Abstract
Members of the ERF transcription factor family play important roles in regulating gene expression in response to biotic and abiotic stresses. In soybean (Glycine max L.), however, only a few ERF genes have been studied so far. In this study, 98 unigenes that contained a complete AP2/ERF domain were identified from 63 676 unique sequences in the DFCI Soybean Gene Index database. The phylogeny, gene structures, and putative conserved motifs in soybean ERF proteins were analysed, and compared with those of Arabidopsis and rice. The members of the soybean ERF family were divided into 12 subgroups, similar to the case for Arabidopsis. AP2/ERF domains were conserved among soybean, Arabidopsis, and rice. Outside the AP2/ERF domain, many soybean-specific conserved motifs were detected. Expression analysis showed that nine unigenes belonging to six ERF family subgroups were induced by both biotic/abiotic stresses and hormone treatment, suggesting that they were involved in cross-talk between biotic and abiotic stress-responsive signalling pathways. Overexpression of two full-length genes from two different subgroups enhanced the tolerances to drought, salt stresses, and/or pathogen infection of the tobacco plants. These results will be useful for elucidating ERF gene-associated stress response signalling pathways in soybean.
Keywords: ERF family, gene function, phylogeny, soybean, stress response
Introduction
Drought, high salinity, low temperature, and pathogen attack are the most common stress factors that influence plant growth and development. To overcome these limitations, plants respond and adapt to stresses at the physiological and biochemical levels. AP2/ERF transcription factors, characterized by the presence of the AP2/ERF DNA-binding domain, play significant roles in regulating plant biotic and abiotic stress-responsive gene expression, (Sakuma et al., 2002). AP2/ERF genes constitute a large superfamily, which has been divided into three groups named the AP2, ERF, and RAV families based on their sequence similarities and numbers of AP2/ERF domains (Nakano et al., 2006). AP2 proteins contain two AP2/ERF domains, and genes in this family participate in the regulation of developmental processes (Elliott et al., 1996; Chuck et al., 1998; Boutilier et al., 2002). RAV family proteins contain one AP2/ERF domain and a B3 domain, and have different biological functions compared with members in other families. Recently, members of the RAV family were shown to be involved in the ethylene response (Alonso et al., 2003), the brassinosteroid response (Hu et al., 2004), and biotic and abiotic stress responses (Sohn et al., 2006). ERF family proteins contain a single AP2/ERF domain, and are sometimes further divided into two major subfamilies, the CBF/DREB subfamily and the ERF subfamily (Sakuma et al., 2002). Genes in the CBF/DREB subfamily play a crucial role in the response of plants to abiotic stresses by recognizing the dehydration-responsive element (DRE) with a core motif of A/GCCGAC (Yamaguchi-Shinozaki and Shinozaki, 1994; Thomashow, 1999). The ERF subfamily genes are mainly involved in response to biotic stresses such as pathogenesis by recognizing the cis-acting element AGCCGCC, known as the GCC box (Hao et al., 1998).
ERF and CBF/DREB subfamily transcription factors have been identified in various plant species, including Arabidopsis (Liu et al., 1998; Oñate-Sánchez and Singh, 2002), rice (Cao et al., 2006), and cotton (Huang et al., 2007; Jin and Liu, 2008). The roles of ERF and CBF/DREB proteins in the response to biotic and abiotic stress have also been extensively documented (Gutterson and Reuber, 2004; Agarwal et al., 2006). The sequenced Arabidopsis genome contains 145 distinct genes encoding AP2/ERF-type proteins classified into five groups, that is APETALA2 (AP2) subfamily (17 genes), RAV subfamily (six genes), CBF/DREB subfamily (56 genes), ERF subfamily (65 genes), and one very specific gene, AL079349, based on similarities of their AP2/ERF DNA-binding domains (Sakuma et al., 2002). The proteins of the CBF/DREB subfamily were further divided into six subgroups, A-1 to A-6, among which A-1 and A-2 were the two largest (Sakuma et al., 2002). Expression of the DREB1A/CBF3 (A-1) genes is induced by low temperature stress, but not by drought or high salt stresses, whereas DREB2A (A-2) genes are induced by drought and high salt, but not by low temperature (Liu et al., 1998). Overexpression of DREB1A/CBF3 under control of the cauliflower mosaic virus (CaMV) 35S promoter increased tolerance to drought, high salt, and freezing stresses (Liu et al., 1998, Kasuga et al., 1999, Gilmour et al., 2000). Overexpression of constitutively active DREB2A resulted in significant drought stress tolerance, but only slight freezing tolerance in transgenic Arabidopsis plants (Sakuma et al., 2006). Other DREB proteins such as TINY2 (A-4), GhDBP1 (A-5), GmDREB2 (A-5), and ZmDBF1 (A-6) were also characterized as stress-inducible proteins (Kizis and Pages, 2002; Wei et al., 2005; Huang and Liu, 2006; Chen et al., 2007). The proteins of the ERF subfamily were also divided into six groups termed B-1 to B-6. The expression and biological functions of genes in the ERF subfamily were summarized by Nakano et al. (2006). As an example, transcription of tobacco Tsi1 (for Tobacco stress-induced gene1) was induced by salt, ethephon (ET), and salicylic acid (SA). Overexpression of Tsi1 improved the tolerance to salt and pathogen attack (Park et al., 2001). Nakano et al. (2006) reported 147 and 157 genes in Arabidopsis and rice, respectively, which were classified as members of the AP2/ERF superfamily. Among the Arabidopsis ERF genes, 122 were considered as members of the CBF/DREB subfamily and the ERF subfamily (Sakuma et al., 2002). In rice, there are 139 genes in the ERF family (Nakano et al., 2006). The phylogeny, gene structures, and conserved motifs of ERF genes in these two species were also analysed (Nakano et al., 2006).
Soybean is one of the most economically important crop species in the world. Only a few members of the ERF and CBF/DREB subfamily have been characterized in this species (Mazarei et al., 2002; Li et al., 2005), and most of their functions remain to be determined. Recently, four CBF/DREB homologous genes (GmDREBa/b/c and GmDREB2), and one ERF homologous gene (GmEREBP1) were isolated from soybean and their expression characteristics were investigated (Mazarei et al., 2002; Li et al., 2005; Chen et al., 2007). To gain further information about the AP2/ERF superfamily in soybean, the DFCI Soybean Gene Index database was surveyed and 148 unigenes were identified in this superfamily, including 120 ERF family unigenes, 26 AP2 family unigenes, and two RAV family unigenes. Phylogenetic and protein motif structural analyses of the ERF and CBF/DREB subfamily were undertaken. The expression patterns of some genes belonging to the ERF subfamily were also characterized. The biological functions of the two full-length ERF genes were investigated in transgenic tobacco plants. The results from this study, reported herein, form a basis for future functional analyses of the soybean ERF family genes.
