Abstract
The gaseous hormone ethylene is perceived by a family of ethylene receptors which interact with the Raf-like kinase CTR1. SlTPR1 encodes a novel TPR (tetratricopeptide repeat) protein from tomato that interacts with the ethylene receptors NR and LeETR1 in yeast two-hybrid and in vitro protein interaction assays. SlTPR1 protein with a GFP fluorescent tag was localized in the plasmalemma and nuclear membrane in Arabidopsis, and SlTPR1-CFP and NR-YFP fusion proteins were co-localized in the plasmalemma and nuclear membrane following co-bombardment of onion cells. Overexpression of SlTPR1 in tomato resulted in ethylene-related pleiotropic effects including reduced stature, delayed and reduced production of inflorescences, abnormal and infertile flowers with degenerate styles and pollen, epinasty, reduced apical dominance, inhibition of abscission, altered leaf morphology, and parthenocarpic fruit. Similar phenotypes were seen in Arabidopsis overexpressing SlTPR1. SlTPR1 overexpression did not increase ethylene production but caused enhanced accumulation of mRNA from the ethylene responsive gene ChitB and the auxin-responsive gene SlSAUR1-like, and reduced expression of the auxin early responsive gene LeIAA9, which is known to be inhibited by ethylene and to be associated with parthenocarpy. Cuttings from the SlTPR1-overexpressors produced fewer adventitious roots and were less responsive to indole butyric acid. It is suggested that SlTPR1 overexpression enhances a subset of ethylene and auxin responses by interacting with specific ethylene receptors. SlTPR1 shares features with human TTC1, which interacts with heterotrimeric G-proteins and Ras, and competes with Raf-1 for Ras binding. Models for SlTPR1 action are proposed involving modulation of ethylene signalling or receptor levels.
Keywords: Development, ethylene signalling, SlTPR1, tetratricopeptide repeat protein, tomato
Introduction
Ethylene regulates many aspects of plant growth and development including ripening, senescence, abscission, and responses to biotic and abiotic stresses (Abeles et al., 1992). It also has dramatic effects on plant growth habit, such as the classic ethylene triple response of exaggerated apical hook, swollen hypocotyls, and inhibited root growth displayed by etiolated seedlings (Guzman and Ecker, 1990). Ethylene biosynthesis occurs via the Yang pathway (Yang and Hoffmann, 1984) involving two key biosynthetic enzymes, 1-aminocyclopropane-1-carboxylate (ACC) synthase and ACC oxidase, encoded by differentially expressed multigene families (Kende, 1993; Zarembinski and Theologis, 1994; Barry et al., 1996). Mutants that are defective in ethylene biosynthesis or perception also exhibit altered morphology, fertility, and final organ size. For example, the Arabidopsis ethylene-overproducer1 (eto1) and constitutive triple response1 (ctr1) mutations, which cause ethylene overproduction and constitutive activation of ethylene signalling, respectively, result in reduced stature, small inflorescences and flowers, and low fertility (Guzman and Ecker, 1990; Kieber et al., 1993).
In Arabidopsis, ethylene is perceived by a family of five receptors (ETR1, ETR2, ERS1, ERS2, and EIN4) that possess sequence similarity with bacterial two-component His kinases (Bleecker et al., 1998; Chang and Shockey, 1999; Wang et al., 2002). The receptors are divided into two subfamilies. The subfamily I receptors, ETR1 and ERS1 contain three transmembrane domains and a conserved histidine kinase domain, and have been shown to function as homodimers, whereas the subfamily II receptors ETR2, EIN4, and ERS2 have an additional N-terminal hydrophobic region and a degenerate histidine kinase domain (Wang et al., 2002). The receptors act as redundant negative regulators of ethylene signalling to suppress ethylene responses (Hua and Meyerowitz, 1998; Hall and Bleecker, 2003). Multiple loss-of-function receptor mutants show enhanced ethylene responses, grow slowly, have reduced organ size and are infertile (Hua and Meyerowitz, 1998). Recently, it has been shown that null mutations in either of the subfamily I ethylene receptors ETR1 or ERS1 result in increased sensitivity to ethylene and double null mutations show strong constitutive ethylene-response phenotypes (Qu et al., 2007). Ethylene binding to the receptors results in the inactivation of the receptor signalling to CTR1, a negative regulator with similarity to Raf-like protein kinases, that interacts with the receptors at the ER (Kieber et al., 1993; Gao et al., 2003). Receptors have also been shown to occur at different subcellular locations and to be regulated by other proteins. For example, RTE1 (REVERSION-TO-ETHYLENE SENSITIVITY 1) is a positive regulator of the ETR1 receptor (Resnick et al., 2006; Zhou et al., 2007) and the two proteins are found to co-localize predominantly at the Golgi apparatus but also at the ER (Dong et al., 2008). In addition, the tobacco ethylene receptor NTHK1 appears to localize at the plasma membrane (Xie et al., 2003) and it has been suggested that the different subcellular localizations of the ethylene receptors may have functional relevance (Dong et al., 2008).
A family of six putative ethylene receptor genes has been identified in tomato: LeETR1, LeETR2, Never ripe (NR), LeETR4, LeETR5, and LeETR6 (Wilkinson et al., 1995; Zhou et al., 1996; Lashbrook et al., 1998; Tieman and Klee, 1999), and the expression of some of these is differentially regulated during development (Tieman and Klee 1999; Klee, 2002). The NR gene is regulated at the transcription level by ethylene and during ripening (Payton et al., 1996; Lashbrook et al., 1998). The NR mutation confers ethylene insensitivity and fruit fail to ripen. Antisense knockout of the mutated NR gene restores fruit ripening, consistent with the receptor inhibition model of ethylene action (Hackett et al., 2000). In contrast to Arabidopsis, which has a single CTR1 gene, tomato has a small family of CTRs, which complement the ctr1 mutant and bind to the receptor NR at the ER (Adams-Phillips et al., 2004; Zhong et al., 2008a). Although in Arabidopsis there is evidence for a degree of receptor redundancy, it has also been observed that knocking out a single tomato receptor, for example, LeETR4 or LeETR6, results in constitutive ethylene responses (Tieman et al., 2000; Kevany et al., 2007), indicating that some receptors are functionally more significant than others in specific situations. It has recently been reported that the tomato receptors are rapidly degraded in the presence of ethylene, probably through a 26S proteasome-dependent pathway, and this receptor degradation is thought to be an important aspect of developmental control (Kevany et al., 2007).
