Abstract
Motivation: Phospholipid scramblases (PLSCRs) constitute a family of cytoplasmic membrane-associated proteins that were identified based upon their capacity to mediate a Ca2+-dependent bidirectional movement of phospholipids across membrane bilayers, thereby collapsing the normally asymmetric distribution of such lipids in cell membranes. The exact function and mechanism(s) of these proteins nevertheless remains obscure: data from several laboratories now suggest that in addition to their putative role in mediating transbilayer flip/flop of membrane lipids, the PLSCRs may also function to regulate diverse processes including signaling, apoptosis, cell proliferation and transcription. A major impediment to deducing the molecular details underlying the seemingly disparate biology of these proteins is the current absence of any representative molecular structures to provide guidance to the experimental investigation of their function.
Results: Here, we show that the enigmatic PLSCR family of proteins is directly related to another family of cellular proteins with a known structure. The Arabidopsis protein At5g01750 from the DUF567 family was solved by X-ray crystallography and provides the first structural model for this family. This model identifies that the presumed C-terminal transmembrane helix is buried within the core of the PLSCR structure, suggesting that palmitoylation may represent the principal membrane anchorage for these proteins. The fold of the PLSCR family is also shared by Tubby-like proteins. A search of the PDB with the HHpred server suggests a common evolutionary ancestry. Common functional features also suggest that tubby and PLSCR share a functional origin as membrane tethered transcription factors with capacity to modulate phosphoinositide-based signaling.
Contact: agb@sanger.ac.uk
1 INTRODUCTION
Biological membranes have an asymmetric distribution of lipids. A variety of proteins have been identified which create and maintain this distribution. Phospholipid scramblase (PLSCR) proteins were initially identified as being able to mediate the collapse of this asymmetric distribution, by accelerating transbilayer movement of membrane phospholipids in response to elevated [Ca2+]. This activity was first purified in the human PLSCR1 protein (Zhou et al., 1997), named PLSCR1 for phospholipid scramblase 1. Subsequently, further members of the gene family have been identified in the human genome (Wiedmer et al., 2000). More recently members of this family have been shown to have a role in cellular signaling. When palmitoylated, PLSCR1 was shown to be raft-associated, to physically interact with ligand-activated EGF receptors, and to serve as an adapter to promote interaction between c-Src, Shc and the EGFR receptor kinase, thus serving to enhance receptor transactivation (Sun et al., 2002). In rat, PLSCR1 has been shown to be involved in mast cell activation through a Lyn-dependent pathway (Amir-Moazami et al., 2008) with PLSCR2 possibly playing an antagonistic role in mast cell activation (Hernandez-Hansen et al., 2005). It has also been shown that PLSCR1 when not palmitoylated is avidly imported into the nucleus by the importin-α β nuclear chaperones, where it functions as a DNA-binding protein with transcriptional activity (Ben-Efraim et al., 2004). One identified gene target of nuclear PLSCR1 is the inositol 1,4,5-trisphosphate receptor type 1 gene (IP3R1), nuclear PLSCR1 serving to increase both IP3R1 transcription and protein expression (Zhou et al., 2002).
Mouse knockouts of each of PLSCR1 and PLSCR3 have been studied. Deletion of PLSCR1 affects myelopoiesis in response to G-CSF, causing a defect in emergency granulopoiesis (Zhou et al., 2002). In contrast, deletion of PLSCR3 in mouse gives rise to abnormal triglyceride and cholesterol accumulation in white fat, with subsequent manifestations of the metabolic syndrome (Wiedmer et al., 2004). Deletion of PLSCR orthologues in Drosophila was found to cause abnormality in flight that was apparently related to a defect in motor neurons affecting the distribution and size of neutransmitter storage vesicles at the synapse (Acharya et al., 2006). PLSCR3 and other members of the PLSCR family have also been reported to be mitochondrial proteins, and proposed to mediate both lipid transport and pro-apoptotic functions Liu et al., 2003a, b; Van et al., 2007).
The seemingly disparate and enigmatic biology that has been observed for the PLSCR family of proteins has to date precluded the development of a logically consistent molecular model for how these highly conserved and presumably related proteins might collectively function in the cell. In addition to the disparity of molecular interactions and biologic functions that have been ascribed to the PLSCR proteins, a unifying concept of the functional properties of the PLSCR family has also been hindered by the absence of any solved or predicted molecular structure for its members, precluding an attempt to gain insight into protein function from protein structure. Whereas site-directed mutagenesis has revealed selected residues that are either targets of post-translational modification (thiolesterification and phosphorylation), or that are required to observe PLSCR1 binding to lipid, proteins or DNA, there is virtually no information as to how the functional domains might be organized, or how their 3D structure might be related to other proteins with defined function. In order to gain new insight into the potential structure of the PLSCR protein family, we undertook extensive similarity searches to identify structural homologues for the phospholipid scramblase family.
