Abstract
Protein metabolism plays an important role in plant adaptation to heat stress. This study was designed to identify heat-responsive proteins in roots associated with thermotolerance for two C3 grass species contrasting in heat tolerance, thermal Agrostis scabra and heat-sensitive Agrostis stolonifera L. Plants were exposed to 20 °C (control), 30 C (moderate heat stress), or 40 °C (severe heat stress) in growth chambers. Roots were harvested at 2 d and 10 d after temperature treatment. Proteins were extracted and separated by two-dimensional polyacrylamide gel electrophoresis. Seventy protein spots were regulated by heat stress in at least one species. Under both moderate and severe heat stress, more proteins were down-regulated than were up-regulated, and thermal A. scabra roots had more up-regulated proteins than A. stolonifera roots. The sequences of 66 differentially expressed protein spots were identified using mass spectrometry. The results suggested that the up-regulation of sucrose synthase, glutathione S-transferase, superoxide dismutase, and heat shock protein Sti (stress-inducible protein) may contribute to the superior root thermotolerance of A. scabra. In addition, phosphoproteomic analysis indicated that two isoforms of fructose-biphosphate aldolase were highly phosphorylated under heat stress, and thermal A. scabra had greater phosphorylation than A. stolonifera, suggesting that the aldolase phosphorylation might be involved in root thermotolerance.
Keywords: Grass, heat tolerance, phosphoproteomics, protein, proteomics, thermotolerance
Introduction
An increase in temperature associated with global warming is a growing concern, as it limits plant growth and productivity, especially for temperate species. Physiological mechanisms of heat tolerance have been examined extensively in various plant species, but the molecular basis of heat tolerance is not well understood (Wahid et al., 2007). Plant adaptation to environmental stresses is dependent upon the activation of cascades of molecular networks involved in stress perception, signal transduction, and the expression of stress-related proteins. Knowledge of heat-responsive proteins is critical for further understanding of the molecular mechanisms of stress tolerance.
Proteomics offers a powerful approach to discover the proteins and pathways that are crucial for stress responsiveness and tolerance. Two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) in combination with mass spectrometry (MS) allows rapid and reliable protein identification and can provide information about abundance and post-translation modification (PTM). In recent years, proteomic-based technologies have been successfully applied to the systematic study of the proteomic responses in many plant species to a wide range of abiotic stresses, including heat (Ferreira et al., 2006; Lee et al., 2007), drought (Pinheiro et al., 2005), cold (Yan et al., 2006), oxidative stress (Wang et al., 2004), anoxia (Chang et al., 2000), salt (Yan et al., 2005), ultraviolet-B (Xu et al., 2008a), and metal stress (Labra et al., 2006). Lee et al. (2007) found that heat shock proteins (HSPs) and antioxidant enzymes were up-regulated under heat stress in rice (Oryza sativa) leaves, and also the enzymes related to metabolic pathway were differentially accumulated. Ferreira et al. (2006) reported that in Populus euphratica a moderate heat response involves changes in proteins related to lipid biogenesis, cytoskeleton structure, sulphate assimilation, thiamine and hydrophobic amino acid biosynthesis, and nuclear transport. Protein phosphorylation is one of the most prominent PTMs by which cells transduce signals (Kalume et al., 2003; Bentem and Hirt, 2007). It has been inferred that 5% of the Arabidopsis thaliana genome encodes kinases and phosphatases, representing >1000 enzymes controlling the phosphorylation status of thousands of proteins (Arabidopsis Genome Initiative, 2000; Kerk et al., 2002). Previous studies have suggested a critical role for protein phosphorylation in plant stress responses (Mizoguchi et al., 1996; Xiong and Yang, 2003).
In recent years, knowledge of the mechanisms underlying plant responses to heat stress has grown (Wahid et al., 2007). However, most research focuses on stress adaptation mechanisms of the aboveground organs, whereas mechanisms of root tolerance to heat stress are much less investigated. Various studies have demonstrated that roots are more sensitive to heat stress, and suggest that high soil temperature is more detrimental than high air temperature for whole-plant growth (Xu and Huang, 2001; Liu and Huang, 2005). Roots may express different proteomes from leaves, grains, or fruits due to their different sensitivity to heat stress and unique functions. Proteomic profiling associated with root thermotolerance will enable molecular dissection of heat tolerance mechanisms. One approach to understanding the mechanisms of plant tolerance to stresses is to examine plants adapted to extremely stressful environments, since these plants may retain regulatory mechanisms enabling their survival. The dissection of such mechanisms may reveal a set of genes and proteins that may contribute to genetic improvement for stress tolerance in other plants, such as economically important cultivars. Several C3 grass species have been identified growing in geothermally heated areas in Yellowstone National Park (YNP) (Stout and Al-Niemi, 2002; Tercek et al., 2003). Thermal Agrostis scabra (‘thermal’ rough bentgrass) is one of the predominant grass species in thermal areas. This geothermal grass species can survive and even grow at temperatures up to 45–50 °C in soils that are permeated by steam (Tercek et al., 2003). In contrast, the growth temperature for common C3 grass species is between 10 and 18 °C for roots and between 15 and 24 °C for shoots, and physiological injury and death occur in roots of temperate grass species when soil temperatures reach 23 °C (Pote et al., 2006). Previous studies found that thermal A. scabra was able to maintain high root viability and new root production under high temperatures (35–40 °C) whereas severe root death occurred for A. stolonifera (Pote et al., 2006; Rachmilevitch et al., 2006a, b). The fact that thermal A. scabra is able to survive extreme temperatures marks it out as an important plant species to study the mechanisms responsible for survival after heat stress. Investigation into differentially accumulated proteins in the roots of heat-tolerant plants in comparison with heat-sensitive plants may identify specific proteins related to root thermotolerance, which could be used to develop molecular markers to select heat-tolerant germplasm or to create tolerant grasses through genetic manipulation.
The objectives of this study were to compare protein/phosphoprotein profiles of roots between thermal A. scabra and heat-sensitive A. stolonifera, under heat stress conditions, and to identify heat-regulated proteins associated with thermotolerance in roots of cool-season grasses.
Materials and methods
Plant materials and treatments
Thermal A. scabra plants were generated from seeds collected from a geothermal site in YNP, Wyoming, USA. Agrostis stolonifera L. (cv. Penncross) plants were collected from field plots from the turfgrass research farm at Rutgers University (New Brunswick, NJ, USA). Both species were propagated vegetatively in a greenhouse. Clonal plants of approximately 60 d old were then transplanted into plastic pots (20 cm deep and 15 cm in diameter) filled with washed, fine sand. Plants were maintained in a greenhouse for 28 d and then moved to a growth chamber set at 20/15 °C (day/night temperature), 75% relative humidity, 600 mmol m−2 s−1 of photosynthetically active radiation, and a 12 h photoperiod. Plants were allowed to acclimate to the growth chamber conditions for 7 d before being exposed to three air temperature regimes: 20 °C (control), 30 °C (moderate heat stress), and 40 °C (severe heat stress). The soil temperatures were 20.1, 29.5, and 39.3 °C (average of four replicaes), respectively, under control, moderate heat stress, and severe heat stress conditions. Each treatment was repeated three times in three different chambers to minimize chamber effects. During plant establishment and temperature treatment, plants were watered every day until water drained from the bottom of each pot in order to ensure full hydration of plants and avoid the occurrence of water deficit, and fertilized once a week with full-strength Hoagland's nutrient solution (Hoagland and Arnon, 1950).
Evaluation of root thermotolerance
Root viability was determined to evaluate root thermotolerance. At 10 d of temperature treatments, roots were washed free of soil. About 0.4 g (fresh weight) of roots (whole roots with base and tips) was collected for the measurement of root viability using a modified 2,3,5-triphenyltetrazolium chloride (TTC) reduction technique (Knievel, 1973). Roots were incubated in the dark for 24 h in 0.6% TTC at 37 °C, then rinsed with deionized water and placed in 95% ethanol at 60 °C for formazan extraction. The absorbance of the incubation solution was measured at 490 nm with a spectrophotometer (Model U-1100, Hitachi, Tokyo, Japan). Four independent samples were determined for each treatment. Live roots were mixed with different proportions of autoclave-killed roots to construct a standard curve. Root viability was expressed as the percentage of live root biomass to total root biomass.