Although mutant analysis has been highly successful in identifying components of the ethylene perception and signalling pathway, it is possible that certain types of proteins involved in signalling are difficult to identify using this approach. The functional characterization of SlTPR1, a novel tetratricopeptide repeat protein initially isolated as a NR-interacting protein in a yeast two-hybrid screen, is reported here. Further study indicated that SlTPR1 interacts with both NR and LeETR1 ethylene receptors, and SlTPR1 appeared to co-localize with NR at both the plasma and nuclear membranes when transiently co-expressed in onion epidermal cells. Overexpresssion of SlTPR1 in planta resulted in small plants with severely reduced fertility, parthenocarpic fruit, altered leaf and fruit shape, epinasty, and enhanced ethylene responses. The results suggest that SlTPR1 plays an important role in ethylene signalling and is involved in cross-talk between ethylene signalling and auxin responses.
Materials and methods
Plant materials and growth conditions
Seeds of tomato (Solanum lycopersicon cv. Ailsa Craig, formerly Lycopersicon esculentum) and Arabidopsis thaliana (ecotype Columbia) were grown from homozygous lines under general greenhouse conditions.
Constructs and plant transformation
All molecular cloning procedures were carried out using standard methods (Sambrook and Russell, 2001). The full-length coding sequence of SlTPR1 was PCR amplified and cloned into the pENTR/D-TOPO vector (Invitrogen) and confirmed by sequencing. The pENTR-SlTPR1 was recombined into the gateway binary vector pK7FWG2 (Karimi et al., 2002). The resulting construct was sequenced and introduced into Agrobacterium tumefaciens LB4404 cells (Bevan, 1984) and used to transform wild tomato cotyledons, as previously described by Smith et al. (1990). The SlTPR1-GFP construct was introduced into competent Agrobacterium tumefaciens C58 and used to transform Arabidopsis by the floral dip method (Clough and Bent, 1998).
RNA isolation and Northern analysis
RNA extraction and blotting were carried out as described in Griffiths et al. (1999). Probes were synthesized using the Rediprime™II- random prime labelling system following the manufacturer's instructions (GE Healthcare). Pre-hybridization and hybridization was carried out for 16 h at 42 °C in buffer containing 1% (w/v) SDS, 50% (v/v) deionized formamide, 5× SSC, 50 mM sodium phosphate pH 6.8, 0.1% (w/v) sodium pyrophosphate, 10% (w/v) dextran sulphate, and 50 μg ml−1 salmon sperm DNA. Hybridized membranes were finally washed in 0.2× SSC, 0.1% SDS and the signal was detected by autoradiography. Quantitative analysis of Northern blots was carried out using the phosphor screen-K (Kodak) and Molecular Imager FX system (Bio-Rad) following the manufacturer's instructions.
Yeast two-hybrid analysis
The LexA-based interaction trap system described by Golemis and Brent (1997) was used in this study. All plasmids and S. cerevisiae strain EGY48 were kindly supplied by R Brent, Massachusetts General Hospital, Boston. The tomato ripening fruit cDNA library and SlTPR1 cDNA (nt: 25-786) were inserted into the EcoRI/XhoI restriction site of ‘prey’ vector pJG4-5. ‘Bait’ proteins consisting of partial ethylene receptor sequences were constructed by insertion of cDNA sequences into the EcoRI/XhoI or BamHI/XhoI restriction sites of plasmid pEG202, downstream of and in-frame with the bacterial LexA DNA-binding domain sequence (DB). All the constructs were confirmed by sequencing. The homeodomain of bicoid protein fused to the LexA DNA-binding domain, encoded in plasmid pRFHM1, was used as a negative control, while pSH17-4, which encodes the LexA DNA-binding domain fused to the Gal4 activation domain, was used as a positive control.
Preparation of GST-fusion proteins and in vitro pull-down assay
The cDNAs encoding the full-length NR protein (aa 1–754) and LeETR1132–754 were amplified by PCR, inserted into the BamHI site of vector pESP-2 in frame with the GST tag (Stratagene). Constructs were confirmed by sequencing and then transformed into yeast S. pombe strain SP-Q01. Total proteins were extracted in PBST plus 0.5% N-lauryl sarcosine with proteinase inhibitors (137 mM NaCl, 2 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 1% Triton® X-100, 1 mM PMFS, and 100 μM leupeptin). GST-fusion proteins were purified on GST affinity resin (BD ClonTech) according to the manufacturers’ recommendations. Proteins were separated by SDS-PAGE and visualized by Coomassie blue staining (Bio-Rad) or verified using an anti-GST antibody (GE Healthcare). The coding sequence of SlTPR1 was inserted into pEG202 to form DB-SlTPR1. This construct was introduced into yeast S. cerevisiae strain EGY48 and grown in minimal media lacking histidine (Golemis and Brent, 1997) at 29 °C overnight. Total proteins were extracted in PBST plus proteinase inhibitors as described above and quantified using the Bio-Rad Protein Assay kit. The expression of the LexA fusion proteins was detected using an anti-LexA antibody (Invitrogen) following immunoblotting. For in vitro pull-down assays, 1 μg of each purified GST-receptor fusion protein was bound to GST affinity resin and 200 μg of total yeast extracts containing DB-SlTPR1, or the control protein, were added. Samples were maintained in 1 ml of PBST buffer plus protein inhibitors (2 mM PMSF, 1 μM leupeptin) and rotated for 1 h at 4 °C. After washing five times, samples were subjected to SDS-PAGE (10%) and detected with anti-LexA or anti-GST antibodies.