Materials and methods
Data set and data treatment
The soybean unigene set and the expressed sequence tags (ESTs) used as a primary sequence data set are available on the TIGR Web site (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb = soybean) as the DFCI Soybean Gene Index, from which 63,676 unique sequences were download including 31 928 TCs (tentative consensus sequences), 31 636 singleton ESTs, and 112 singleton expressed transcript sequences. These data were released on 20 September 2004.
The Transeq program from the EMBOSS package was used to translate DNA sequences into protein sequences. The amino acid sequences of the longest open reading frame (ORF) in six ORFs were selected, and amino acid sequences whose lengths <100 were excluded for the following analysis. Based on the HMMER User's Guide (http://hmmer.wustl.edu/ Version 2.3.2; Oct 2003), the Hmmpfam program was then used to annotate various kinds of domains in the query sequence, then the hmmfetch program was used to retrieve an HMM as a seed model from an HMM database, including the AP2/ERF domain. Finally, the hmmalign program was used to align multiple TC/EST sequences to the seed profile HMM, and 148 TC/EST sequences containing the AP2/ERF domain were obtained.
The Arabidopsis gene set is available through the Arabidopsis Information Resource (http://www.Arabidopsis.org). The rice genes of the ERF family were downloaded from the National Center for Biotechnology Information (NCBI) and TIGR rice genome annotation databases (http://rice.plantbiology.msu.edu/).
Alignment, phylogenetic analysis, and motif detection
All similarity searches were executed locally using the BlastN, BlastX, or BlastP tools at the NCBI, TIGR, and TAIR web sites. Conserved domain searches were performed against the conserved domain database at NCBI using the reversed position-specific blast algorithm with translated unigenes. A phylogenetic tree was constructed with the aligned soybean AP2/ERF protein sequences using MEGA (version 4.0; http://www.megasoftware.net; Tamura et al., 2007) and the Neibhbor-Joining (NJ) method with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed). The amino acid variation rates were also obtained. Motif detection was performed with MEME (Bailey et al., 2006) (MEME version 3.5.7, http://meme.sdsc.edu/meme/meme.html).
Plant materials, stress treatments and RT-PCR analysis
Soybean cultivars ‘Tiefeng 8’, resistant to salinity stress, and ‘Zhongpin 95-5383’, resistant to soybean mosaic virus (SMV), were used throughout the study. Plant treatments were performed at the two-true leaf stage. ‘Tiefeng 8’ was used for salt, drought, cold stresses, and abscisic acid (ABA) treatments. ‘Zhongpin 95-5383’ was used for SA, ET, jasmonate (JA), and SMV infection treatments. Stress treatments were performed as follows: for salt stress, the roots of seedlings were dipped into 200 mM NaCl solutions; for chilling treatment, seedlings were put into a 4 °C growth chamber; for drought, the root systems of intact plants were washed gently with water to remove soil and then put on filter paper for rapid dehydration; for ABA, SA, and Me-JA treatments, soybean seedlings were sprayed with 200 μM ABA dissolved in 0.01% ethanol, 2 mM SA in water, and 100 μM JA in 0.01% ethanol, respectively. ET treatment was performed in a gas-tight plexiglass chamber by dissolving 2 ml of 40% ET and 1 g of NaHCO3 in 200 ml of H2O (under these conditions ET will liberate ethylene gas); and for SMV treatment, mechanical inoculation was carried out by rubbing leaves with a brush dipped in a mixture of carborundum and an extract of infected leaves ground in the phosphate buffer (pH 7.2). After inoculation, leaves were rinsed with tap water. After exposure to these stresses, soybean seedlings were harvested at various time points, frozen in liquid nitrogen, and kept at –80°C for further analysis.
Total RNA was isolated from plant materials at various time points with Trizol (Tiangen Biotech., Beijing, China) according to the manufacturer's instructions. Poly(A)+ RNA was used as the template for synthesis of first-strand cDNA. cDNA was generated with reverse transcriptase (TaKaRa, Dalian, China). Gene-specific primers, listed in Supplementary Table S1 available at JXB online, were designed to avoid the conserved region. The specific primer pair (5′-AACCTCCTCCTCATCGTACT- 3′ and 5′-GACAGCATCAGCATGTTCA-3′) for soybean tubulin was used as an internal control. Reactions were performed in a PTC-200 Peltier Thermal Cycler (MJ RESEARCH).
Plant transformation and stress tolerance analyses of transgenic tobacco plants
To analyse gene functions, full-length transcripts of GmERF057 in the B-2 subgroup and GmERF089 in the B-5 subgroup of the ERF subfamily were amplified using the specific primer pair 5′-GCTCTAGATTCATCTGAGATGTGTGGAGG-3′ and 5′-CGGAGCTCAAGCGGTTCAGAAAACTCCA-3′ for GmERF057, and 5′-GCTCTAGATGGCTTCATCTTCCATCAAAAACAC-3′ and 5′-CGGAGCTCTCAAAGAGCGACAAGAGGATCCCA-3′ for GmERF089, and then cloned into the polylinker site of the binary vector, pBI121. The constructs were transferred into tobacco W38 using the Agrobacterium-mediated method (Hoekema et al., 1983). Transgenic plants were selected on MS medium containing 200 μg ml−1 kanamycin and 500 μg ml−1 carbenicillin.
For salt stress, shoot tips of both wild-type and GmERF057 transgenic tobacco were excised from aseptic seedlings and were transferred to 1/2 MS medium containing 200 mM NaCl for 30 d. Eight-week-old, soil-grown GmERF089 plantlets and the wild-type control were watered once every 5 d using 600 mM NaCl solution. For drought stress, surface-sterilized seeds of wild-type and T2 transgenic GmERF089 tobacco plants were plated on MS medium for seed germination, then transferred to MS medium containing 2% polyethylene glycol (PEG) for 30 d. Plantlet growth was evaluated for drought tolerance.
For bacterial resistance experiments, Ralstonia solanacearum strain was grown in LB broth. The bacterial cells were collected, washed, and resuspended in 10 mM MgCl2. Bacterial cells in suspension (2×107 cfu ml−1) were infiltrated into fully expanded tobacco leaves using a 10 ml plastic syringe without a needle, following the method of Thilmony et al. (1995). Bacterial populations in leaves were measured by grinding four leaf discs in 10 mM MgCl2, plating serial dilutions on LB plates, and counting colony-forming units.