Protein extraction
Roots were harvested at 2 d and 10 d of temperature treatment, immediately frozen in liquid nitrogen, and then stored at –80 °C prior to analysis. Four independent samples were harvested from each treatment. Root protein extraction followed the procedure described by Xu et al. (2008b). A 1 g aliquot of root sample was ground to powder with liquid nitrogen, homogenized, and incubated with 10 ml of precipitation solution [10% trichloroacetic acid (TCA) and 0.07% 2-mercaptoethanol in acetone] for 2 h at –20 °C. The precipitated proteins were pelleted and washed with ice-cold acetone containing 0.07% 2-mercaptoethanol until the supernatant was colourless. The pellet was vacuum-dried, resuspended in resolubilization solution [8 M urea, 2 M thiourea, 2% CHAPS, 1% dithiothreitol (DTT), 1% pharmalyte], and sonicated to extract proteins. Insoluble tissue was removed by centrifugation at 21 000 g for 20 min. Protein concentration was determined according to Bradford (1976) using a commercial dye reagent (Bio-Rad Laboratories, Hercules, CA, USA) with bovine serum albumin (BSA) as a standard.
Two-dimensional PAGE
An IPGPhor apparatus (GE Healthcare, Piscataway, NJ, USA) was used for isoelectric focusing (IEF) with immobilized pH gradient (IPG) strips (pH 3.0–10.0, linear gradient, 13 cm). The IPG strips were rehydrated for 12 h at 20 °C with 250 μl of rehydration buffer [8 M urea, 2 M thiourea, 2% (w/v) CHAPS, 1% (v/v) IPG buffer, 1% DTT, and 0.002% bromophenol blue] containing 300 mg of proteins. The voltage settings for IEF were 500 V for 1 h, 1000 V for 1 h, and 8000 V to a total 56.50 kVh. Following IEF, the protein in the strips was denatured with equilibration buffer (50 mM Tris–HCl pH 8.8, 6 M urea, 30% glycerol, 2% SDS, 0.002% bromophenol blue, 1% DTT) and then incubated with the same buffer containing 2.5% iodoacetamide instead of DTT for 20 min. The second dimension electrophoresis was performed on a 12.5% gel using a Hoefer SE 600 Ruby electrophoresis unit (GE Healthcare). For phosphoprotein detection, 2-D gels were stained with a modified protocol using Pro-Q Diamond Phosphoprotein Stain (Pro-Q DPS; Molecular Probes) (Agrawal and Thelen, 2005). Following scanning of Pro-Q DPS-stained gels, gels were stained with colloidal Coomassie brilliant blue (CBB) G-250 to detect total proteins (Newsholme et al., 2000).
Gel images were analysed with Progenesis (version 4.01) (non-linear) software. Image analysis included the following procedures: spot detection, spot measurement, background subtraction, and spot matching. Only spots that were detected on all the four replicate gels were analysed further. To correct the variability due to staining, the spot volumes were normalized as a percentage of the total volume of all spots on the gel. Data were subjected to analysis of variance (ANOVA) to test for the effects of species, heat, and their interactions. Means were separated by least significance difference test (P <0.05).
Protein identification
The gel spots were excised and washed with 30% acetonitrile (ACN) in 50 mM ammonium bicarbonate prior to DTT reduction and iodoacetamide alkylation. Trypsin was used for digestion at 37 °C overnight. The resulting peptides were extracted with 30 ml of 1% trifluoracetic acid (TFA) followed by C18 Ziptip desalting. For the MS analysis, the peptides were mixed with 7 mg ml−1 α-cyano-4-hydroxy-cinnamic acid matrix in a 1:1 ratio and spotted onto a matrix-assisted laser desorption/ionization (MALDI) plate. The peptides were analysed on a 4800 MALDI TOF/TOF analyser (Applied Biosystem, Framingham, MA, USA). Mass spectra (m/z 880–3200) were acquired in positive ion reflector mode. The 25 most intense ions were selected for subsequent MS/MS sequencing analysis in 1 kV mode. Protein identification was performed by searching the combined MS and MS/MS spectra against the green plant NCBI database using a local MASCOT search engine (V.1.9) on a GPS (V. 3.5, ABI) server. Proteins containing at least two peptides with confidence interval (CI) values no less than 95% were considered as being identified.
Experimental design and statistical analysis
The experimental design was a split-plot design with temperature as the main plot and grass species as the subplot, and each treatment had four replicates. Root viability was subjected to ANOVA to test for the effects of heat and species. Treatment means were separated by the least significant difference test at a P-value of <0.05.
Results
Changes in root viability in response to heat stress
Root viability of thermal A. scabra did not change as temperature increased from 20 °C to 30 °C, but decreased at 40 °C (Table 1). A significant decline in root viability was observed at both 30 °C and 40 °C, compared with the control at 20 °C for A. stolonifera. The root viability of the two species did not differ at 20 °C, but thermal A. scabra had significantly higher root viability than A. stolonifera at 30 °C and 40 °C.
Table 1.
Root viability of thermal A. scabra and A. stolonifera as affected by heat stress (30 °C and 40 °C) at 10 d of treatment
| Species | Root viability (% live roots) |
||
| 20 °C | 30 °C | 40 °C | |
| A. scabra | 83.8 Aa | 79.5 Aa | 55.4 Ab |
| A. stolonifera | 85.9 Aa | 65.2 Bb | 33.8 Bc |
Data are the means of four replicates. Means followed by the same letters were not statistically different based on the least significance test at P=0.05. Uppercase letters are for comparison between two grass species at a given temperature treatment. Lowercase letters are for comparisons between temperature treatments for a given grass species.
Proteomic responses to heat stress between grass species
The 2-D polyacrylamide gels were reproducible and exhibited clearly separated protein spots. Root protein profiles of the two grass species exposed to 20 °C were similar, except that A. stolonifera had higher intensities of spots 52, 53, and 33, and lower intensities of spots 34, 35, and 36, than A. scabra. However, the response patterns of proteins to heat stress varied between the two species. A representative gel image stained by CBB is presented in Fig. 1. Protein spots that were significantly affected by heat stress at one or both sampling times in at least one species were analysed further. A total of 70 protein spots exhibited differential accumulation under heat stress, and four regions of differentially expressed proteins are presented in Fig. 2.
Fig. 1.
Coomassie-stained 2-D polyacrylamide gel of separated proteins from A. scabra roots grown at 20 °C. Proteins were separated in the first dimension on an IPG strip (pH 3.0–10.0) and in the second dimension on a 12.5% polyacrylamide gel. The numbered spots were affected by heat stress.
Fig. 2.
Selected differentially expressed protein spots in two species growing under different temperatures.
Among the 70 protein spots, one spot (spot 52) exhibited increases in intensity or up-regulation in thermal A. scabra, but decreases in the intensity or down-regulation in A. stolonifera at moderate or severe heat stress. The intensity of 47 spots (spots 1–47; Fig. 1; Table 2) decreased and that of 22 spots (spots 48–51 and 53–70; Fig. 2; Table 2) increased under moderate or severe heat stress in at least one species. More protein spots exhibited down-regulation than those showing up-regulation under heat stress. In the group of up-regulated spots, 13 spots (spots 48, 53, 54, 56, 58–60, 62, 63, 65–67, and 70) were increased in both species and nine spots (spots 49–51, 55, 57, 61, 64, 68, and 69) were increased only in thermal A. scabra (Table 2; Fig. 3). Thermal A. scabra had more up-regulated protein spots than A. stolonifera under moderate and severe heat stress. Among the 47 down-regulated spots, 25 spots (spots 1–5, 7, 8, 15, 18, 21, 22, 25–27, 29, 32, 33, 36–42, and 44) were decreased in both species, nine spots (spots 6, 11–14, 16, 17, 24, and 45) were decreased only in A. stolonifera, and 13 spots (spots 9, 10, 19, 20, 23, 28, 30, 31, 34, 35, 43, 46, and 47) were decreased only in A. scabra (Table 2; Fig. 3). Eleven protein spots (spots 9, 10, 12, 13, 17, 24, 30, 34, 35, 46, and 68) were responsive only to short-term heat stress (2 d), while 18 (spots 6, 8, 16, 18–20, 43, 45, 47, 48–51, 57, 59, 67, and 70) were responsive only to long-term heat stress (10 d). The remaining 41 protein spots were responsive to both short-term and long-term heat stress (Table 2; Fig. 3). Most spots were responsive to heat stress in both species, nine spots (spots 6, 11–14, 16, 17, 24, and 45) only in A. stolonifera, and 23 (spots 9, 10, 19, 20, 23, 28, 30, 31, 34, 35, 43, 46, 47, 49–52, 55, 57, 61, 64, 68, and 69) only in A. scabra (Table 2). Most of the differentially accumulated protein spots were regulated by both moderate and severe heat stress. Twenty-seven spots were only affected by severe heat stress, while one spot (spot 13, down-regulated only in A. stolonifera) was only affected by moderate heat stress (Table 2).