Ethylene treatment of fruit and measurement of ethylene production from leaves
Mature green fruit was collected, placed in a 250 ml glass jar, sealed with a Subaseal vaccine cap and treated with 10 ppm ethylene. After 6 h, fruit was harvested for RNA analysis. All experiments were carried out in triplicate. Ethylene production was measured according to Smith et al. (1986). Three compound leaves from the tops of the main stem were collected, weighed, and placed in a 250 ml glass jar and sealed with a Subaseal vaccine cap. After 2 h, 1 ml of gas from the headspace was withdrawn and ethylene was analysed on a Pye Unicam gas chromatography apparatus. Ethylene production was calculated as nl g−1 h−1.
Transient expression in onion epidermal cells
The full-length coding sequence of SlTPR1 and NR were inserted in the transient expression vector pDH51-GW-CFP (Zhong et al., 2008a). Transient gene expression in onion epidermal cells was carried out using a Biolistic PDS-1000/He Particle Delivery System (Bio-Rad) as previously described (Zhong et al., 2008b). Gold particles (0.8–1.5 μm, AlfaAesar), coated with 2 μg of each plasmid DNA, were bombarded into onion epidermal peels placed on MS medium using a 1100 psi rupture disc (Bio-Rad) under a vacuum of 28 in (71 cm) Hg. The Petri dishes containing the onion peels were then incubated in the dark at room temperature overnight prior to imaging.
Florescence microscopy
All images were obtained by using a Leica TCS SP2 AOBS confocal scanning microscope. CFP variant Cerulean was excited using a 458 nm laser and emissions were collected from 465–505 nm. For GFP, 488 nm was used for excitation and the emissions collected from 495–550 nm. YFP variant Venus was excited by 514 nm laser and the emission collected from 525–600 nm.
Results
SlTPR1 interacts with the ethylene receptors NR and LeETR1
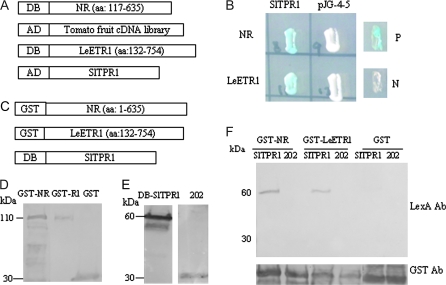
In order to identify novel ethylene signalling components, a tomato ripening fruit cDNA library constructed in the prey vector pJG4-5 containing the activation domain (AD) of B42 was used for an interaction screen with the ethylene receptor NR. The NR partial cDNA sequence (NR117–635, lacking the N-terminal membrane spanning region), was inserted in the yeast two-hybrid bait vector pEG202 (containing the LexA DNA-binding domain, DB) (Fig. 1A). In the first round, 2×106 independent yeast colonies were screened and 47 positive interacting cDNA clones belonging to four families were recovered and sequenced. BLAST searches showed the encoded proteins (INT proteins; Table 1) have homologies to a TPR protein (initially called INT106; Arciga et al., 2003; named here as SlTPR1: Solanum lycopersicum tetratricopeptide repeat protein 1), and INT clones 129 (zinc metalloproteinase), 119 (ubiquitin fusion degradation protein 1), and 22 (dTDP-glucose 4,6-dehydratase). SlTPR1 also interacted with the tomato LeETR1 ethylene receptor, but not with LeETR2, 4, 5, and 6 ethylene receptors in yeast two-hybrid assays (Fig. 1B; see Supplementary Fig. S1 at JXB online). (Previously named tomato genes are referred to by their original Le- prefix, for Lycopersicon esculentum.)
Fig. 1.
Assays for the interaction of SlTPR1 with the ethylene receptors NR and LeETR1. (A) Constructs used for yeast two-hybrid screening. The partial NR cDNA encoding amino acids 117–635 and the partial LeETR1 cDNA encoding aa 132–754 were inserted in the bait vector pEG202 (DB), and the tomato ripening fruit cDNA library and SlTPR1 cDNA were constructed in the prey vector pJG4-5 (AD). (B) Activation assay of LacZ reporter by interaction of SlTPR1 and the receptors by growing yeast on galactose containing 5-bromo-4-chloro-3-indolyl-β-D-galactopyrano-side (X-gal) medium. P, positive control pSH17-4; N, Negative control pRFHM1. (C) Constructs for in vitro pull-down assays. NR1–635 and LeETR1132–754 were fused to GST, whereas SlTPR1 was inserted in pEG202 downstream of LexA DNA binding domain (DB). (D) Immunoblotting to verify the expression of GST-receptor fusions purified from S. pombe using anti-GST antibody. GST-NR = NR fused to GST; DB-SlTPR1 = Tomato TPR1 fused to the LexA DNA binding domain; 202 = LexA DNA binding domain alone. (E) Immunoblotting to verify DB-SlTPR1 expression in yeast, using anti-LexA antibody. (F) Protein–protein interactions of SlTPR1 with NR and LeETR1 in vitro assays. Anti-LexA antibody was used to detect whether DB-SlTPR1 or LexA-DB (202) was co-precipitated with either GST-receptor fusions or the GST control. DB-SlTPR1 (60 kDa) was only detected in the lanes containing GST-NR/B-SlTPR1 and GST-LeETR1/DB-SlTPR1. Anti-GST antibody was used to verify the input of the GST fusions (lower panel).
Table 1.