2 METHODS
Profile–profile comparisons were carried out using the Pfam database of protein families (release 23). The profile for the Scramblase family (accession PF03803) was compared with the 10 339 other family profiles using the SCOOP software (Bateman and Finn, 2007) and the PRC software using default parameters. The remote homology detection server HHpred (Soding, 2005) was used to search for homologs of human PLSCR1 in the Protein Data Bank with default parameters. An alignment of human PLSCR1 with the DUF567 family member At5g01750 from Arabidopsis thaliana (PDB-code: 1zxu) was generated with HHpred (Soding, 2005) using its maximum accuracy (MAC) alignment algorithm (Soding et al., 2005) and adjusted manually. Structural models were built with the Modeller software (Sali and Blundell, 1993) with default parameters. Loops were modeled with the MODloop server (Fiser and Sali, 2003). Verify3d (Luthy et al., 1992) was used to check the model quality.
3 RESULTS
Profile–profile comparisons provide a powerful way to link homologous protein families. Using the Pfam database of protein families we were able to scan for similarities of the Scramblase family to over 10 000 other protein families. A potential relationship of the Pfam Scramblase family to the uncharacterized Pfam protein family DUF567 was initially identified with the SCOOP software (Bateman and Finn, 2007) with a score of 47.7. The next highest scoring match was DUF512 with a score of 15.3 which is not considered significant. In addition the PRC (Profile Comparer) software identified the relationship between the same two Pfam families with an E-value of 0.0267. The next highest scoring match was to the RdRP_2 family (PF00978) with a non-significant E-value of 2.39. We also removed the N-terminal proline-rich stretch of 70 amino acids from human PLSCR1 and searched through the Protein Database with the HHpred server (Soding et al., 2005). The first match was to At5g01750 (PDB-code: 1zxu), a member of the DUF567 family from A.thaliana. At E-value 2e-11 and probability for homology of 99.5% this result is strongly indicative of a homologous relationship. The second match was again to 1zxu, but with a different alignment, indicating the presence of internal sequence symmetry (E-value 0.04, 95%). The third hit was to Tubby-related protein 1 (PDB: 2fim_A), with E-value 14 and probability for homology of 42%.
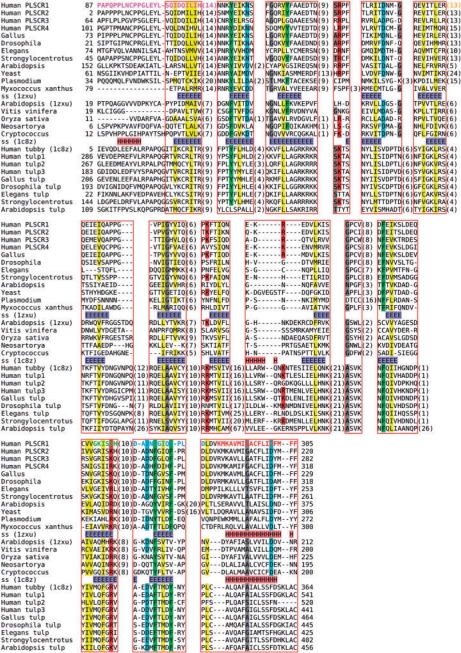
The DUF567 family, defined by the Pfam database (Finn et al., 2008), contains only uncharacterized proteins. It is found in plants, fungi, eubacteria and some archaea. Within the plants DUF567 is greatly expanded, with A.thaliana containing 21 members of the family. Despite being functionally uncharacterized, the family has had a representative structure solved by the CESG structural genomic project (http://www.uwstructuralgenomics.org/) for the Arabidopsis protein At5g01750 (UniProt:Q9LZX1) (PDB-code: 1zxu). Although no function is known for this protein, taking its sequence and PSI-BLAST searching (Altschul et al., 1997) it against the NR database at NCBI with default E-values we find that by round four several members of the Scramblase family are identified, including PLSCR1, PLSCR3 and PLSCR5. This confirms that the scramblase family is related to the DUF567 family and therefore identifies a structural template for modeling of the scramblases. An alignment of representatives of the scramblases to the DUF567 family is shown in Figure 1.
Fig. 1.
Multiple alignment showing members of the scramblase and DUF567 families (upper block) and the tubby-like family (lower block). Members within each family were aligned with the multiple alignment program PROMALS (Pei and Grishin, 2007). The scramblase and DUF567 sequences were aligned with each other using HHpred (Soding, 2005) in local MAC alignment mode (Soding et al., 2005), while keeping the family alignments frozen. The resulting alignment was merged with the alignment of tubby-like proteins by using the structural alignment of 1zxu and 1c8z from TMalign (Zhang et al., 2005) as a guide and again keeping sub-alignments frozen. Red boxes represent regions that are part of the structural alignment. Alignment columns for which scramblase and tubby family members exhibit similar amino acids are colored according to the chemical nature of the residue class. Various sequence features of the PLSCR1 sequence are indicated by colored, bold letters: transcriptional activation domain (magenta), Cysteine palmitoylation motif (orange), non-classical nuclear localization signal (green), Ca2+-binding motif (blue), predicted transmembrane helix (red).