Table 2.
Differentially expressed proteins identified by mass spectrometry between thermal A. scabra (ecotype ‘NTAS’, N) and A. stolonifera (cultivar ‘Penncross’, P) under heat stress (30 °C and 40 °C) compared with those at normal temperature (20 °C)
| ID | Protein identification [source] | H. pI/MW | Accession no. | PS | PM | Heat stress treatment |
|||
| 2 d |
10 d |
||||||||
| 30 °C | 40 °C | 30 °C | 40 °C | ||||||
| Protein spots decreased by heat stress | |||||||||
| Category 01 Metabolism | |||||||||
| 1 | Methionine synthase protein (EC 2.1.1.14) [Catharanthus roseus] | 6.10/84 857 | S57636 | 190 | 14 | N*** | N*, P* | P*** | N**, P*** |
| 2 | Methionine synthase protein (EC 2.1.1.14) [Sorghum bicolor] | 5.93/83 788 | Q8W0Q7 | 351 | 9 | N*** | P** | P*** | N**, P*** |
| 3 | Methionine synthase protein (EC 2.1.1.14) [Sorghum bicolor] | 5.93/83 788 | Q8W0Q7 | 211 | 15 | P** | P** | N**, P** | N***, P*** |
| 4 | Cytosolic glutamine synthetase (EC 6.3.1.2) [Populus alba×Populus tremula] | 6.61/18 429 | gi|37956277 | 209 | 5 | N**, P** | N**, P** | N**, P** | |
| 5 | Serine hydroxymethyltransferase (SHMT) (EC 2.1.2.1) [Arabidopsis thaliana] | 7.12/51 797 | Q9FPJ3 | 214 | 5 | N**, P** | N**, P** | N**, P** | N**, P** |
| 6 | SHMT (EC 2.1.2.1) [Arabidopsis thaliana] | 7.12/51 797 | Q9FPJ3 | 196 | 6 | P** | P** | ||
| 7 | Nucleotide-sugar dehydratase [Arabidopsis thaliana] | 8.58/38 621 | F84688 | 504 | 10 | N***, P** | N***, P*** | P*** | N***, P*** |
| Category 02 Energy | |||||||||
| 8 | Cytoplasmic aconitate hydratase (EC 4.2.1.3) [Arabidopsis thaliana] | 6.72/10 8201 | B84471 | 186 | 8 | N**, P*** | N**, P** | ||
| 9 | Fumarase (EC 4.2.1.2) [Solanum tuberosum] | 8.01/52 999 | gi|1488652 | 268 | 5 | N*** | N*** | ||
| 10 | Malate dehydrogenase (EC 1.1.1.37) [Oryza sativa] | 8.74/35 460 | Q94JA2 | 132 | 5 | N** | |||
| 52 | Sucrose synthase (EC 2.4.1.13) Ss1 [Hordeum vulgare] | 5.94/92 211 | S29242 | 354 | 23 | P*** | P*** | P** | P*** |
| 11 | Pyrophosphate-dependent phosphofructokinase alpha subunit (EC 2.7.1.90) [Citrus paradise Grapefru] | 6.71/67 373 | Q9ZST2 | 162 | 11 | P*** | P*** | ||
| 12 | Pyruvate kinase (EC 2.7.1.40) [Glycine max] | 7.50/55 302 | T07787 | 176 | 5 | P** | P*** | ||
| 13 | Pyruvate kinase (EC 2.7.1.40) cytosolic [Solanum tuberosum] | 6.64/55 170 | P22200 | 238 | 7 | P** | |||
| 14 | Fructose-bisphosphate (FBP) aldolase (EC 4.1.2.13) [Oryza sativa] | 6.55/38 719 | Q40676 | 692 | 11 | P** | P*** | P** | |
| 15 | FBP aldolase (EC 4.1.2.13) [Oryza sativa] | 6.55/38 719 | Q40676 | 545 | 10 | P*** | N**, P*** | N***, P*** | |
| 16 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (EC 1.2.1.12) cytosolic (fragment) [Hordeum vulgare] | 6.20/33 235 | A24159 | 885 | 8 | P** | |||
| 17 | Non-symbiotic (non-legume) haemoglobin [Gossypium hirsutum] | 8.97/18 442 | gi|3913789 | 203 | 4 | P*** | P*** | ||
| 18 | Phosphogluconate dehydrogenase (decarboxylating) (EC 1.1.1.44) cytosolic [Zea mays] | 5.92/53 055 | T01658 | 744 | 13 | N**, P** | N**, P** | ||
| 19 | Phosphogluconate dehydrogenase (decarboxylating) (EC 1.1.1.44) (fragment) [Zea mays] | 10.00/8528 | T01660 | 186 | 2 | N** | |||
| 20 | Ferredoxin-NADP reductase (EC 1.18.1.2) precursor [Zea mays] | 8.37/36 375 | S53305 | 210 | 13 | N* | N*** | ||
| 21 | Ferredoxin-NADP reductase (EC 1.18.1.2) precursor root (fragment) [Zea mays] | 8.37/36 375 | S53305 | 432 | 12 | P*** | N** | ||
| 22 | NADH2 dehydrogenase (ubiquinone) (EC 1.6.99.3) flavoprotein 1 precursor [Solanum tuberosum] | 8.45/53 618 | S52261 | 117 | 8 | P** | N**, P** | ||
| Category 06 Protein destination and storage | |||||||||
| 23 | Mitochondrial processing peptidase (MPP) (EC 3.4.24.64) alpha-chain [Dactylis glomerata] | 6.53/53 377 | Q9FNU9 | 250 | 5 | N** | N** | N** | N** |
| 24 | Putative disulphide-isomerase (EC 5.3.4.1) [Oryza sativa] | 5.01/56 854 | Q53LQ0 | 257 | 4 | P** | |||
| 19 | 26S protease regulatory subunit 7 [Oryza sativa] | 6.03/47 682 | Q9FXT9 | 176 | 9 | N** | |||
| Category 07 Transporters | |||||||||
| 25 | H+-transporting two-sector ATPase (EC 3.6.3.14) alpha chain mitochondrion [Triticum aestivum] | 5.70/55 306 | Q36567 | 604 | 14 | N*** | N***, P*** | N***, P*** | |
| Category 09 Cell structure | |||||||||
| 26 | Reversibly glycosylated polypeptide [Triticum aestivum] | 5.82/41 498 | gi|4158232 | 517 | 12 | N**, P** | N**, P** | N**, P** | |
| 27 | Putative oxidase [Oryza sativa] | 8.93/74 298 | Q9ZQP2 | 147 | 2 | N***, P** | N***, P** | ||
| 28 | Putative oxidase [Oryza sativa] | 8.93/74 298 | Q9ZQP2 | 133 | 2 | N*** | N** | ||
| Category 10 Signal transduction | |||||||||
| 29 | GTP-binding protein [Oryza sativa] | 8.39/68 030 | Q8W315 | 260 | 12 | N*** | N***, P*** | N** | N***, P** |
| 30 | GTP-binding protein beta chain homologue curled-leaved [Nicotiana tabacum] | 7.02/36 006 | T16970 | 251 | 4 | N* | |||
| 31 | GTP-binding protein beta chain Nicotiana tabacum] | 7.02/36 006 | T16970 | 94 | 6 | N** | N** | ||
| 32 | Nucleoside diphosphate kinase (EC 2.7.4.6) [Pinus pinaster] | 8.38/26 144 | Q8RVI6 | 164 | 2 | N** | N** | N** | N***, P*** |