NR-interacting proteins from yeast two-hybrid screen
| INT family | Number of clones | Homology |
| INT22 | 22 | dTDP-glucose 4-6-dehydratase |
| SlTPR1 | 17 | Tetratricopeptide repeat protein |
| INT119 | 2 | Ubiquitin fusion degradation protein 1 |
| INT129 | 6 | Zinc metalloproteinase |
Protein–protein interactions between SlTPR1 and the tomato ethylene receptors NR and LeETR1 were further tested by in vitro pull-down assays. Full-length NR and LeETR1132–745 were expressed as glutathione-S-transferase (GST) translational fusions in Schizosaccharomyces pombe (Fig. 1C, D) and the SlTPR1 coding sequence was fused to the LexA DNA binding domain (DB-SlTPR1) in the yeast two-hybrid vector pEG202 (DB-SlTPR1) (Fig. 1C–E). For pull-down assays, purified GST-receptor fusions bound to GST resin were incubated with yeast crude extract containing DB-SlTPR1 (see Materials and methods). Protein extracts from cells with either the GST vector (GST) or pEG202 (202) were used as negative controls (Fig. 1D, E). Protein input was verified by immunoblotting using Anti-GST antibody (Fig. 1F, lower panel). Immunoblotting using anti-LexA antibody detected DB-SlTPR1 with a molecular weight of approximately 60 kDa (SlTPR1 29 kDa, LexA 30 kDa) only in the lanes containing GST-NR/DB-SlTPR1 and GST-LeETR1132–745/DBSlTPR1 (Fig. 1F). No bands were detected in the controls (lanes containing GST-NR/202, GST-LeETR1/202, GST/DB-SlTPR1, and GST/202).
SlTPR1 encodes a TPR motif-containing protein
The coding sequence of SlTPR1 cDNA is 786 nucleotides in length and encodes a putative protein of 261 amino acid residues with a molecular weight of 29 kDa. The protein contains three TPR motifs (Blatch and Lässle, 1999) from amino acids 89 to 197 (Fig. 2A). TPR is a structural motif present in a wide range of proteins from bacteria to humans. Individual TPR domains are composed of two anti-parallel alpha helices separated by a turn. Multiple TPR domains are often arranged to form a large surface area available for ligand binding (Das et al., 1998). They usually mediate protein–protein interactions or the assembly of multi-protein complexes, and are involved in cell cycle regulation, interaction with chaperones, transcription control, and protein degradation (Das et al., 1998).
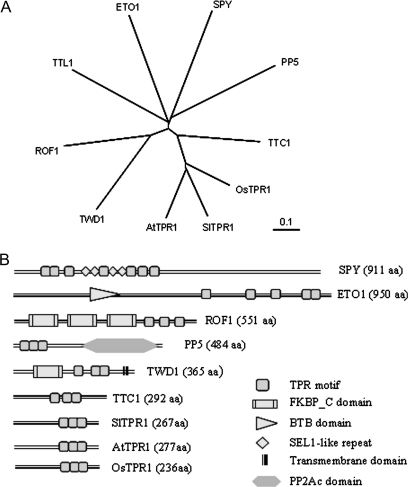
Fig. 2.
Sequence analysis of SlTPR1. (A) The nucleotide sequence and the deduced protein sequence of SlTPR1. TPR motifs are underlined. (B) Alignment of SlTPR1 with the homologues from rice (OsTPR1, accession number: AAP54347), Arabidopsis (AtTPR1, At4g30480), and human (TTC1, accession number: NM_003314). The consensus sequence is boxed.
Sequence comparison indicates that the SlTPR1 protein shares 50% similarity to the human TETRATRICOPEPTIDE REPEAT DOMAIN1 (TTC1, accession number: NM_003314) protein (Fig. 2B). TTC1 is a 292-amino-acid protein with three TPR motifs that interacts with heterotrimeric G-proteins and Ras, and is also able to compete with Raf-1 for Ras-binding (Marty et al., 2003). Interestingly, plant ethylene receptors are known to interact with a downstream Raf-like kinase CTR1. SlTPR1 shares high similarity to two other plant TPR proteins: one from rice (accession number: AAP54347, referred to here as OsTPR1) and another from Arabidopsis (At4g30480, referred to here as AtTPR1) with 74% and 72% similarity, respectively, over the entire sequences (Fig. 2B). However, there are no mutant phenotypes documented or functional studies on either of these genes. AtTPR1 is the only orthologue to SlTPR1 from Arabidopsis and no other SlTPR1-like sequences have been found so far in the tomato EST database. Phylogenetic analysis using the protein sequences of SlTPR1, TTC1, OsTPR1, and Arabidopsis TPR motif-containing proteins AtTPR1, SPY (At3g11540), ETO1 (At3g52770), TTL1 (At1g53300), ROF1 (At3g25230), TWD1 (At3g21640), and PP5 (At2g42810) also indicated that SlTPR1, AtTPR1, OsTPR1, and TTC1 are the most closely related sequences (Fig. 3A). TPR domains can be dispersed in a protein sequence (Fig. 3B) and these proteins play important roles in signalling events, for example, TWD1 (TWISTED DWARF1) is a plasma membrane-anchored immunophilin-like protein that physically interacts with the multidrug resistance/P-glycoprotein ATP-binding cassette transporter PGP1 and PGP19 and controls PGP-mediated auxin transport (Bouchard et al., 2006). Other plant TPR proteins, such as SPINDLEY (SPY), ETO1, and TTL1 are reported to play important roles in GA, ethylene, and ABA signalling (Tseng et al., 2001; Wang et al., 2004; Greenboim-Wainberg et al., 2005; Rosado et al., 2006), although SlTPR1 shares little overall similarity to these proteins (Fig. 3).
Fig. 3.
Structural comparison of SlTPR1 and TPR proteins. (A) Comparison of SlTPR1 with other TPR proteins. The structures were produced in SMART programme (Simple Modular Architecture Research Tool). The structures are drawn approximately to scale and individual TPR, FKBP_C (FK506-binding protein), BTB (Bric-a-brac, Tramtrack, Broad-complex), SEL1-like repeats, Transmembrane, and PP2Ac (Protein phosphatase 2A homologues, catalytic domain) domains are shown. (B) Phylogenetic analysis using the protein sequences of SlTPR1, OsTPR1 (AAP54347), TTC1 (NM_003314), and Arabidopsis AtTPR1 (At4g30480), ETO1 (At3g52770), SPY (At3g11540), TTL1 (At1g53300), PP5 (A22g42810), ROF1 (At3g25230), and TWD1 (At3g21640) indicating that SlTPR1, AtTPR1, and OsTPR1 are most closely related to human TTC1. Phylogenetic tree was produced using ClustalW2 (http://www.ebi.ac.uk).