Results
Identification of unigenes containing the AP2/ERF domain in soybean
A total of 148 unigenes were identified as possibly containing AP2/ERF domain(s) (Table 1). The individual unigenes are listed in Supplementary Tables S2 and S3 at JXB online. Among them, eight unigenes were predicted to encode two complete or incomplete AP2/ERF domains and were assigned to the AP2 family. Eighteen unigenes were predicted to encode a complete or incomplete AP2/ERF domain, whereas their AP2/ERF domains were distinct from those of members of the ERF family and more closely related to those of the AP2 family. Thus, these unigenes were also assigned to the AP2 family. TC218282 was predicted to encode one AP2/ERF domain and one B3 domain. Although BE659979 was predicted to encode one AP2/ERF domain, it differed from the ERF type and was more closely related to the RAV type; two such unigenes were assigned to the RAV family. Of 120 unigenes encoding a single AP2/ERF domain, and assigned to the ERF family, 98 were predicted to encode a complete AP2/ERF domain and the remaining 22 encoded an incomplete AP2/ERF domain. The 98 unigenes were further subclassified into two groups on the basis of similarity of the amino acid sequences of the AP2/ERF domains; 36 unigenes encoding CBF/DREB-like protein were assigned to the CBF/DREB subfamily and 62 unigenes encoding ERF-like protein were assigned to ERF subfamily.
Table 1.
Summary of the structure of the AP2/ERF superfamily in the soybean compared with Arabidopsis
| Group | Conserved domain | Soybean | Arabidopsisa |
| AP2 family | 26 | 17 | |
| Double complete or incomplete AP2/ERF domain | 8 | ||
| Single complete or incomplete AP2/ERF domain | 18 | ||
| RAV family | 2 | 6 | |
| Single AP2/ERF domain and one B3 domain | 1 | ||
| Single AP2/ERF domain | 1 | ||
| ERF family | 120 | 121 | |
| Single incomplete AP2/ERF domain | 22 | ||
| CBF/DREB subfamily | Single complete AP2/ERF domain | 36 | 56 |
| ERF subfamily | Single complete AP2/ERF domain | 62 | 65 |
| AL079349 | 1 | ||
| AP2/ERF superfamily | Total | 148 | 145 |
Totals for each family are in bold.
From Sakuma et al. (2002).
The 98 unigenes of the ERF family containing a complete AP2/ERF domain were analysed further. Generic names (GmERF001–GmERF098) were assigned to distinguish each one for the purposes of the study (Supplementary Table S3 at JXB online).
Phylogenetic relationships between members of the ERF family in soybean and Arabidopsis
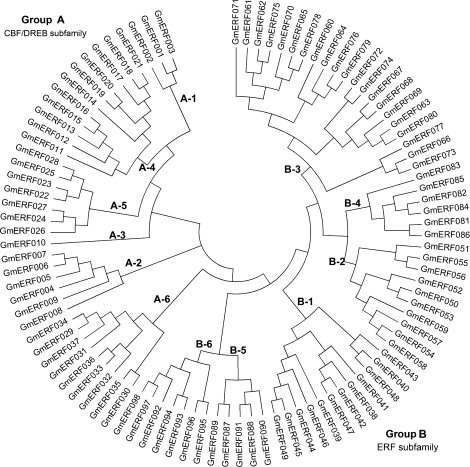
To confirm classifications and to analyse the phylogenetic relationships, multiple alignment analyses of the amino acid sequences of the AP2/ERF domain in the 98 soybean ERF proteins were performed using the 122 Arabidopsis ERF proteins described by Nakano et al. (2006) (Supplementary Fig. S1 at JXB online) for comparison. Residues Glu17, Trp36, Leu37, and Gly38 were completely conserved among all 220 proteins in both species (Supplementary Fig. S1). In addition, >95% of the ERF family members contain Gly4, Arg6, Arg8, Gly12, Ile18, Arg19, Arg34, Ala46, Ala47, Asp51, and Asn65 residues. Based on alignment, an NJ phylogenetic tree was generated with bootstrap analysis (1000 replicates). As shown in Fig. 1 and Supplementary Fig. S2 at JXB online, the phylogenetic tree divided the ERF family proteins of Arabidopsis and soybean into 12 subgroups, designated A-1 to A-6 and B-1 to B-6, in accordance with the classification described by Sakuma et al. (2002). For example, CBF2/DREB1C (At4g25470) and DDF1/DREB1E (At1g63030) in subgroup A-1 in Arabidopsis (Sakuma et al., 2002) was also placed in subgroup A-1 in the present study. Subgroups A-1 to A-6 represent the CBF/DREB subfamily, and subgroups B-1 to B-6, the ERF subfamily. Comparative analyses of the phylogenetic tree suggested that the classification of the soybean ERF family was similar and applicable to the Arabidopsis ERF family.
Fig. 1.
An unrooted phylogenetic tree of soybean ERF proteins. The amino acid sequences of the AP2/ERF domain of 98 soybean ERF family proteins were aligned by Clustal W and the phylogenetic tree was constructed using MEGA 4.0 and the NJ method. The proteins are named according to GmERF numbers (see Supplementary Table S3 at JXB online).
Conserved motifs outside of the AP2/ERF domain in soybean and Arabidopsis
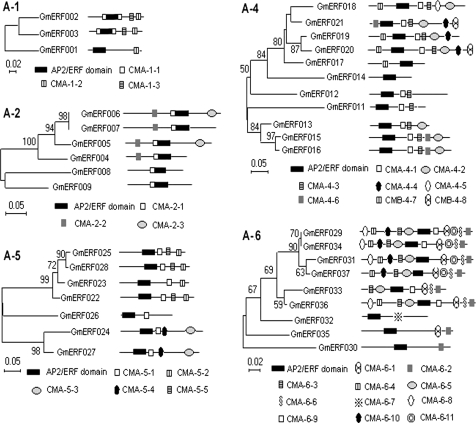
The conserved motifs in ERF family proteins in both soybean and Arabidopsis were investigated using MEME version 3.5.4, and the results for both species are listed in Supplementary Table S4 at JXB online. Most members in the same group shared one or more motifs outside the AP2/ERF domain (Figs 2, 3). For example, the A-1 subgroup consisted of three unigenes (GmERF001 to 003) and contained three conserved motifs (Fig. 2). All of the unigenes in this subgroup contain the CMA-1-2 motif in the C-terminal region; this was reported as an LWSY conserved motif in OsDREB1A/B/C and in AtCBF3/DREB1A (Dubouzet et al., 2003). In addition, this is the CMA-1-1 motif located on both sides of the AP2/ERF domain in the proteins of GmERF002 and GmERF003 (Fig. 2).