| Category 11 Disease/defence | |||||||||
| 33 | Probable peroxidase (EC 1.11.1.-) 1 precursor anionic [Zea mays] | 5.41/37 774 | T04360 | 68 | 4 | P*** | N*, P*** | N* | |
| 34 | Probable peroxidase (EC 1.11.1.-) 1 precursor anionic [Zea mays] | 5.41/37 774 | T04360 | 69 | 3 | N** | |||
| 35 | Probable peroxidase (EC 1.11.1.-) 1 precursor anionic [Zea mays] | 5.41/37 774 | T04360 | 69 | 3 | N*** | |||
| Category 20 Secondary metabolism | |||||||||
| 36 | Phenylalanine ammonia-lyase (PAL) (fragment) (EC 4.3.1.5) [Hordeum vulgare] | 5.73/54 073 | T05968 | 427 | 11 | N** | N*** | N**, P* | N***, P* |
| 37 | PAL (EC 4.3.1.5) [Hordeum vulgare] | 5.73/54 073 | T05968 | 285 | 6 | N*** | P** | P*** | N**, P*** |
| 38 | dDTP-glucose 4–6-dehydratases-like protein [Arabidopsis thaliana] | 7.09/38 389 | T45701 | 297 | 7 | P** | P*** | N***, P*** | |
| 39 | Adenosylhomocysteinase (EC 3.3.1.1) [Triticum aestivum] | 5.65/53 436 | T06764 | 560 | 11 | N**, P** | N**, P** | N**, P** | |
| 40 | S-Adenosylmethionine synthase (SAMS) (EC 2.5.1.6) [Arabidopsis thaliana] | 5.51/42 795 | Q9LUT2 | 437 | 8 | N***, P*** | N***, P*** | N***, P*** | |
| 41 | SAMS (EC 2.5.1.6) [Dendrobium crumenatum] | 5.42/43 209 | Q944U4 | 906 | 12 | N***, P*** | N***, P*** | N***, P*** | |
| Category 12 Unclear classification | |||||||||
| 42 | AB019533 NID [Oryza sativa] | 6.68/41 341 | BAA77337 | 445 | 9 | N**, P*** | N** | N**, P*** | N**, P** |
| 43 | AY135561 NID [Arabidopsis thaliana] | 8.02/43 358 | AAN15218 | 191 | 6 | N*** | N** | ||
| 44 | No confident ID | N***, P*** | N***, P*** | ||||||
| 45 | No confident ID | P** | |||||||
| 46 | No confident ID | N*** | N*** | ||||||
| 47 | No confident ID | N** | N** | ||||||
| Protein spots increased by heat stress | |||||||||
| Category 01 Metabolism | |||||||||
| 48 | Phosphoserine aminotransferase (EC 2.6.1.52) [Oryza sativa] | 8.53/44 931 | Q8LMR0 | 243 | 7 | N**, P*** | |||
| 49 | Phosphoserine aminotransferase (EC 2.6.1.52) [Oryza sativa] | 8.53/44 931 | Q8LMR0 | 380 | 7 | N*** | |||
| 50 | Phosphoserine aminotransferase (EC 2.6.1.52) [Oryza sativa] | 8.53/44 931 | Q8LMR0 | 420 | 8 | N*** | |||
| 51 | Plastidic ATP sulphurylase (APS) (EC 2.7.7.4) [Oryza sativa] | 9.00/52 354 | Q9ZWM0 | 320 | 13 | N** | |||
| Category 02 Energy | |||||||||
| 52 | Sucrose synthase (EC 2.4.1.13) Ss1 [Hordeum vulgare] | 5.94/92 211 | S29242 | 354 | 23 | N** | N* | ||
| 53 | GAPDH (phosphorylation) (EC 1.2.1.12) [Hordeum vulgare] | 6.20/33 235 | P08477 | 850 | 8 | N*** | N** | N***, P** | |
| 54 | GAPDH (phosphorylating) (EC 1.2.1.12) [Hordeum vulgare] | 6.20/33 235 | P08477 | 880 | 11 | P** | N***, P*** | ||
| 55 | Cytoplasmic FBP aldolase (EC 4.1.2.13) [Oryza sativa] | 6.55/38 719 | Q40676 | 237 | 10 | N*** | N*** | N*** | |
| 56 | Cytoplasmic FBP aldolase. (EC 4.1.2.13) [Oryza sativa] | 6.55/38 719 | Q40676 | 217 | 10 | N*** | N***, P*** | ||
| 67 | Mitochondrial aldehyde dehydrogenase (EC 1.2.1.3) [Secale cereale] | 6.58/59 323 | Q8LST6 | 190 | 6 | N* | N*, P** | ||
| Category 05 Protein biosynthesis | |||||||||
| 57 | Putative asparagine-tRNA ligase (EC 6.1.1.22) [Oryza sativa] | 5.68/62 588 | Q93WM3 | 281 | 10 | N** | |||
| Category 06 Protein destination and storage | |||||||||
| 58 | Cyclophilin A-2 (EC 5.2.1.8) (peptidyl-prolyl cis–trans isomerase) [Triticum aestivum] | 8.52/18 379 | Q93XQ6 | 108 | 3 | P** | P*** | N***, P*** | |
| 65 | Stress-induced protein (Os02g0644100) [Oryza sativa] | 6.03/64 914 | gi|115447567 | 369 | 16 | N***, P*** | P** | N**, P*** | |
| 66 | Sti (stress-inducible protein) [Glycine max] | 5.81/63 585 | Q43468 | 178 | 4 | N***, P*** | N** | N***, P*** | |
| Category 08 Intracellular traffic | |||||||||
| 59 | Ran (Small GTP-binding protein) (Ran2) [Oryza sativa] | 6.66/25 038 | Q9XJ45 | 601 | 10 | N**, P** | |||
| 60 | GTP-binding nuclear protein Ran2 [Arabidopsis thaliana] | 6.38/25 062 | P41917 | 189 | 6 | P*** | N***, P*** | ||
| Category 11 Disease/defence | |||||||||
| 61 | Glutathione S-transferase GST 34 (EC 2.5.1.18) [Zea may] | 5.63/24 573 | Q9FQA5 | 80 | 4 | N*** | N*** | N*** | |
| 62 | GST (EC 2.5.1.18) [Triticum aestivum] | 5.79/23 338 | Q9SP56 | 238 | 5 | N*** | N***, P*** | ||
| 63 | GST (EC 2.5.1.18) [Triticum aestivum] | 5.79/23 338 | Q9SP56 | 176 | 4 | P** | N**, P*** | N***, P*** | |
| 64 | Superoxide dismutase (EC 1.15.1.1) (Mn) 3.2 precursor [Zea mays] | 6.71/25 238 | B48684 | 282 | 5 | N*** | |||
| Category 20 Secondary metabolism | |||||||||
| 68 | UDP-glucose 6-dehydrogenase (EC 1.1.1.22) [Glycine max] | 5.74/52 941 | T08818 | 290 | 12 | N** | |||
| Category 12 Unclear classification | |||||||||
| 69 | r40c1 protein [Oryza sativa] | 6.30/38 822 | Q40705 | 162 | 7 | N* | N** | ||
| 70 | Os03g0737000 [Oryza sativa] | 9.18/22 307 | gi|115455195 | 293 | 6 | N*, P* | |||
ID, spot ID (corresponding to Fig. 1); H pI/MW, hypothetical isoelectrical point/molecular weight; PS, protein score; PM, the number of unique peptides matched.
*0.05>P≥0.01; **0.01>P≥0.001; ***0.001>P.
Fig. 3.
Venn diagram illustrating the expression patterns of heat-responsive proteins in the roots of Agrostis grass.