SlTPR1 transcripts are abundant in developing and ripening fruit, leaves, and flower buds
Northern analysis indicated that the SlTPR1 mRNA was present in roots, stems, young and mature leaves, flower buds, and fully opened flowers (Fig. 4). It was highly expressed in developing fruits at immature green and mature green stages, increased at the onset of fruit ripening (breaker stage), and accumulated to high levels at later stages of ripening. SlTPR1 mRNA slightly increased following exogenous ethylene treatment, and it was also highly expressed in mature and ripening fruits of ripening mutants Never-ripe (Nr), which is ethylene insensitive, and ripening inhibitor (rin), which is deficient in a MADS box transcription factor required for ripening (Vrebalov et al., 2002).
Fig. 4.
Expression pattern of SlTPR1 by Northern analysis. RNA samples were isolated from a range of tissues at different developmental stages from the wild-type tomato. YL, young leaves; ML, mature leaves; SL, senescent leaves; 1, unopened flower buds; 2, opened flower buds; 3, fully-opened flowers; 4, early senescing flowers; 5, senescent flowers; IM, immature green fruit; MG, mature green fruit; ME, ethylene-treated mature green fruit; B, fruit at breaker stage; +3 + 6 +9, fruits at 3, 6, 9 d after start of colour change; ST, stem; RT, root; Nr, Never ripe; rin, ripening inhibitor. 10 μg total RNA was used for the blotting and the full-length SlTPR1 cDNA was used as the probe. The ethidium bromide-stained rRNA gel below indicates the equal sample loading.
Overexpression of SlTPR1 in tomato resulted in dwarf plants
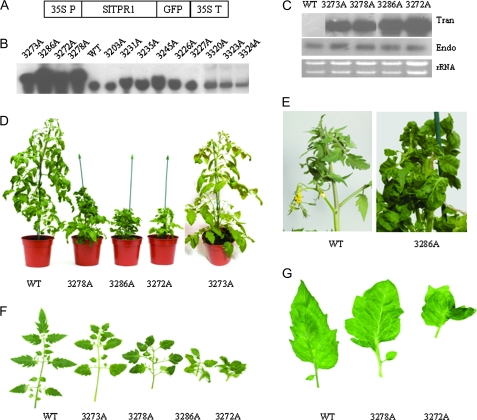
The role of SlTPR1 in tomato was addressed by expressing the SlTPR1 full-length coding sequence using the 35S cauliflower mosaic virus (CaMV) promoter in the gateway binary vector pK7FWG2 (named SlTPR1-GFP) (Karimi et al., 2002) (Fig. 5A). The construct was introduced into the wild-type tomato cultivar Ailsa Craig by Agrobacterium-mediated transformation. Thirty primary transformants (T0 generation) were regenerated on selective media and grown to maturity. Northern blot assay identified four independent T0 lines (3273A, 3278A, 3286A, and 3272A) that clearly over-expressed the SlTPR1 transgene mRNA (Fig. 5B). Three lines 3278A, 3286A, and 3272A, each with high levels of transgene expression, exhibited phenotypic affects on plant habit, with marked dwarfism, reduced apical dominance, epinasty, and altered leaf shape (Fig. 4C–F). Line 3273A displayed intermediate phenotypes with effects on branching, leaf and fruit morphology, and reproductive development, although the plant reached a similar size to the wild type (Fig. 5D, E). Northern analysis using RNA from vegetative buds showed no reduction of the endogenous SlTPR1 mRNA in the transgenics (Fig. 5C), suggesting that the phenotypic effects were caused by overexpression of the SlTPR1-GFP transgene rather than co-suppression of the endogenous SlTPR1.
Fig. 5.
Characterization of SlTPR1-overexpressing tomato plants. (A) Overexpression construct of SlTPR1. (B) Northern analysis to determine the SlTPR1 transgene expression in tomato primary transformants. Total RNA from both transgenic and wild-type plants was blotted and probed with the SlTPR1 cDNA. (C) Northern analysis to determine the mRNA levels of both endogenous and transgene SlTPR1 in the transgenic lines (the transgene SlTPR1 has a GFP-tag and is larger than the endogenous mRNA). 10 μg total RNA from the vegetative buds was blotted and probed with the SlTPR1 cDNA. The ethidium bromide-stained rRNA below indicates sample loading. All the samples were run in the same gel, but the order of the lanes was rearranged to correspond to the expression level of the transgene and to aid comparison with Fig. 5D. (D) Phenotypes of SlTPR1 transgenic plants. Photographs were taken of plants 80 d after transferring from tissue culture to compost. (E) A close-up of plants in (d) to show inhibited apical dominance and altered leaves of line 3286A compared with the wild type. (F, G) Leaf morphology of the transgenic plants compared with the wild type.
The dwarf plants resulting from strong overexpression of SlTPR1 were less than one-third of the wild-type height at 60 cm after 80 d, with shorter internodes (1–3 cm versus wild type 7–8 cm), inhibited apical dominance (Fig. 5E) and numerous side shoots (Fig. 5D, E; Table 2). Anatomical analysis indicated that the reduced height was related to smaller cell sizes (data not shown). Notably, the higher the transgene expression the more severe the phenotypic abnormality (Fig. 5C, D), suggesting that an appropriate level of SlTPR1 in planta is very important for growth and development. Applying 5 μM exogenous gibberellin to the cuttings of the dwarf lines: 3278A, 3286A, and 3272A had no effect on the growth habit measured after 3 weeks (data not shown).