Fig. 2.
Phylogenetic relationships among soybean CBF/DREB subfamily (group A) unigenes. Bootstrap values from 1000 replicates were used to assess the robustness of the trees. The phylogenetic tree and a schematic diagram of the protein structures of every group are shown. Each box represents the AP2/ERF domain. Conserved motifs are summarized in Supplementary Table S4 at JXB online. These motifs were defined by multiple alignments with an MEME search.
Fig. 3.
Phylogenetic relationships among the soybean ERF subfamily (group B) unigenes. Bootstrap values from 1000 replicates were used to assess the robustness of the trees. The phylogenetic tree and a schematic diagram of the protein structures of every group are shown. Each box represents the AP2/ERF domain. Conserved motifs are summarized in Supplementary Table S4 at JXB online. These motifs were defined by multiple alignments with an MEME search.
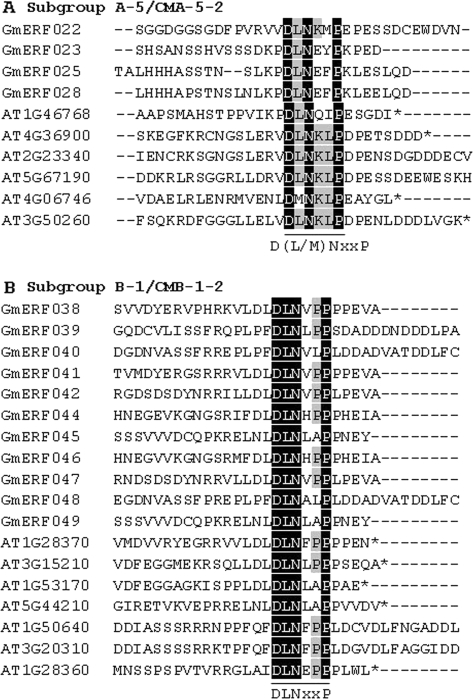
Some conserved motifs identified in the Arabidopsis ERF family were also examined in the deduced amino acid sequences of GmERF unigenes. For example, the ERF-associated amphiphilic repression (EAR) motif was found in members of subgroup A-5 as the CMA-5-2 motif, and in subgroup B-1 as the CMB-1-2 motif in both Arabidopsis and soybean (Supplementary Table S4 at JXB online, Figs 2–4). In addition, alanine-rich (in subgroup A-4 as the CMA-4-1 motif), glutamine-rich (in subgroup A-6 as the CMA-6-3 motif), and serine-rich (in subgroup A-4 as the CMA-4-7 motif, and in subgroup B-1 as the CMB-1-1 and CMB-1-3 motifs) amino acid sequences were detected in both Arabidopsis and soybean (Figs. 2, 3, Supplementary Table S4). These previously were often designated as transcriptional activation domains (Liu et al., 1999), but their functions were not rigorously demonstrated. An MCGGAII/L motif, designated as CMB-1-1, was a characteristic feature of subgroup B-1 in both species (Fig. 3, Supplementary Table S4).
Fig. 4.
EAR motif-like sequences conserved in the C-terminal region of subgroup A-5 and B-1 proteins in soybean and Arabidopsis. (A) Amino acid sequence alignment of the C-terminal region of subgroup A-5 proteins. (B) Amino acid sequence alignment of the C-terminal region of subgroup B-1 proteins. Conserved motifs are underlined. Black and grey shading indicate identical and conserved amino acid residues present in >50% of the aligned sequences, respectively. Consensus amino acid residues are given below the alignment. ‘X’ in the sequences indicates no conservation at that position.
In addition to the conserved motifs between soybean and Arabidopsis, there were also some soybean-specific motifs in the ERF family (Fig. 5, Supplementary Table S4). For example, the CMA-5-5 motif in subgroup A-5, the CMA-6-10 and CMA-6-11 motifs in subgroup A-6, the CMB-1-4, CMB-1-8, and CMB-1-9 motifs in subgroup B-1, the CMB-2-2 and CMB-2-8 motifs in subgroup B-2, and the CMB-3-9 motif in subgroup B-3 occurred only in soybean ERF proteins, but the functions of these motifs remain unknown.
Fig. 5.
Soybean-specific sequence motifs conserved in subgroups A-6, B-1, and B-2 of the ERF family. (A) Amino acid sequence alignment of subgroup A-6 proteins. (B) Amino acid sequence alignment of subgroup B-1 proteins. (C) Amino acid sequence alignment of subgroup B-2 proteins. The conserved motifs are underlined. Black and grey shading indicate identical and conserved amino acid residues present in >50% of aligned sequences, respectively.
Comparative analysis of the ERF gene family between soybean and rice
A total of 139 rice ERF family members were downloaded from the NCBI and TIGR rice genome annotation databases according to the report of Nakano et al. (2006). To determine the phylogenetic relationships of the ERF family genes in soybean and rice, a multiple sequence alignment was performed using amino acid sequences in the AP2/ERF domain. This analysis revealed that those amino acid residues which might be involved in some form of physical contact with DNA are also conserved among most of the soybean ERF proteins and rice ERF proteins (Supplementary Fig. S3 at JXB online). Residues Arg8 and Gly39 were completely conserved among all 237 proteins in both species (Supplementary Fig. S3). In addition, >95% of the ERF family members contain Gly4, Arg6, Arg23, Arg33, Trp37, Leu38, Ala48, Ala50, Asp52, Asn75, and Phe76 residues (Supplementary Fig. S3). The phylogenetic tree containing soybean and rice ERF genes was constructed, and the phylogram were classed into 15 groups, namely groups I–XIV and a solo group VI-L (Supplementary Fig. S4 at JXB online), according to the report of Nakano et al. (2006). Among these groups, groups I–X were relevant to A1–A6 and B1–B6 shown in Supplementary Fig. S4 and Table S5 at JXB online. No soybean ERFs were assigned to groups XI–XIV and VI-L (Supplementary Fig. S4) that were specific to rice. Similarly, no Araidopsis ERFs fell in the groups XI–XIV either (Nakano et al., 2006).