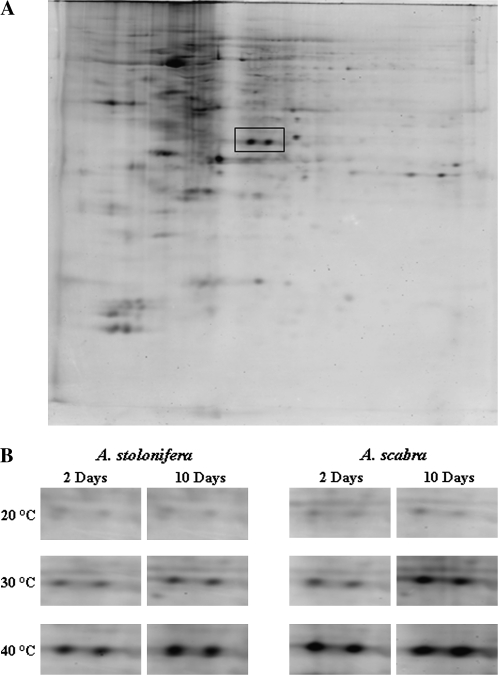
Root phosphoproteomic responses to heat stress were also investigated; a representative image is presented in Fig. 4A. The phosphorylation level of two proteins increased under heat stress, to a greater extent in A. scabra than in A. stolonifera. The magnified regions of these two spots from different treatments are presented in Fig. 4B. The comparison of two images from the same gel by different staining methods showed that these two spots corresponded to spots 55 and 56 in the image of CBB-stained gels. These two protein spots exhibited a higher intensity by the Pro-Q DPS staining method than by the CBB staining method.
Fig. 4.
Pro-Q DPS-stained 2-D polyacrylamide gel of separated proteins from A. scabra roots growing under 40 °C at 10 d of treatment (A), and magnified regions of differentially phosphorylated protein spots in two species growing under different temperatures (B).
The 70 differentially accumulated protein spots were digested with trypsin, subjected to MALDI TOF/TOF MS, and 66 protein spots were identified. The results are listed in Table 2. Most spots contained only one protein, while one spot contained two proteins (spot 19: phosphogluconate dehydrogenase and 26S protease regulatory subunit 7). The identified proteins were classified according to the functional categories described by Bevan et al. (1998): they belonged to the categories of metabolism, energy, protein destination/storage, protein synthesis, transporters, intracellular traffic, disease/defence, and secondary metabolism (Tables 2 and 3).
Table 3.
Functional distribution of protein spots responsive to heat stress
 |
Proteins were grouped according to the functional categories described by Bevan et al. (1998). Protein spot 19, containing two proteins, and spot 52, which was decreased in A. stolonifera and increased in A. scabra, were each counted twice.
Discussion
Higher root viability in thermal A. scabra under heat stress suggests that A. scabra had superior thermotolerance to A. stolonifera. This result is in agreement with results from previous studies (Rachmilevitch et al., 2006a, b). Superior root thermotolerance in thermal A. scabra could be associated with the expression of certain heat-responsive proteins. In fact, the proteomic response to heat stress varied between the two species, and the differentially accumulated proteins have diverse functions, as shown in Table 3 and discussed below.
Metabolism category
This category included 11 protein spots regulated by heat stress in at least one species. The down-regulated proteins are cytosolic glutamine synthetase (GS; spot 4), methionine synthase (spots 1–3), serine hydroxymethyltransferase (SHMT; spots 5 and 6), and nucleotide-sugar dehydratase (spot 7). All seven spots were decreased in both species, except spot 6 which decreased only in A. stolonifera. GS catalyses the assimilation of ammonium to glutamine using glutamic acid as its substrate (Chen and Silflow, 1996). Reduction of GS under stress conditions has been reported, and this may be a protective mechanism because nitric oxide, an intermediate of nitrogen assimilation, is an active radical (Wang et al., 2004; Yan et al., 2005; Xu et al., 2008a). However, Sahu et al. (2001) reported that GS activities increased and decreased under salt stress in tolerant and sensitive rice leaves, respectively. Plomion et al. (2006) also found that GS protein was increased by drought in leaves of poplar (Populus alba L.). El-Khatib et al. (2004) reported that overexpression of cytosolic GS in poplar enhanced photorespiration during drought and could contribute to the protection of photosynthesis. Methionine synthase catalyses the transfer of a methyl group from 5-methyltetrahydrofolate to homocysteine, resulting in the formation of methionine. SHMT catalyses interconversion of serine and glycine. The down-regulation of these proteins suggests that heat stress suppressed amino acid synthesis, including methionine, serine, and glycine in roots of the two cool-season grass species. One spot (spot 48) of phosphoserine aminotransferase was up-regulated in both species. Phosphoserine aminotransferase is the enzyme catalysing the second step in serine biosynthesis.
There are variations in the response of phosphoserine aminotransferase (spots 49 and 50) and plastidic ATP sulphurylase (APS; spot 51) to heat stress between the two species. APS catalyses the first step in sulphur assimilation. The up-regulation of APS at the transcription and protein levels under metal stress was reported (Roth et al., 2006; Weber et al., 2006). Enhanced sulphur assimilation may increase glutathione generation for active oxygen species scavenging. APS and two spots (spots 49 and 50) of phosphoserine aminotransferase were up-regulated only in roots of thermal A. scabra following 10 d of moderate or severe heat stress, suggesting the importance of serine and sulphur metabolism in root thermotolerance, particularly during prolonged periods of stress.
Energy category
In this category, 21 protein spots were altered by heat stress. Among the 21 proteins, four spots [spots 53 and 54, glyceraldehyde-3-phosphate dehydrogenase (GAPDH); spots 55 and 56, fructose-bisphosphate (FBP) aldolase] were up-regulated while 16 spots were down-regulated (Table 2). Thirteen protein spots involved in carbon degradation and the electron transport chain in mitochondria were down-regulated by heat stress, including aconitate hydratase (spot 8), fumarase (spot 9), malate dehydrogenase (spot 10), sucrose synthase (spot 52), pyrophosphate-dependent phosphofructokinase (spot 11), pyruvate kinase (spots 12 and 13), FBP aldolase (spots 14 and 15), GAPDH (spot 16), phosphogluconate dehydrogenase (spots 18 and 19), and NADH2 dehydrogenase (spot 22). The down-regulation of these proteins involved in respiration may contribute to root adaptation to heat stress by lowering respiratory energy consumption (Rachmilevitch et al., 2006a, b). In addition, these results suggest the sensitivity of root respiration to heat stress.
Agrostis scabra and A. stolonifera had different response patterns of sucrose synthase (SS) and GAPDH to heat stress. The SS was down-regulated in A. stolonifera while it was up-regulated in thermal A. scabra by heat stress. SS catalyses both synthesis and degradation of sucrose (Geigenberger and Stitt, 1993), but the degradation process dominates in vivo. SS in the cytosol is thought to supply UDP-glucose and fructose produced by sucrose cleavage for glycolysis, and possibly starch synthesis. The expression of SS was enhanced under low O2 or low temperature, and the increase in the activity of SS was suggested to contribute to low O2 or low temperature tolerance (Crespi et al., 1991; Harada and Ishizawa, 2003; Harada et al., 2005). The increased accumulation of SS in thermal A. scabra may contribute to superior root thermotolerance by regulating sucrose metabolism. In this study, GAPDH was present in three spots (spots 16, 53, and 54). The intensity of spot 16 decreased only in A. stolonifera while that of another two spots (spots 53 and 54) increased in both grass species. All three spots are abundant proteins. Interestingly, thermal A. scabra had a higher level of spot 54 and a lower level of spot 53 than A. stolonifera, which may be due to the different cellular locations and functions of these isoforms. In addition to catalysing a reaction in glycolysis, GAPDH has been shown to exhibit protein kinase activity (Duclos-Vallee et al., 1998), bind RNA (Nagy and Rigby, 1995), suppress the production of active oxygen species (Baek et al., 2008), and enhance ribozyme (Sioud and Jespersen, 1996) and phosphotransferase activities (Engel et al., 1998). In leaves of P. euphratica, GAPDH increased under heat stress (Ferreira et al., 2006). Although many studies indicated that GAPDH was up-regulated under different stress conditions (Yang et al., 1993; Chang et al., 2000), little is known about how GAPDH in involved in the defence mechanism against heat stress. Elucidation of the multifaceted properties of this protein during heat stress would help to understand how this protein regulates thermotolerance.