Table 2.
Effect of SlTPR1 overexpression on development
| Lines | Internodes (cm) | True leaves before first inflorescence | Total inflorescences on main stem | Total fruits produced |
| Wild type | 6.5±3.5 | 9 | 7 | 67 |
| 3278A | 3.2±1.9 | 13 | 2 | 0 |
| 3273A | 8±5.0 | 12 | 3 | 18 |
| 3272A | 1.8±2.2 | 13 | 3 | 0 |
| 3286A | 1±0.5 | 13 | 3 | 0 |
SlTPR1 overexpression resulted in epinasty and altered leaf morphology
Overexpression of SlTPR1 in tomato resulted in marked leaf and petiole epinasty (Fig. 5E), a phenotype associated with responses to ethylene or auxin (Abeles et al., 1992; Barry et al., 2001). The leaf morphology of the SlTPR1 overexpressors was remarkably altered. Wild-type Ailsa Craig tomato leaves are unipinnately compound with a terminal leaflet and three pairs of lobed major lateral leaflets with pinnate venation, and the terminal leaflet points straight forwards. Smaller leaflets are often seen between the major leaflets (Fig. 5E, F). By contrast, the compound leaves of the transgenic plants were smaller, sometimes with an asymmetrical arrangement, the terminal leaflet often pointed side-wards, leaf-like structures were seen to replace the larger leaflets, and frequently two or four pairs of large leaflets were seen instead of three pairs in the wild type (Fig. 5G). Leaf margins were twisted, and sometimes vascular patterning was changed (Fig. 5F). Petioles often exhibited epinasty; leaflets were wider and more rounded with reduced lobes; the lamina was often wrinkled; the texture of the leaves was thicker and harder (data not shown).
Overexpression of SlTPR1 affected reproductive growth
Overexpression of SlTPR1 had a severe impact on reproductive growth and development (Fig. 6). The transgenic lines showed delayed inflorescence development and the numbers and sizes of the inflorescences were also reduced. Wild-type tomato plants normally produce the first inflorescence after forming nine compound leaves. In the SlTPR1 transgenic lines, however, the first inflorescence was produced after 12–13 leaves (Table 2). The number of inflorescences was also reduced in three strong overexpressing lines (3278A, 3286A, and 3272A) with three to five in the main stem versus seven in the wild type, whereas the intermediate overexpressor 3273A was less affected (Table 2). The first inflorescence of the wild-type plants had 10 flowers with four distinct whorls: six sepals, five petals, fused stamens, and the carpel (Fig. 6D, E). By contrast, the first inflorescences of three dwarfed lines were dramatically altered, with five or six very tightly arranged small flowers (3–4 mm versus the wild type 15–20 mm) with unusually large trichomes (Fig. 6A, B, F), which lacked a style and failed to open (Fig. 6F, arrow). Flowers from later inflorescences did open, but sometime petals and stamens were fused together with misplaced style-like structures (Fig. 6G, arrow shows a misplaced style), or sometimes the stamens were open rather than fused (Fig. 6H, arrows). These flowers often failed to abscise (data not shown). As a consequence of these abnormalities, the three strong overexpressing lines were infertile (Table 2).
Fig. 6.
Flower morphology of tomato plants overexpressing SlTPR1. (A–C) Comparison of inflorescences and flowers from wild-type and transgenic plants, and (B) diagram of the flower arrangement from (A). (D, E) The normal appearance of wild-type flowers. (F) Underdeveloped flower from line 3278A with enlarged trichomes and a retarded style (arrow). (G) Abnormal flower from the transgenic line 3278A with fused sepals and stamens and a misplaced style-like structure (arrow). (H) Abnormal flower from line 3278A with open rather than fused stamen filaments (arrow). (I) A shortened and half-opened style (arrow) from line 3273A. (J) Pollen from the transgenic line 3273A that overexpressed SlTPR1 compared with the wild type. Pollen grains released from fully-opened flowers were stained with Alexander's stain and examined on a Nikon Microscope with white light. Full pollen grains containing cytoplasm from the wild type stained red, in contrast, pollen grains from line 3273A were either empty or nearly empty, and deformed, consisting mainly of exine walls stained green.
Line 3273A with the lowest SlTPR1 overexpression level and intermediate phenotypes (Fig. 5) displayed normal inflorescence size and produced more flowers than other transgenic lines (Table 2). The style, however, was sometimes shorter and half open (Fig. 6I, arrow), and the pollen grains had reduced cytoplasm or were empty and some were deformed (Fig. 6J). These abnormalities prevented pollination and fertilization although line 3273A produced parthenocarpic fruits (Fig. 7A). Fruits were distinct from wild type, being ovate, often with deeper crevices and a beak-like structure, increased locules, and an enlarged columella (Fig. 7A, arrow). Often the style and petals remained attached to the fruits when ripe (Fig. 7A, B, D, arrows), indicating inhibited abscission. The pedicels were longer than the wild type and the knuckle (abscission zone) was sometimes absent, but clearly visible in the wild type (Fig. 7C, E). Ripening was not noticeably different from the wild type, but fruits remained firmly attached to the pedicels and did not abscise even 80 d after ripening (Fig. 7E, F). Fused multiple parthenocarpic fruits were also found in cuttings derived from this line (Fig. 7G, H).
Fig. 7.
Altered fruit morphology in line 3273A overexpressing SlTPR1. (A) Control and ovate transgenic fruit with enlarged columella (arrow) and no seeds. (B) Transgenic fruits with a beak-like structure and an attached style (arrows). (C) Elongated pedicel without the knuckle (abscission zone) in the transgenic fruit compared with the wild-type fruit which has a shorter pedicel with an obvious knuckle (arrowed). (D) Senescing petals attached to the mature fruits. (E) Comparison of the transgenic fruits with the wild type at different developmental stages. (F) Transgenic fruits on line 3273A firmly attached to the pedicel 80 d after ripening. (G, H) Fused multiple parthenocarpic fruits produced by plants developed from cuttings of line 3273A.
