Z. Intracellular trafficking by Star regulates cleavage of the Drosophila EGF receptor ligand Spitz. Genes Dev. 2002;16:222–234. doi: 10.1101/gad.214202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culi J, Springer T A, Mann R S. Boca-dependent maturation of beta-propeller/EGF modules in low-density lipoprotein receptor proteins. EMBO J. 2004;23:1372–1380. doi: 10.1038/sj.emboj.7600132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J L. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation) Biochem J. 2007;404:353–363. doi: 10.1042/BJ20061890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piiper A, Elez R, You S J, Kronenberger B, Loitsch S, Roche S, Zeuzem S. Cholecystokinin stimulates extracellular signal-regulated kinase through activation of the epidermal growth factor receptor, Yes, and protein kinase C. Signal amplification at the level of Raf by activation of protein kinase Cepsilon. J Biol Chem. 2003;278:7065–7072. doi: 10.1074/jbc.M211234200. [DOI] [PubMed] [Google Scholar]
- Krysan K, Reckamp K L, Dalwadi H, Sharma S, Rozengurt E, Dohadwala M, Dubinett S M. Prostaglandin E2 activates mitogen-activated protein kinase/Erk pathway signaling and cell proliferation in non-small cell lung cancer cells in an epidermal growth factor receptor-independent manner. Cancer Res. 2005;65:6275–6281. doi: 10.1158/0008-5472.CAN-05-0216. [DOI] [PubMed] [Google Scholar]
- Chatzistamou I, Schally A V, Szepeshazi K, Groot K, Hebert F, Arencibia J M. Inhibition of growth of ES-2 human ovarian cancers by bombesin antagonist RC-3095, and luteinizing hormone-releasing hormone antagonist Cetrorelix. Cancer Lett. 2001;171:37–45. doi: 10.1016/s0304-3835(01)00543-2. [DOI] [PubMed] [Google Scholar]
- Lango M N, Dyer K F, Lui V W, Gooding W E, Gubish C, Siegfried J M, Grandis J R. Gastrin-releasing peptide receptor-mediated autocrine growth in squamous cell carcinoma of the head and neck. J Natl Cancer Inst. 2002;94:375–383. doi: 10.1093/jnci/94.5.375. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Lamharzi N, Schally A V, Halmos G, Szepeshazi K, Groot K, Cai R Z. Inhibition of growth of MDA-MB-231 human breast cancer xenografts in nude mice by bombesin/gastrin-releasing peptide (GRP) antagonists RC-3940-II and RC-3095. Eur J Cancer. 1998;34:710–717. doi: 10.1016/s0959-8049(97)10123-x. [DOI] [PubMed] [Google Scholar]
- Radulovic S, Schally A V, Reile H, Halmos G, Szepeshazi K, Groot K, Milovanovic S, Miller G, Yano T. Inhibitory effects of antagonists of bombesin/gastrin releasing peptide (GRP) and somatostatin analog (RC-160) on growth of HT-29 human colon cancers in nude mice. Acta Oncol. 1994;33:693–701. doi: 10.3109/02841869409121784. [DOI] [PubMed] [Google Scholar]
- Xiao D, Qu X, Weber H C. GRP receptor-mediated immediate early gene expression and transcription factor Elk-1 activation in prostate cancer cells. Regul Pept. 2002;109:141–148. doi: 10.1016/s0167-0115(02)00197-0. [DOI] [PubMed] [Google Scholar]