The comparative analysis of conserved motifs indicated that most of the motifs conserved in the soybean and Arabidopsis ERF families also existed in the rice ERF families (Supplementary Table S5 at JXB online). However, some motifs, for example CMA-5-4 in group A-5 (identical to group II), CMA-4-4, CMA-4-5, and CMA-4-8 in group A-4 (belonging to group III), CMA-2-3 in group A-2 (belonging to group IV), CMB-5-5 in group B-5 (identical to group VI), CMB-1-6, CMB-1-9, and CMB-1-10 in group B-1 (identical to group VIII), CMB-3-8, CMB-3-10, CMB-3-11, and CMB-3-12 in group B-3 (identical to IX), and CMB-4-2,CMB-4-3, and CMB-4-4 in group B-4 (identical to group X), existing in both soybean and Arabidopsis ERF families, were not found in the rice ERF family. In contrast, the motifs CMX-1 and CMX-2, identified in both the soybean and rice ERF families, were not found in the Arabidopsis ERF family.
Expression pattern of certain unigenes belonging to the ERF subfamily in soybean
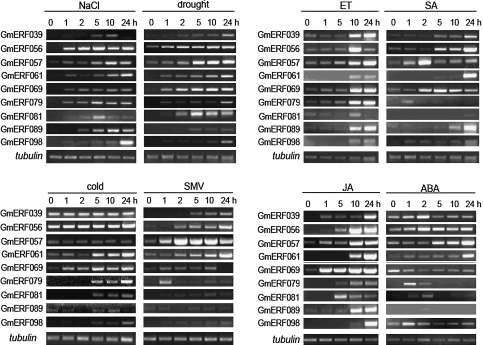
Previous reports indicated that the roles of the A group (CBF/DREB subfamily) of transcription factors were predominantly in regulation of the abiotic stress response, while those of the B group (ERF subfamily) were involved in biotic stress responses, abiotic stress responses, or both. Some members belonging to the B group were chosen for further study. The expression patterns of nine unigenes, namely GmERF039 in subgroup B-1, GmERF056 and GmERF057 in subgroup B-2, GmERF061, GmERF079, and GmERF069 in subgroup B-3, GmERF081 in subgroup B-4, GmERF089 in subgroup B-5, and GmERF098 in subgroup B-6, were investigated using RT-PCR under various stress conditions. As shown in Fig. 6, for high salt treatment, expression levels of GmERF056, GmERF079, and GmERF081 increased after initiation of the treatment, reached maxima at 5 h, and then decreased. The expression of GmERF039, GmERF057, and GmERF089 reached maxima at 10 h after salt treatment, and expression levels of GmERF061, GmERF069, and GmERF098 increased gradually over the 24 h period of treatment. For drought treatment, the expression levels of all nine unigenes increased gradually for at least 24 h, except for GmERF081 which peaked at 5 h. For cold treatment, the expression levels of GmERF098, GmERF081, GmERF061, and GmERF079 showed rapid increases at 5 h and remained at high levels until 24 h. The expression level of GmERF069 peaked at 5 h, and those of GmERF056, GmERF039, GmERF057, and GmERF089 were not affected. Following SMV inoculation, expression levels of GmERF039, GmERF056, and GmERF061 increased gradually for at least 24 h, whereas that of GmERF079 rapidly accumulated at 1 h, and then decreased to a low level. The expression of GmERF069 rapidly accumulated at 1 h, and was maintained for at least 10 h, but at 24 h its expression had declined to the level of uninoculated leaves. The expression level of GmERF057 reached a maximum at 2 h, and those of GmERF081, GmERF089, and GmERF098 were not affected.
Fig. 6.
Expression patterns of nine unigenes under various stresses. Total RNAs were isolated from soybean seedlings exposed to NaCl, drought, cold, SMV, ET, SA, Me-JA, and ABA for the indicated times. A 5 μg aliquot of total RNA was reverse-transcribed into first-strand cDNA for RT-PCR. The tubulin was amplified as a control.
Applications of ET, SA, and JA induced the expression of all nine unigenes. With ET treatment, the expression levels of all of unigenes peaked at 10 h or 24 h. For SA treatment, the expression levels were all induced and increased gradually until at least 24 h, except that GmERF057 peaked at 2 h, GmERF079 peaked at 1 h, and GmERF069 peaked at 5 h. For JA treatment, expression of GmERF069 rapidly increased at 1 h after treatment, and remained at this high level of expression for at least 24 h. The expression of GmERF081 was rapidly accumulated until 5 h after treatment, and subsequently declined. The expression profiles of the other seven unigenes reached a high level at 10 h or 24 h after treatment. For ABA treatment, GmERF089 transcript levels were slightly increased at 2 h after treatment, and otherwise they were similar to non-stressed levels. Transcript levels of GmERF039 and GmERF081 peaked at 2 h and then returned to pre-stressed levels. Expression levels of GmERF079 and GmERF098 reached peaks at ∼1 h after treatment. The expression of GmERF056 peaked at ∼2 h, and was maintained at a high level for up to 24 h. The GmERF061 and GmERF057 transcript levels gradually increased over the entire 24 h period. GmERF069 was subjected to negative regulation by ABA, with transcription after treatment being lower than pre-treatment levels.
Overexpression of GmERF057 and GmERF089 confers increased tolerance to biotic and abiotic stresses in transgenic tobacco plants
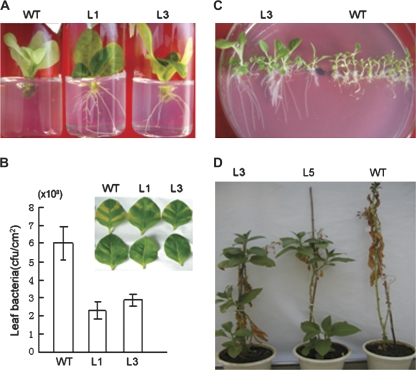
To date, most research has focused on the B-3 subgroup of the ERF subfamily, and the functions of members from other subgroups remain largely known. To verify the functions of members of the B-2 and B-5 subgroups, GmERF057 in the B-2 subgroup and GmERF089 in the B-5 subgroup were overexpressed in tobacco plants under the control of the CaMV35S promoter. For salt tolerance analyses of GmERF057 transgenic plants, significant phenotypic differences between wild-type and transgenic plants were observed after 30 d (Fig. 7A). During that period, leaves of wild-type plants gradually lost greenness, and root elongation was severely retarded, whereas leaves of the transgenic plants remained green and root development was vigorous, indicating that transgenic plants displayed tolerance against salt stress. The effects of overexpression of GmERF057 on the response to bacterial infection are shown in Fig. 7B. The transgenic lines exhibited significantly reduced disease lesions, and bacterial numbers were significantly reduced relative to wild-type plants. Bacterial numbers in the transformed plants were ∼50% of those in wild-type plants after 7 d of incubation. For GmERF089, increased drought and salt tolerances of transgenic tobacco plants were also observed in the seedling and mature growth stages, respectively (Fig. 7C, D). However, GmERF089 transgenic plants did not exhibit detectable tolerances to bacterial infection (data not shown).