Altered expression of ethylene and auxin responsive genes in SlTPR1 overexpressing lines
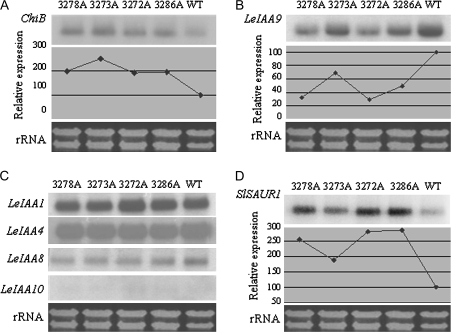
To test whether the phenotypes could result from altered ethylene synthesis or signalling, ethylene evolution was measured from transgenic and wild-type plants using the first four fully-expanded compound leaves on the main stem 8 weeks from propagation, commencing 2 h after excision to allow wound ethylene to subside. The results indicated that there was no evidence for major changes in ethylene evolution compared with the wild type (Table 3). Studies on the ethylene responsive gene ChitB (Danhash et al., 1993) showed a 2–2.5-fold increase in expression in transgenic lines compared to the wild type (Fig. 8A), although E4, another ethylene responsive gene (Lincoln et al., 1987) only showed a slight increase (data not shown).
Table 3.
Ethylene production by SlTPR1 overexpression lines
| Lines | Ethylene production (nl g−1 h−1) |
| Wild type | 2.9±0.2 |
| 3278A | 2.3±0.8 |
| 3273A | 3.3±1.3 |
| 3272A | 2.0±0.9 |
| 3286A | 2.7±1.4 |
Ethylene production was measured from leaves of both wild type (AC++) and transgenic lines. The data are the mean of three samples from each line.
Fig. 8.
Northern analysis to determine the expression of ethylene/auxin responsive genes in SlTPR1-overexpressing tomato plants. 10 μg total RNA from the vegetative buds of the transgenic and wild-type plants was used for the assays, and radiolabelled 32P emission was detected using a Molecular Imager FX (BioRad). The ethidium bromide-stained rRNA below each panel indicates the sample loading. The coding sequences of ChiB, LeIAA9, LeIAA1, LeIAA4, LeIAA8, and LeIAA10 were used as probes.
The reduced apical dominance, altered leaf and fruit morphology, and production of parthenocarpic fruits were reminiscent of auxin-related responses. Therefore, the expression of a set of early auxin response genes (LeIAA genes) was examined by northern analysis. mRNA from LeIAA9 (Wang et al., 2005), an auxin early responsive gene that is repressed by ethylene (Jones et al., 2002), was decreased to 30% in some lines (Fig. 8B). By contrast, LeIAA1, LeIAA4, LeIAA8, and LeIAA10 mRNAs did not show significant changes (Fig. 8C), whereas a small auxin up-regulated RNA gene, SlSAUR1-like (EST: TC181903), was increased 2–3-fold in different lines (Fig. 8D). These experiments were repeated with biological replicates and similar results obtained. The degree of alteration in the levels of LeIAA9 and SlSAUR1-like expression corresponded to the phenotype and the level of the SlTPR1 transgene expression in planta, with the least change in line 3273A.
The responses of the SlTPR1 transgenic lines to auxin were also examined. Cuttings about 8 cm in length from the side shoots of wild-type and transgenic plants were dipped in talcum powder containing 0 or 1000 μg g−1 indole-3-butyric acid (IBA) (Clark et al., 1999). Ten cuttings were taken from plants of each line, and propagated in 5 cm2 pots containing perlite and maintained in the greenhouse in high humidity. After 3 weeks, the total number of root initials for each cutting was examined. Without IBA treatment the transgenic cuttings had poor adventitious root formation compared to the wild type; without exogenous IBA the wild-type cuttings typically formed 15 adventitious roots, whereas the transgenic lines 3278A, 3286A, 3272A, and 3273A averaged 8, 2.6, 2.2, and 3.7 adventitious roots, respectively, with a reduced number of cuttings rooted (see Supplementary Fig. S2 and Supplementary Table S1 at JXB online). Although application of exogenous IBA stimulated transgenic cuttings to form adventitious roots, the total number of roots formed was still much lower than the wild type (see Supplementary Fig. S2 and Supplementary Table S1 at JXB online), suggesting altered auxin sensitivity or responses.
Overexpression of SlTPR1 in Arabidopsis resulted in stunted plants with altered leaf and silique morphology
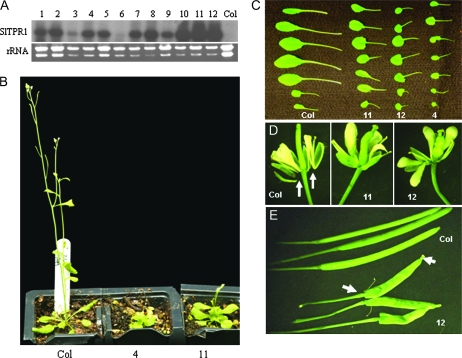
Transgenic Arabidopsis plants over-expressing SlTPR1 with a fluorescent GFP tag were generated by the floral dip method (Clough and Bent, 1998). The T2 progeny of nine independent transgenic lines displayed a similar range of phenotypic alterations, including reduced stature, and small, rounded rosette leaves (Fig. 9B, C). Flower parts were abnormal and sepals and petals abscised later than in the wild type (Fig. 9). The morphology of the siliques was noticeably altered, and even during early development they appeared swollen, shortened, and twisted, and abscission of the sepals and petals was inhibited (Fig. 9D). The mature transgenic siliques were often lanceolate with narrow, elongated regions at their proximal and distal ends. These distorted siliques contained a reduced number of seeds and the stamens remained attached to the siliques during fruit maturation (Fig. 9E).
Fig. 9.