The levels of FBP aldolase phosphorylation under heat stress were also different between the two species. FBP aldolase catalyses a glycolysis reaction in which FBP is broken down into glyceraldehyde 3-phosphate and dihydroxyacetone phosphate. Higher plants contain two isoforms, one in the cytosol and the other in the chloroplasts (Lebherz et al., 1984). Riccardi et al. (1998) reported that FBP aldolase was increased by water deficit in maize (Zea mays) leaves. In the present study, four protein spots were identified as FBP aldolase. Two abundant spots (spots 14 and 15) exhibited down-regulation while two weak spots (spots 55 and 56) showed up-regulation under heat stress. Interestingly, aldolase in these two weak spots was greatly phosphorylated in both species by heat stress. Also, the phosphorylation occurred early during 2 d of heat stress and thermal A. scabra had a greater level of phosphorylation than A. stolonifera. The phosphorylation of these two FBP aldolase isoforms might be related to the defence mechanism against heat stress. However, little is known about the function of FBP aldolase in plant response to stresses. It would be interesting to identify the kinase that phosphorylates FBP aldolase, and find out how aldolase and carbon metabolism are regulated in plants by FBP aldolase phosphorylation.
Protein destination and storage category
This category had six protein spots regulated by heat stress. Spots 19 (26S protease regulatory subunit 7) and 23 [mitochondrial processing peptidase (MPP)] were down-regulated only in thermal A. scabra, and spot 24 (disulphide-isomerase) was down-regulated only in A. stolonifera, while spots 65 and 66 (HSP Sti) and spot 58 [peptidyl-prolyl cis–trans isomerase (PPIase)] were up-regulated in both grass species. However, spot 19 contained two proteins (phosphogluconate dehydrogenase and 26S protease regulatory subunit 7). Heat stress may affect one or both of the proteins contained in this spot. PPIase accelerates the folding of proteins. It catalyses the cis–trans isomerization of proline imidic peptide bonds in oligopeptides. Little is known about the function of PPIases under heat stress conditions.
Interestingly, MPP was down-regulated only in thermal A. scabra, and disulphide-isomerase was down-regulated only in A. stolonifera. Most mitochondrial proteins encoded in the nucleus are synthesized as precursor proteins with extension peptides and are targeted to the mitochondria. After import of the precursors into the mitochondria, the extension peptides are cleaved off by MPP. This protein was also decreased by drought in roots of poplar (Plomion et al., 2006). In this study, MPP was decreased only in thermal A. scabra under both moderate and severe heat stress. How changes in the expression of MPP under heat stress are involved in root thermotolerance requires further investigation. Disulphide-isomerase catalyses the rearrangement of -S–S- bonds in proteins and participates in the folding of proteins containing disulphide bonds. The down-regulation of this protein only in A. stolonifera indicates that heat damage in roots may be related to the disruption of protein folding associated with the degradation of disulphide-isomerase. HSP Sti, also known as stress-inducible protein Sti, contains two heat shock chaperonin-binding motif (STI1), three tetratricopeptide repeat (TPR), and two Sti1 domains. It is believed that the function of TPR-containing proteins is mediated through protein–protein interaction to modulate diverse cellular processes, including Hsp90 signalling and interaction (Flom et al., 2006), protein transport across mitochondria (Chan et al., 2006), regulation of meristem cellular organization (Guyomarc'h et al., 2004), and gibberellin signalling (Izhaki et al., 2001). This protein was up-regulated in response to salt stress (Dooki et al., 2006). In this study, it was also up-regulated by heat stress, and thermal A. scabra had a higher level of this protein than heat-sensitive A. stolonifera, suggesting its positive relationship with root thermotolerance.
Stress defence category
Seven protein spots in this category were altered by heat stress (Table 3). Spots 34 and 35 were down-regulated only in thermal A. scabra and spot 33 was down-regulated in both species; all three spots were identified as peroxidase. Spots 61 [glutathione S-transferase (GST)] and 64 [superoxide dismutase (SOD)] were increased only in thermal A. scabra, and spots 62 and 63 (GST) were up-regulated in both species. GST is an abundant protein and has functions in conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. Recent studies have also suggested GSTs as components of ultraviolet-inducible cell signal pathways and as potential regulators of apoptosis (Dixon et al., 2002). The plant-specific phi class might counteract the consequences of generation of reactive oxygen species during photosynthesis (Edwards et al., 2000). The increased expression of GSTs has been identified in several proteomics or transcription analyses of plants that were exposed to different stresses (Dixon et al., 2002; Roth et al., 2006; Gazanchian et al., 2007; Yang et al., 2007), although Plomion et al. (2006) reported that it was reduced by drought in poplar roots. Hajheidari et al. (2007) reported that drought stress increased GST in a tolerant cultivar of sugar beet (Beta vulgaris L.) while it decreased it in a sensitive cultivar.
In this study, one spot (spot 61) of GST was induced by heat stress only in thermal A. scabra, and another two spots (spots 62 and 63) had higher intensity in thermal A. scabra than in A. stolonifera under heat stress. The higher GST level in A. scabra may lead to lower production of active oxygen species, resulting in superior root thermotolerance. Also, the two species had variation in the levels of SOD and peroxidase. SOD acts as the first line of defence converting superoxide to the less toxic hydrogen peroxide molecule. In the present study, thermal A. scabra had a higher level of SOD than A. stolonifera under heat stress. In addition to H2O2 detoxification, peroxidases are also implicated in various physiological processes such as auxin catabolism, liginfication, suberization, stress response, and senescence (Hiraga et al., 2001; Passardi et al., 2005). Three differentially accumulated spots (spots 33–35) were identified as peroxidase and all were decreased by heat stress. Interestingly, A. stolonifera only had spots 33 and 34, and the intensity of spot 33 was higher, while the intensity of spot 34 was lower in A. stolonifera than in thermal A. scabra under both control and stress conditions, indicating that the peroxidase isoforms presented in spots 34 and 35 might be important for heat tolerance. The higher level of SOD and some isoforms of peroxidase in roots of thermal A. scabra may contribute to the superior thermotolerance by suppressing the production of active oxygen species.
Secondary metabolism category
In this category, seven protein spots were affected by heat stress, of which one exhibited up-regulation (spot 68, UDP-glucose 6-dehydrogenase) only in thermal A. scabra and six [spots 36 and 37, phenylalanine ammonia-lyase (PAL); spot 38, dDTP-glucose 4–6-dehydratases-like protein; spot 39, adenosylhomocysteinase; spots 40 and 41, S-adenosylmethionine synthase (SAMS)] showed down-regulation in both species. PAL is a key enzyme in plant secondary metabolism, catalysing the first reaction in the biosynthesis from L-phenylalanine to a wide variety of natural products based on the phenylpropane skeleton. SAMS catalyses the production of S-adenosyl-L-methionine (SAM) from L-methionine and ATP. SAM serves as a methyl group donor in numerous transmethylation reactions and is the precursor for the biosynthesis of polyamines and ethylene among other metabolites. Several authors have shown that the SAMS gene and/or enzyme activity are stimulated under different stress conditions, suggesting the induction of lignification during stress (Chang et al., 1995; Yan et al., 2006). However, other studies indicated that the protein and transcript levels of SAMS were decreased under salt and mental stress (Jiang et al., 2007; Yang et al., 2007). The roles of dDTP-glucose 4–6-dehydratases-like protein, adenosylhomocysteinase, and UDP-glucose 6-dehydrogenase in plant tolerance of heat stress are unclear.
Other proteins
Nucleoside diphosphate kinase (NDPK) is believed to use ATP to maintain cellular levels of CTP, GTP, and UTP. It is also associated with H2O2-mediated mitogen-activated protein kinase (MAPK) signalling (Moon et al., 2003). The up-regulation of NDPK has been reported in response to drought (Salekdeh et al., 2002; Hajheidari et al., 2005), cold (Imin et al., 2004), heat, and salt stress (Dooki et al., 2006; Lee et al., 2007). However, in this study it was down-regulated by heat stress. Ran is an evolutionarily conserved eukaryotic GTPase, which is likely to be involved in protein import into the nucleus and RNA export from the nucleus, in chromatin condensation, and in cell cycle control (Kahana and Cleveland, 1999; Yang, 2002). However, little is known about the function of Ran in plant response to stresses. It was found that its abundance was increased under salt and heat stress (Ferreira et al., 2006; Jiang et al., 2007). In this study it was also increased by heat stress, and thermal A. scabra had a higher level than A. stolonifera, suggesting that Ran could play roles in nucleocytoplasmic interactions under heat stress.