Fig. 7.
Abiotic and biotic tolerance analyses of transgenic tobacco plants. (A) Salt tolerance of GmERF057 transgenic tobacco seedlings. Shoot tips excised from aseptic seedlings of wild-type and GmERF057 transgenic tobacco plants were transferred to 1/2 MS medium containing 200 mM NaCl for 30 d. (B) Bacterial pathogen responses of two independent transgenic tobacco lines (L1 and L3) and wild-type (WT) plants. The bacterial population was measured 7 d after inoculation with 107 cfu ml−1 of Ralstonia solanacearum. Disease symptoms in wild-type and GmERF057 plants are shown in the inset. Photographs are of leaves inoculated with bacteria (upper) and mock-inoculated (lower) with MgCl2 solution. Bar charts show bacterial counts for the wild-type and transformed plants. Results are averages of three replicates ±SD. (C) Drought tolerance in GmERF089 transgenic tobacco. Surface-sterilized seeds of the wild-type and T2 transgenic GmERF089 tobacco plants were plated on MS medium for seed germination, then moved to MS medium containing 2% PEG for 30 d. (D) Salt tolerance of GmERF089 transgenic tobacco. Eight-week-old, soil-grown GmERF089 plantlets and a wild-type control were watered once every 5 d by addition of 50 mM NaCl to a final concentration of 600 mM NaCl, and then photographed.
Discussion
Nakano et al. (2006) systematically surveyed the gene structures, phylogeny, and conserved motifs of the ERF gene family in Arabidopsis and rice, but relatively few soybean ERF genes were studied previously. To gain further information about the ERF family in soybean, 148 members of the AP2/ERF superfamily were identified from the soybean DFCI Soybean Gene Index database, including 120 members in the ERF family. These numbers were similar to those in Arabidopsis (147 members of the AP2/ERF superfamily) and rice (157 members), and included 122 and 139 members, respectively, in the ERF family (Nakano et al., 2006), indicating that although soybean has a large genome of 1115 Mb (Arumuganathan and Earle, 1991) compared with Arabidopsis (145 Mb) and rice (420 Mb), the structure and phylogeny of the AP2/ERF superfamily are similar in the three species. The presence of most subgroups in the three species also suggests that many of the genes pre-date the species divergence. Likewise, some groups/subgroups are present in only one species; for example, groups XI–XIV existed only in the rice ERF family but not in the Arabidopsis or soybean ERF families (Supplementary Fig. S4 at JXB online), suggesting that these groups had evolved or been lost in one species after this divergence. However, this comparison alone provides limited functional information, whereas queries with Arabidopsis or rice ERF genes of known function could identify candidate soybean orthologs with functional similarities. Some incompletely full-length ERF genes were missed in the present study, decreasing the likely number of ERF family members in soybean. However, a comparative analysis of soybean and Arabidopsis indicated that the phylogenetic analysis in soybean was reliable. Some of the unigenes have high sequence similarities with registered proteins in the NCBI web site (Table 2). For example, GmERF005, GmERF027, GmERF028, and GmERF076 encode proteins sharing 100% amino acid sequence identities with registered soybean proteins (GenBank accession nos AAT12423, AAP47161, ABB36646, and AAM45475, respectively). Therefore, the unigenes of the ERF family acquired in this study reflect the general status of ERF family members in soybean, and can be subjected to further analyses.
Table 2.
Soybean unigenes whose amino acid sequences have significant matches against the GenBank protein database
| Generic name | Match description | GenBank accession no. | E-value | Length query (amino acids) | Identities |
| GmERF005 | DREBa transcription factor | AAT12423 | 7.00E-99 | 215 | 100% |
| GmERF006 | DREB | AAP83131 | 2.00E-78 | 241 | 98% |
| GmERF007 | DREB | AAP83131 | 2.00E-68 | 219 | 98% |
| GmERF023 | DREB protein | AAY89658 | 4.00E-66 | 172 | 94% |
| GmERF025 | Dehydration-responsive element-binding protein | AAP47161 | 6.00E-77 | 178 | 90% |
| GmERF027 | Dehydration-responsive element-binding protein 3 | ABB36646 | 4.00E-104 | 229 | 100% |
| GmERF028 | Dehydration-responsive element-binding protein | AAP47161 | 3.00E-89 | 178 | 100% |
| GmERF029 | Dehydration-responsive element-binding protein 3 | AAZ03388 | 6.00E-130 | 316 | 99% |
| GmERF036 | DREB2 | AAQ57226 | 6.00E-146 | 313 | 99% |
| GmERF054 | Ethylene-responsive protein | AAQ10777 | 0 | 383 | 97% |
| GmERF058 | Ethylene-responsive protein | AAQ10777 | 1.00E-110 | 251 | 94% |
| GmERF076 | Ethylene-responsive element-binding protein 1 | AAM45475 | 1.00E-97 | 203 | 100% |
Comparative analysis of amino acid residues of the AP2/ERF domains in the soybean ERF family proteins with those of Arabidopsis and rice suggested the AP2/ERF domains were well conserved among the three species. These conserved amino acid residues probably indicate crucial roles for ERF family genes involved in different forms of physical contact with DNA. According to Allen et al. (1998), the AP2/ERF domain recognizes its target DNA via the conserved arginine and tryptophan residues in the β-sheet. Ala37 might play a crucial role in DNA binding or the stability of the AP2/ERF domain (Liu et al., 2006). The conserved motif analysis of the ERF family demonstrated that most motifs were conserved in soybean, Arabidopsis, and rice. Proteins within a subgroup that share these conserved motifs are likely to have similar functions. For example, the EAR motif is essential for gene repression (Ohta et al., 2001; Yang et al., 2005). Tobacco NtERF3, and Arabidopsis AtERF3 and AtERF4, containing the conserved EAR motif, repress the expression of a GCC-box-containing reporter gene (Fujimoto et al., 2000; Ohta et al., 2000, 2001). Mutations within the EAR motif eliminated this capacity for repression (Ohta et al., 2001). In addition to common conserved motifs in soybean, Arabidopsis, and rice, there are soybean-specific ERF family motifs, which may have important roles in regulating biological processes in soybean; those functions need to be be demonstrated further. The comparative analysis of conserved motifs in soybean, Arabidopsis, and rice suggested that protein functions have been both conserved and diverged during evolution of the ERF gene family.