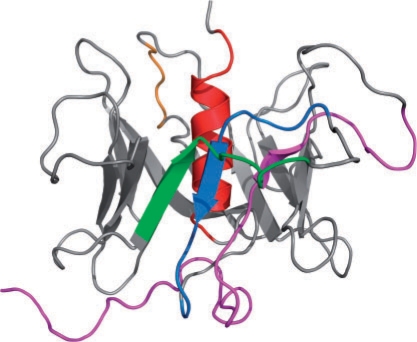
The structure of At5g01750, and therefore by similarity the Scramblase family, is a 12-stranded β-barrel that encloses a central C-terminal α-helix, see Figure 2. This C-terminal helix is thought to be a transmembrane helix. However, the structural model suggests that the hydrophobic nature of this helix is due to its packing in the core of the protein domain and that this is not a transmembrane helix. It can be seen that most of the sequence conservation lies within the secondary structures as well as the β-hairpin turns between strands 2–3 and 4–5 providing further support for the model.
Fig. 2.
3D structural model of PLSCR1 computed by homology modeling. PLSCR1 forms a closed, symmetric β -barrel of 12 β-strands wrapped around a very hydrophobic C-terminal helix. Various sequence features of PLSCR1 are highlighted in color: transcriptional activation domain (magenta), Cysteine palmitoylation motif (orange), non-classical nuclear localization signal (green), Ca2+-binding motif (blue), predicted transmembrane helix (red).
The structural model allows an improved understanding of previous results. For example, the DNA-binding motif defined by deletion experiments (Zhou et al., 2005) was identified as spanning residues 86–118 in human PLSCR1. This deletion would have removed the first β-strand of the domain potentially leading to misfolding of the β-barrel with observed loss of DNA binding. Thus, it remains unresolved whether the observed results from truncation mutation identify the actual DNA-binding residues in PLSCR1, or, cause misfolding of the DNA-binding motif. Also it has been hypothesized that PLSCR contains an EF-hand like Ca2+-binding motif (D273–D284), and mutation of select residues within that motif were observed to abrogate both Ca2+ binding to PLSCR1 and the expression of its Ca2+-dependent PL scramblase activity (Zhou et al., 1998). The proposed motif overlaps with one of the core β-strands of the β-barrel and thus is structurally incompatible with an EF-hand structure.
As well as providing a structural model for the scramblases, the structure of At5g01750 shows the same fold as found in the C-terminal domain of the Tubby protein (Boggon et al., 1999). A possible homologous relationship between the tubby-like proteins and phospholipid scramblases is indicated by the fact that a tubby family protein (PDB-code: 2fimA) appears as second best match in a database search of PLSCR1 through the PDB using HHpred, albeit with marginal statistical significance. The tubby family of proteins is only found in eukaryotic species, whereas the scramblase/DUF567 family are found in eukaryotes and eubacteria, which suggests that the scramblases have a more ancient evolutionary origin. We hypothesize that the tubby family of proteins evolved from an ancestral scramblase-like protein. The scramblase structure appears to have fewer elaborations between the β-strands compared with the tubby family, which also suggests their more basic ancestral nature.
If scramblases and tubby proteins are distantly related, we might expect to find some common functional features. There are a number of interesting similarities between the two families of proteins that support an evolutionary connection. First, most members of the scramblase and tubby family possess an N-terminal stretch of 100–250 natively unfolded residues that might function as activation domains or protein–protein interaction domains. Both scramblases and tubby are localized to the inner side of the cell membrane, in the case of scramblases by palmitoylation and in the case of Tubby by phosphoinositol binding. In both cases these localizations appear to be reversible and disruption of the membrane binding gives rise to a nuclear localization. Members of each family have been shown to be DNA binding (Boggon et al., 1999; Zhou et al., 2005), although no specific target has yet been identified for Tubby. Figure 1 shows two positions that conserve positively charged residues between the scramblases and tubby proteins, one between strands 3 and 4 and one at the end of strand 11. These two positions are both at the same end of the barrel and might form part of a DNA-binding site. One of the binding targets of PLSCR1 is the promoter of the inositol 1,4,5-trisphosphate receptor. It is notable that calcium release mediated by inositol 1,4,5-trisphosphate is part of the signaling cascades mediated by this protein. Furthermore, it has been shown that Tubby can bind to phosphatidylinositol 4,5-bisphosphate. We tentatively suggest that scramblase genes may also share inositol polyphosphate-binding activity. It is also possible that tubby proteins may possess scramblase activity.