In summary, different proteomic profiles were detected between thermal A. scabra and heat-sensitive A. stolonifera under heat stress, and more proteins were up-regulated in A. scabra than in A. stolonifera. The higher levels of SS, GST, SOD, Sti, and some peroxidase isoforms in thermal A. scabra could be related to its superior root thermotolerance relative to A. stolonifera. In addition, phosphorylation of FBP aldolase isoforms may also contribute to better root thermotolerance in A. scabra. Genes encoding these differentially regulated proteins between the two grass species may be further investigated using molecular approaches, which may provide the molecular basis of root thermotolerance in cool-season grass species.
Acknowledgments
The authors thank Yan Xu for experiment assistance, and Dr PE Thomas for providing instruments and software with image acquisition and analyses. Thanks go to Emily Merewitz, Yan Xu, and Dr Yan Zhang for critical review of the manuscript.
References
- Agrawal GK, Thelen JJ. Development of a simplified economical polyacrylamide gel staining protocol for phosphoproteins. Proteomics. 2005;5:4684–4688. doi: 10.1002/pmic.200500021. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Baek D, Jin Y, Jeong JC, et al. Suppression of reactive oxygen species by glyceraldehyde-3-phosphate dehydrogenase. Phytochemistry. 2008;69:333–338. doi: 10.1016/j.phytochem.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Bentem SF, Hirt H. Using phosphoproteomics to reveal signalling dynamics in plants. Trends in Plant Science. 2007;12:404–411. doi: 10.1016/j.tplants.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent E, et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chan NC, Likić VA, Waller RF, Mulhern TD, Lithgow T. The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. Journal of Molecular Biology. 2006;358:1010–1022. doi: 10.1016/j.jmb.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear JD, Dias MADL, Funkhouser EA, Newton RJ, Cairney J. Gene expression under water deficit in loblolly pine (Pinus taeda L.): isolation and characterization of cDNA clones. Physiologia Plantarum. 1995;95:1–10. [Google Scholar]
- Chang WWP, Huang L, Shen M, Webster C, Burlingame AL, Roberts JKM. Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment, and identification of proteins by mass spectrometry. Plant Physiology. 2000;122:295–317. doi: 10.1104/pp.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Silflow CD. Isolation and characterization of glutamine synthetase genes in Chlamydomonas reinhardtii. Plant Physiology. 1996;112:987–996. doi: 10.1104/pp.112.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi MD, Zabaleta EJ, Pontis HG, Salerno GL. Sucrose synthase expression during cold acclimation in wheat. Plant Physiology. 1991;96:887–891. doi: 10.1104/pp.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R. Plant glutathione transferases. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-3-reviews3004. REVIEWS3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooki AD, Mayer-Posner FJ, Askari1 H, Zaiee A, Salekdeh GH. Proteomic responses of rice young panicles to salinity. Proteomics. 2006;6:6498–6507. doi: 10.1002/pmic.200600367. [DOI] [PubMed] [Google Scholar]
- Duclos-Vallee JC, Capel F, Mabit H, Petit MA. Phosphorylation of the hepatitis B virus core protein by glyceraldehyde-3-phosphate dehydrogenase protein kinase activity. Journal of General Virology. 1998;79:1665–1670. doi: 10.1099/0022-1317-79-7-1665. [DOI] [PubMed] [Google Scholar]
- Edwards R, Dixon D, Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends in Plant Science. 2000;5:193–198. doi: 10.1016/s1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- El-Khatib RT, Hamerlynck EP, Gallardo F, Kirby EG. Transgenic poplar characterized by ectopic expression of a pine cytosolic glutamine synthetase gene exhibits enhanced tolerance to water stress. Tree Physiology. 2004;24:729–736. doi: 10.1093/treephys/24.7.729. [DOI] [PubMed] [Google Scholar]
- Engel M, Seifert M, Theisinger B, Seyfert U, Welter C. Glyceraldehyde-3-phosphate dehydrogenase and Nm23-H1/nucleoside diphosphate kinase A: two old enzymes combine for the novel Nm23 protein phosphotransferase function. Journal of Biological Chemistry. 1998;273:20058–20065. doi: 10.1074/jbc.273.32.20058. [DOI] [PubMed] [Google Scholar]
- Ferreira S, Hjernø K, Larsen M, Wingsle G, Larsen P, Fey S, Roepstorff P, Salomé Pais M. Proteome profiling of Populus euphratica Oliv. upon heat stress. Annals of Botany. 2006;98:361–377. doi: 10.1093/aob/mcl106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom G, Weekes J, Williams JJ, Johnson JL. Effect of mutation of the tetratricopeptide repeat and asparatate–proline domains of Sti1 on Hsp90 signaling and interaction in Saccharomyces cerevisiae. Genetics. 2006;172:41–51. doi: 10.1534/genetics.105.045815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazanchian A, Hajheidari M, Sima NK, Salekdeh GH. Proteome response of Elymus elongatum to severe water stress and recovery. Journal of Experimental Botany. 2007;58:291–300. doi: 10.1093/jxb/erl226. [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M. Sucrose synthase catalyses a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta. 1993;189:329–339. doi: 10.1007/BF00194429. [DOI] [PubMed] [Google Scholar]
- Guyomarc'h S, Vernoux T, Traas J, Zhou D, Delarue M. MGOUN3, an Arabidopsis gene with tetratrico-peptide-repeat-related motifs, regulates meristem cellular organization. Journal of Experimental Botany. 2004;55:673–684. doi: 10.1093/jxb/erh069. [DOI] [PubMed] [Google Scholar]
- Hajheidari M, Salekdeh GH, Heidari M, Abdollahian-Noghabi M, Sadeghian SY. Proteome analysis of sugar beet leaves under drought stress. Proteomics. 2005;5:950–960. doi: 10.1002/pmic.200401101. [DOI] [PubMed] [Google Scholar]
- Hajheidari M, Eivazi A, Buchanan BB, Wong JH, Majidi I, Salekdeh GH. Proteomics uncovers a role for redox in drought tolerance in wheat. Proteome Research. 2007;6:1451–1460. doi: 10.1021/pr060570j. [DOI] [PubMed] [Google Scholar]
- Harada T, Ishizawa K. Starch degradation and sucrose metabolism during anaerobic growth of pondweed (Potamogeton distinctus A. Benn.) turions. Plant and Soil. 2003;253:125–135. [Google Scholar]
- Harada T, Satoh S, Yoshioka T, Ishizawa K. Expression of sucrose synthase genes involved in enhanced elongation of pondweed (Potamogeton distinctus) turions under anoxia. Annals of Botany. 2005;96:683–692. doi: 10.1093/aob/mci220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H. A large family of class III plant peroxidases. Plant and Cell Physiology. 2001;42:462–468. doi: 10.1093/pcp/pce061. [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. California Agricultural Experiment Station Circular. 1950 247. [Google Scholar]
- Imin N, Kerim T, Weinman JJ, Rolfe BG. Effect of early cold stress on the maturation of rice anthers. Proteomics. 2004;4:1873–1882. doi: 10.1002/pmic.200300738. [DOI] [PubMed] [Google Scholar]
- Izhaki A, Swain SM, Tseng T, Borochov A, Olszewski NE, Weiss D. The role of SPY and its TPR domain in the regulation of gibberellin action throughout the life cycle of Petunia hybrida plants. The Plant Journal. 2001;28:181–190. doi: 10.1046/j.1365-313x.2001.01144.x. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yang B, Harris NS, Deyholos MK. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. Journal of Experimental Botany. 2007;58:3591–3607. doi: 10.1093/jxb/erm207. [DOI] [PubMed] [Google Scholar]
- Kahana JA, Cleveland DW. Beyond nuclear transport. RAN-GTP as a determinant of spindle assembly. Journal of Cell Biology. 1999;146:1205–1210. doi: 10.1083/jcb.146.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalume DE, Molina H, Pandey A. Tackling the phosphoproteome: tools and strategies. Current Opinion in Chemical Biology. 2003;7:64–69. doi: 10.1016/s1367-5931(02)00009-1. [DOI] [PubMed] [Google Scholar]