Expression patterns of ERF subfamily unigenes under stress treatments
Plants undergo a range of environmental stresses in their natural environments and have evolved a wide range of mechanisms to cope with them. There are multiple stress perception and signalling pathways, some of which are specific whereas others cross-talk at various steps. This signalling cross-talk occurs in biotic stress signalling (Kunkel and Brooks, 2002), abiotic stress signalling (Chinnusamy et al., 2004), or both (Fujita et al., 2006). Recent studies have revealed ERF subfamily transcription factors as promising candidates for proteins involved in cross-talk between stress signalling pathways. In this study, the expression of nine unigenes from different subgroups of the ERF subfamily following various stress treatments was analysed. Inoculation with SMV as a biotic stress increased the transcript levels of six unigenes in soybean plants. The abiotic stresses, drought, low temperature, and high salinity, induced the expression of nine, five, and nine unigenes, respectively. The expression of all nine unigenes was induced by treatments with SA, ET, JA, and ABA. ABA is a phytohormone that is extensively involved in responses to abiotic stresses such as drought and low temperature, as well as osmotic stress. ABA also governs a variety of growth and developmental processes, including seed development, dormancy, germination, and stomatal movement. In contrast, the phytohormones SA, JA, and ET play central roles in biotic stress signalling following pathogen infection. These signalling molecules primarily regulate the protective responses of plants against both biotic and abiotic stresses via synergistic and antagonistic actions, commonly described as signalling cross-talk (Mauch-Mani and Mauch, 2005). The present results suggest that there was significant cross-talk in the expression of the nine unigenes under abiotic and biotic stress conditions. As in other studies (Park et al., 2001; Lee et al., 2004), it is speculated that cross-talk of signalling pathways in plants is a common phenomenon, allowing the formation of elaborate networks to regulate both abiotic stress tolerance and disease resistance. Hence the ERF subfamily of transcriptional factors may be connecting elements involved in cross-talk between stress signalling pathways.
Overexpression of soybean ERF subfamily genes enhanced tolerance to biotic and/or abiotic stress
The ERF subfamily genes have been characterized in tobacco (Park et al., 2001; Fischer and Droge-Laser 2004), Arabidopsis (Broun et al., 2004; Yang et al., 2005), pepper (Lee et al., 2004; Yi et al., 2004), tomato (Wang et al., 2004), corn (Chuck et al., 2002), and rice (Cao et al., 2006). Overexpression of some ERF genes enhanced resistance to biotic and abiotic stresses (He et al., 2001; Berrocal-Lobo et al., 2002; Fischer and Dröge-Laser, 2004). So far, only one ERF subfamily gene (GmEREBP1) has been isolated and characterized from soybean. The transcript abundance decreased in soybean cyst-nematode-infected roots of a susceptible cultivar, whereas it increased in infected roots of a resistant cultivar (Mazarei et al., 2002). Furthermore, ET treatment repressed GmEREBP1 mRNA accumulation in both susceptible and resistant cultivars, whereas wounding increased expression in both cultivars (Mazarei et al., 2002). GmEREBP1 transgenic soybean and Arabidopsis plants inoculated with cyst nematodes did not display significantly altered responses to nematode infection (Mazarei et al., 2007). According to the classification of Sakuma et al. (2002), GmEREBP1 belongs to the B-3 subgroup of the ERF subfamily. In the present study, GmERF057 in subgroup B-2 and GmERF089 in subgroup B-5 were further characterized by overexpression in tobacco plants. Whereas the expression of GmERF057 was induced in soybean by salinity, drought, ET, SA, JA, ABA, and SMV treatments, but not by cold stress, its expression in transgenic tobacco plants conferred enhanced tolerance to salt and pathogen stress. The expression of GmERF089 was induced by salinity, drought, ET, SA, JA, and ABA treatments, but not by cold and SMV stresses, and GmERF089 transgenic plants had enhanced tolerance to salt and drought stresses, but not to pathogen stress. The results suggested that the ERF genes in different subgroups of the ERF subfamily have distinct functions dealing with specific environmental stress conditions using both different and common signal transduction pathways. The mechanism whereby ERF subfamily genes confer different stress tolerances when subjected to biotic and abiotic stresses needs further investigation.
Supplementary data
Table S1. The primer sequences used for RT-PCR amplification of nine selected target unigenes under different stress treatments.
Table S2. Unigene list of the AP2/ERF superfamily in soybean.
Table S3. Ninety-eight ERF family unigenes in soybean.
Table S4. Summary of conserved motifs within the ERF family by comparative analysis of soybean and Arabidopsis.
Table S5. Summary of conserved motifs within the ERF family by comparative analysis of soybean and rice.
Figure S1. The deduced amino acid sequence alignment of the AP2/ERF DNA-binding domains from the 98 soybean ERF proteins in this study and 122 Arabidopsis ERF proteins described by Nakano et al. (2006) using ClustalW.
Figure S2. An unrooted phylogenetic tree of the ERF family of soybean and Arabidopsis.
Figure S3. The deduced amino acid sequence alignment of the AP2/ERF DNA-binding domains from the 98 soybean ERF proteins in this study and 139 rice ERF proteins described by Nakano et al. (2006) using ClustalW.
Figure S4. An unrooted phylogenetic tree of the ERF family of soybean and rice.
Supplementary Material
Acknowledgments
The authors are grateful to Dr RA McIntosh (Plant Breeding Institute, University of Sydney, Australia) for critical review of this manuscript. We thank Dr Lijuan Qiu (Soybean Molecular Breeding Group, Institute of Crop Sciences, CAAS) for the gift of soybean cultivars. This work was funded by the National HITECH Research and Development Program of China (‘863’ program, #2008AA10Z124 and #2006AA10A104), the National Natural Science Foundation of China (#30700508), the National Key Project for Research on Transgenic Plant and Foundation of Institute of Crop Science, Chinese Academy of Agricultural Sciences (#082060302-09).