4 CONCLUSIONS
In conclusion, we have demonstrated a structural model for the family of scramblase proteins and have identified an evolutionary link with the tubby-like proteins. The members of this new superfamily are both endofacial plasma membrane and nuclear distributed, with distinct plasma membrane and nuclear functions. Nuclear trafficking of these proteins requires liberation from membrane attachment and import by nuclear chaperones, and results in binding to chromatin and altered gene transcription. In addition to common structural ancestory, the possibility of conserved function as modifiers of polyphosphoinositide-based cell signaling is also suggested.
Funding: This work was supported by the Wellcome Trust [grant number WT077044/Z/05/Z]; Heart, Lung, Blood Institute, National Institutes of Health (HL036946, HL063819 and HL076215 to P.J.S. and T.W.).
Conflict of Interest: none declared.
REFERENCES
- Acharya U, et al. Drosophila melanogaster Scramblases modulate synaptic transmission. J. Cell Biol. 2006;173:69–82. doi: 10.1083/jcb.200506159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir-Moazami O, et al. Phospholipid scramblase 1 modulates a selected set of IgE receptor-mediated mast cell responses through LAT-dependent pathway. J. Biol. Chem. 2008;283:25514–25523. doi: 10.1074/jbc.M705320200. [DOI] [PubMed] [Google Scholar]
- Bateman A, Finn RD. SCOOP: a simple method for identification of novel protein superfamily relationships. Bioinformatics. 2007;23:809–814. doi: 10.1093/bioinformatics/btm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim I, et al. Phospholipid scramblase 1 is imported into the nucleus by a receptor-mediated pathway and interacts with DNA. Biochemistry. 2004;43:3518–3526. doi: 10.1021/bi0356911. [DOI] [PubMed] [Google Scholar]
- Boggon TJ, et al. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286:2119–2125. doi: 10.1126/science.286.5447.2119. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- Hernandez-Hansen V, et al. Increased expression of genes linked to FcepsilonRI signaling and to cytokine and chemokine production in Lyn-deficient mast cells. J. Immunol. 2005;175:7880–7888. doi: 10.4049/jimmunol.175.12.7880. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. Phospholipid scramblase 3 is the mitochondrial target of protein kinase C delta-induced apoptosis. Cancer Res. 2003;63:1153–1156. [PubMed] [Google Scholar]
- Liu J, et al. Phospholipid scramblase 3 controls mitochondrial structure, function, and apoptotic response. Mol. Cancer Res. 2003;1:892–902. [PubMed] [Google Scholar]
- Luthy R, et al. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23:802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TA. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Soding J, et al. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, et al. Plasma membrane phospholipid scramblase 1 is enriched in lipid rafts and interacts with the epidermal growth factor receptor. Biochemistry. 2002;41:6338–6345. doi: 10.1021/bi025610l. [DOI] [PubMed] [Google Scholar]
- Van Q, et al. Phospholipid scramblase-3 regulates cardiolipin de novo biosynthesis and its resynthesis in growing HeLa cells. Biochem. J. 2007;401:103–109. doi: 10.1042/BJ20060373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmer T, et al. Adiposity, dyslipidemia, and insulin resistance in mice with targeted deletion of phospholipid scramblase 3 (PLSCR3) Proc. Natl Acad. Sci. USA. 2004;101:13296–13301. doi: 10.1073/pnas.0405354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmer T, et al. Identification of three new members of the phospholipid scramblase gene family. Biochim. Biophys. Acta. 2000;1467:244–253. doi: 10.1016/s0005-2736(00)00236-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, et al. Molecular cloning of human plasma membrane phospholipid scramblase. A protein mediating transbilayer movement of plasma membrane phospholipids. J. Biol. Chem. 1997;272:18240–18244. doi: 10.1074/jbc.272.29.18240. [DOI] [PubMed] [Google Scholar]
- Zhou Q, et al. Identity of a conserved motif in phospholipid scramblase that is required for Ca2+-accelerated transbilayer movement of membrane phospholipids. Biochemistry. 1998;37:2356–2360. doi: 10.1021/bi972625o. [DOI] [PubMed] [Google Scholar]
- Zhou Q, et al. Normal hemostasis but defective hematopoietic response to growth factors in mice deficient in phospholipid scramblase 1. Blood. 2002;99:4030–4038. doi: 10.1182/blood-2001-12-0271. [DOI] [PubMed] [Google Scholar]
- Zhou Q, et al. Phospholipid scramblase 1 binds to the promoter region of the inositol 1,4,5-triphosphate receptor type 1 gene to enhance its expression. J. Biol. Chem. 2005;280:35062–35068. doi: 10.1074/jbc.M504821200. [DOI] [PubMed] [Google Scholar]