- Kerk K, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiology. 2002;129:908–925. doi: 10.1104/pp.004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knievel DP. Procedures for estimating ratio of live or dry matter in root samples. Crop Science. 1973;13:124–126. [Google Scholar]
- Labra M, Gianazza E, Waitt R, Eberini I, Sozzi A, Grassi F, Agradi E. Zea mays L. protein changes in response to potassium dichromate treatments. Chemosphere. 2006;62:1234–1244. doi: 10.1016/j.chemosphere.2005.06.062. [DOI] [PubMed] [Google Scholar]
- Lebherz HG, Leadbetter MM, Bradshaw RA. Isolation and characterization of the cytosolic and chloroplast form of spinach leaf fructose diphosphate aldolase. Journal of Biological Chemistry. 1984;259:1011–1017. [PubMed] [Google Scholar]
- Lee DG, Ahsan N, Lee S, Kang KY, Bahk JD, Lee I, Lee B. A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics. 2007;7:3369–3383. doi: 10.1002/pmic.200700266. [DOI] [PubMed] [Google Scholar]
- Liu X, Huang B. Root physiological factors involved in creeping bentgrass response to high soil temperatures. Environmental and Experimental Botany. 2005;53:233–245. [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1996;93:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H, Lee B, Choi G, et al. NDP kinase interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proceedings of the National Academy of Sciences, USA. 2003;100:358–363. doi: 10.1073/pnas.252641899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Rigby WFC. Glyceraldehyde-3-phosphate dehydrogenase selectively binds Au-rich RNA in the NAD+-binding region (Rossmann fold) Journal of Biological Chemistry. 1995;270:2755–2769. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- Newsholme SJ, Maleeft BF, Steiner S, Anderson NL, Schwartz LW. Two-dimensional electrophoresis of liver proteins: characterization of a drug-induced hepatomegaly in rats. Electrophoresis. 2000;21:2122–2128. doi: 10.1002/1522-2683(20000601)21:11<2122::AID-ELPS2122>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Passardi F, Cosio C, Penel C, Dunand C. Peroxidases have more functions than a Swiss army knife. Plant Cell Reporter. 2005;24:255–265. doi: 10.1007/s00299-005-0972-6. [DOI] [PubMed] [Google Scholar]
- Pinheiro C, Kehr J, Ricardo CP. Effects of water stress on lupin stem protein analysed by two-dimensional gel electrophoresis. Planta. 2005;221:716–728. doi: 10.1007/s00425-004-1478-0. [DOI] [PubMed] [Google Scholar]
- Plomion C, Lalanne C, Claverol S, Meddour H, Kohler A, Bogeat-Triboulot M. Mapping the proteome of poplar and application to the discovery of drought-stress responsive proteins. Proteomics. 2006;6:6509–6527. doi: 10.1002/pmic.200600362. [DOI] [PubMed] [Google Scholar]
- Pote J, Wang Z, Huang B. Timing and temperature of physiological decline for creeping bentgrass. Journal of the American Society of Horticultural Science. 2006;131:608–615. [Google Scholar]
- Rachmilevitch S, Huang B, Lambers H. Assimilation and allocation of carbon and nitrogen of thermal and nonthermal Agrostis species in response to high soil temperature. New Phytologist. 2006a;170:479–490. doi: 10.1111/j.1469-8137.2006.01684.x. [DOI] [PubMed] [Google Scholar]
- Rachmilevitch S, Lambers H, Huang B. Root respiratory characteristics associated with plant adaptation to high soil temperature for geothermal and turf-type Agrostis species. Journal of Experimental Botany. 2006b;57:623–631. doi: 10.1093/jxb/erj047. [DOI] [PubMed] [Google Scholar]
- Riccardi F, Gazeau P, Vienne D, Zivy M. Protein changes in response to progressive water deficit in maize. Quantitative variation and polypeptide identification. Plant Physiology. 1998;117:1253–1263. doi: 10.1104/pp.117.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth U, Roepenack-Lahaye E, Clemens S. Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+ Journal of Experimental Botany. 2006;57:4003–4013. doi: 10.1093/jxb/erl170. [DOI] [PubMed] [Google Scholar]
- Sahu AC, Sahoo SK, Sahoo N. NaCl-stress induced alteration in glutamine synthetase activity in excised senescing leaves of a salt-sensitive and a salt-tolerant rice cultivar in light and darkness. Plant Growth Regulation. 2001;34:287–292. [Google Scholar]
- Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J. Proteomic analysis of rice leaves during drought stress and recovery. Proteomics. 2002;2:1131–1145. doi: 10.1002/1615-9861(200209)2:9<1131::AID-PROT1131>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Sioud M, Jespersen L. Enhancement of hammerhead ribozyme catalysis by glyceraldehyde-3-phosphate dehydrogenase. Journal of Molecular Biology. 1996;257:775–789. doi: 10.1006/jmbi.1996.0201. [DOI] [PubMed] [Google Scholar]
- Stout RG, Al-Niemi TS. Heat tolerant flowering plants of active geothermal areas in Yellowstone National Park. Annals of Botany. 2002;90:259–267. doi: 10.1093/aob/mcf174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercek MT, Hauber D, Darwin SP. Genetic and historical relationships among geothermally adapted Agrostis (Bentgrass) of North America and Kamchatka: evidence for a previously unrecognized thermally adapted taxon. American Journal of Botany. 2003;90:1306–1312. doi: 10.3732/ajb.90.9.1306. [DOI] [PubMed] [Google Scholar]
- Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: an overview. Environmental and Experimental Botany. 2007;61:199–223. [Google Scholar]
- Wang SB, Chen F, Sommerfeld M. Proteomic analysis of molecular response to oxidative stress by the green alga Haematococcus pluvialis (Chlorophyceae) Planta. 2004;220:17–29. doi: 10.1007/s00425-004-1323-5. [DOI] [PubMed] [Google Scholar]
- Weber M, Trampczynska A, Clemens S. Comparative transcriptome analysis of toxic metal responses in Arabidopsis thaliana and the Cd2+-hypertolerant facultative metallophyte Arabidopsis halleri. Plant, Cell and Environment. 2006;29:950–963. doi: 10.1111/j.1365-3040.2005.01479.x. [DOI] [PubMed] [Google Scholar]
- Xiong L, Yang Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. The Plant Cell. 2003;15:745–59. doi: 10.1105/tpc.008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Huang B. Lowering soil temperatures improves creeping bentgrass growth under heat stress. Crop Science. 2001;41:1878–1883. [Google Scholar]
- Xu C, Sullivan JH, Garrett WM, Caperna TJ, Natarajan S. Impact of solar ultraviolet-B on the proteome in soybean lines differing in flavonoid contents. Phytochemistry. 2008a;69:38–48. doi: 10.1016/j.phytochem.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Xu C, Xu Y, Huang B. Protein extraction for 2-dimensional gel electrophoresis of proteomic profiling in turfgrass. Crop Science in press. 2008b [Google Scholar]
- Yan S, Tang Z, Su W, Sun W. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics. 2005;5:235–244. doi: 10.1002/pmic.200400853. [DOI] [PubMed] [Google Scholar]
- Yan S, Zhang Q, Tang Z, Su W, Sun W. Comparative proteomic analysis provides new insights into chilling stress responses in rice. Molecular Cellular Proteomics. 2006;5:484–496. doi: 10.1074/mcp.M500251-MCP200. [DOI] [PubMed] [Google Scholar]
- Yang Y, Kwon HB, Peng HP, Shih MC. Stress responses and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiology. 1993;101:209–216. doi: 10.1104/pp.101.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Small GTPases: versatile signaling switches in plants. The Plant Cell. 2002;14:S375–S388. doi: 10.1105/tpc.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Wang Y, Zhang J, Shi W, Qian C, Peng X. Identification of aluminum-responsive proteins in rice roots by a proteomic approach: cysteine synthase as a key player in Al response. Proteomics. 2007;7:737–749. doi: 10.1002/pmic.200600703. [DOI] [PubMed] [Google Scholar]