References
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of CBF/DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Reports. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO Journal. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Molecular Biology Reporter. 1991;9:208–219. [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. The Plant Journal. 2002;29:23–32. doi: 10.1046/j.1365-313x.2002.01191.x. [DOI] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. The Plant Cell. 2002;14:1737–49. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:4706–4711. doi: 10.1073/pnas.0305574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Song F, Goodman RM, Zheng Z. Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. Journal of Plant Physiology. 2006;163:1167–1178. doi: 10.1016/j.jplph.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Chen M, Wang QY, Cheng XG, Xu ZS, Li LC, Ye XG, Xia LQ, Ma YZ. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochemical and Biophysical Research Communications. 2007;353:299–305. doi: 10.1016/j.bbrc.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. Journal of Experimental Botany. 2004;55:225–36. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes and Development. 1998;12:1145–54. doi: 10.1101/gad.12.8.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ. The control of spikelet meristem identity by the branched silklessl gene in maize. Science. 2002;298:1238–1241. doi: 10.1126/science.1076920. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. PThe Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Droge-Laser W. Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Molecular Plant-Microbe Interactions. 2004;17:1162–1171. doi: 10.1094/MPMI.2004.17.10.1162. [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. The Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiology. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors. Current Opinion in Plant Biology. 2004;7:465–471. doi: 10.1016/j.pbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Hao DY, Ohme-Takagi M, Sarai A. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plants. Journal of Biological Chemistry. 1998;273:26857–26861. doi: 10.1074/jbc.273.41.26857. [DOI] [PubMed] [Google Scholar]
- He P, Warren RF, Zhao T, Shan L, Zhu L, Tang X, Zhou JM. Overexpression of Pti5 in tomato potentiates pathogen induced defense gene expression and enhances disease resistance to Pseudomonas syringae pv. tomato. Molecular Plant-Microbe Interactions. 2001;14:1453–1457. doi: 10.1094/MPMI.2001.14.12.1453. [DOI] [PubMed] [Google Scholar]
- Hoekema A, Hirsch P, Hooykaas PJJ, Schilperoort R. A binary plant vector strategy based on separation of virand T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Hu YX, Wang YX, Liu XF, Li JY. Arabidopsis RAVI is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Research. 2004;14:8–15. doi: 10.1038/sj.cr.7290197. [DOI] [PubMed] [Google Scholar]
- Huang B, Jin L, Liu J. Molecular cloning and functional characterization of a DREB1/CBF-like gene (GhDREB1L) from cotton. Science in China Series C—Life Sciences. 2007;50:7–14. doi: 10.1007/s11427-007-0010-8. [DOI] [PubMed] [Google Scholar]
- Huang B, Liu JY. A cotton dehydration responsive element binding protein functions as a transcriptional repressor of DRE mediated gene expression. Biochemical and Biophysical Research Communications. 2006;343:1023–1031. doi: 10.1016/j.bbrc.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Jin LG, Liu JY. Molecular cloning, expression profile and promoter analysis of a novel ethylene responsive transcription factor gene GhERF4 from cotton. Plant Physiology and Biochemistry. 2008;46:46–53. doi: 10.1016/j.plaphy.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology. 1999;17:287291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kizis D, Pages M. Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought responsive element in an ABA-dependent pathway. The Plant Journal. 2002;30:679–689. doi: 10.1046/j.1365-313x.2002.01325.x. [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology. 2002;5:325–31. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Lee JH, Hong JP, Oh SK, Lee S, Choi D, Kim WT. The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequences with different binding affinities: possible biological roles of CaERFLP1 in response to pathogen infection and high salinity conditions in transgenic tobacco plants. Plant Molecular Biology. 2004;55:61–81. doi: 10.1007/s11103-004-0417-6. [DOI] [PubMed] [Google Scholar]
- Li XP, Tian AG, Luo GZ, Gong ZZ, Zhang JS, Chen SY. Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theoretical and Applied Genetics. 2005;110:1355–1362. doi: 10.1007/s00122-004-1867-6. [DOI] [PubMed] [Google Scholar]
- Liu L, White MJ, MacRae TH. Transcription factors and their genes in higher plants functional domains, evolution and regulation. European Journal of Biochemistry. 1999;262:247–257. doi: 10.1046/j.1432-1327.1999.00349.x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain, separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhao TJ, Liu JM, Liu WQ, Liu Q, Yan YB, Zhou HM. The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Letters. 2006;580:1303–1308. doi: 10.1016/j.febslet.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F. The role of abscisic acid in plant–pathogen interactions. Current Opinion in Plant Biology. 2005;8:409–414. doi: 10.1016/j.pbi.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Mazarei M, Elling AA, Maier TR, Puthoff DP, Baum TJ. GmEREBP1 is a transcription factor activating defense genes in soybean and Arabidopsis. Molecular Plant-Microbe Interactions. 2007;20:107–119. doi: 10.1094/MPMI-20-2-0107. [DOI] [PubMed] [Google Scholar]
- Mazarei M, Puthoff DP, Hart JK, Rodermel SR, Baum TJ. Identification and characterization of a soybean ethylene-responsive element-binding protein gene whose mRNA expression changes during soybean cyst nematode infection. Molecular Plant-Microbe Interactions. 2002;15:577–586. doi: 10.1094/MPMI.2002.15.6.577. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. The Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ohme-Takagi M, Shinshi H. Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. The Plant Journal. 2000;22:29–38. doi: 10.1046/j.1365-313x.2000.00709.x. [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Singh KB. Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiology. 2002;128:1313–22. doi: 10.1104/pp.010862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. The Plant Cell. 2001;13:1035–1046. doi: 10.1105/tpc.13.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical and Biophysical Research Communications. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant Cell. 2006;18:1292–309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Molecular Biology. 2006;61:897–915. doi: 10.1007/s11103-006-0057-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thilmony RL, Chen Z, Bressan RA, Martin GB. Expression of the tomato Pto gene in tobacco enhances resistance to Pseudomonas syringae pv tabaci expressing avrPto. The Plant Cell. 1995;7:1529–1536. doi: 10.1105/tpc.7.10.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanism. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang Z, Chen Q, Zhang Z, Zhang H, Wu Y, Huang D, Huang R. Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Molecular Biology. 2004;55:183–192. doi: 10.1007/s11103-004-0113-6. [DOI] [PubMed] [Google Scholar]
- Wei G, Pan Y, Lei J, Zhu YX. Molecular cloning, phylogenetic analysis, expressional profiling and in vitro studies of TINY2 from Arabidopsis thaliana. Journal of Biochemistry and Molecular Biology. 2005;38:440–446. doi: 10.5483/bmbrep.2005.38.4.440. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Molecular Biology. 2005;58:585–596. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]
- Yi SY, Kim JH, Joung YH, Lee S, Kim WT, Yu SH, Choi D. The pepper transcription factor CaPFl confers pathogen and freezing tolerance in Arabidopsis. Plant Physiology. 2004;136:2862–2874. doi: 10.1104/pp.104.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.