Characterization of transgenic Arabidopsis plants overexpressing SlTPR1. (A) Northern analysis to determine the SlTPR1 transgene expression in independent transgenic Arabidopsis plants 1–12. 10 μg total RNA from the young leaves was blotted and probed with the SlTPR1 cDNA. The ethidium bromide-stained rRNA indicates sample loading (rRNA). Col = wild type. (B) Reduced stature of transgenic Arabidopsis plants overexpressing SlTPR1. Transgenic lines 4, 11, and wild type (Col) were photographed at 30-d-old. (C) Leaf morphology. Rosette leaves 1–7 were excised from the transgenic lines 4, 11, 12, and wild type (Col) at 24-d-old and photographed. (D) Immature silique morphology of transgenic lines 11 and 12 compared with the wild type. Note: abscission of the sepals and petals occurred in the wild type (D Col) and not in the transgenic lines (11, 12). (E) Morphology of mature siliques from the transgenics (line 12) compared with the wild type. Arrows show the proximal and distal elongated regions.
Subcellular localization of florescent protein-tagged SlTPR1 and NR
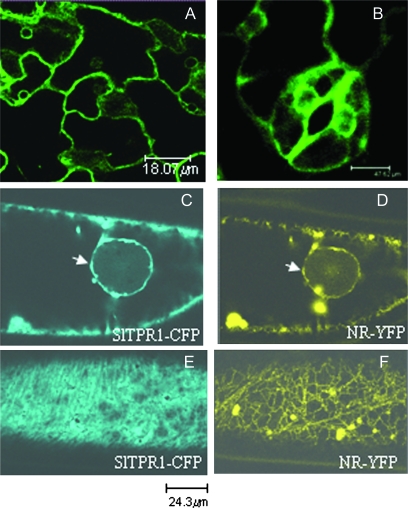
It has been reported that the ethylene receptors can be targeted to several cell membrane systems (Dong et al., 2008). Confocal microscopy of leaf tissue of Arabidopsis plants overexpressing SlTPR1-GFP showed that the fusion protein was localized in the nuclear membrane and plasmalemma, but excluded from the nucleus (Fig. 10A, B). The plasma membrane location of SlTPR1-GFP was confirmed by its co-localization with the red-florescent membrane dye FM 4-64 (see Supplementary Fig. S3 at JXB online). When two fluorescent constructs, SlTPR1 fused to CFP and NR fused to YFP (pDH51-GW-YFP) were transiently expressed in the same onion epidermal cell by co-bombardment, the two florescent fusion proteins were found to co-localize at the plasmalemma and nuclear membranes (Fig. 10C–F). In addition, the NR-YFP fusion protein also appeared to localize at the ER membrane as previously reported (Zhong et al., 2008a).
Fig. 10.
Subcellular localization of fluorescent protein-tagged SlTPR1 and NR. (A, B) SlTPR1-GFP (green fluorescence protein) located in the nuclear membrane and plasma membrane of leaf epidermal cells (A) and guard cells (B) of transgenic Arabidopsis. (C–F) Transient co-expression of SlTPR1-CFP (cyan fluorescence protein-blue) and NR-YFP (yellow fluorescence protein) in onion epidermal cells. The two proteins were localized in the plasma membrane and the nuclear membrane (C, D. arrows), while SlTPR1-CFP was also located in the cytoplasm (E) and NR-YFP in the ER (F). The scale bars are 18 μm (A), 47.62 μm (B), 24.3 μm (C–F).
Discussion
SlTPR1 encodes a novel tomato TPR protein that interacts with the tomato ethylene receptors NR and LeETR1 in yeast two-hybrid and in vitro pull-down assays (Fig. 1). SlTPR1 contains three TPR motifs (Fig. 2), which are frequently found in proteins mediating important regulatory interactions and signalling events (Fig. 3). TPR proteins in plants play important roles in hormone signalling and development, for example, SPY regulates GA/cytokinin signalling cross-talk (Greenboim-Wainberg et al., 2005), and ETO1 interacts with ACS5 (an isoform of ACC synthase) to inhibit its enzyme activity, and also serves as a substrate-specific adaptor protein for ACS5 degradation, by interacting with CUL3, a constituent of the ubiquitin ligase complexes (Wang et al., 2004). ETO1 and SPY are much larger than SlTPR1 (Fig. 3) and they share little similarity outside the TPR motifs. SlTPR1 has no close homologues in tomato EST databases and AtTPR1 also appears as a single sequence in the Arabidopsis genome. SlTPR1 is most closely related to AtTPR1, OsTPR1, and the human TTC1 (Fig. 3), a protein involved in interactions with heterotrimeric G-proteins and Ras (Marty et al., 2003).
In wild-type plants, SlTPR1 mRNA was found in leaves, flowers, stems, and roots, and accumulated to high levels in fruit, peaking during ripening, a process coupled with ethylene evolution. Ethylene treatment of mature green fruit led to a small increase in SlTPR1 mRNA (Fig. 4), but it also accumulated in the Nr mutant, which is largely, but in the Ailsa Craig background not completely, insensitive to ethylene. Overexpression of SlTPR1 in tomato plants resulted in a wide range of developmental responses, including reduced stature and epinasty (Fig. 5); small inflorescences with small, degenerated, and infertile flowers (Fig. 6), and reduced apical dominance (Fig. 5). The severity of the phenotypic effects was directly related to the expression levels of the SlTPR1 transgene mRNA. Higher expression in lines 3272A, 3286A, and 3278A resulted in infertile dwarf plants (Fig. 6), while a line (3273A) that had a lower level of SlTPR1 transgene mRNA had normal stature and less severe effects on side shoot growth, leaf morphology, and flower development. Line 3273A produced degenerated pollen (Fig. 6J), but it was able to produce parthenocarpic fruits (Fig. 7). In addition to these effects, there was a delay in flower bud formation in all the transgenic plants (Table 2). In tomato, flowers are normally produced after nine compound leaves have been formed, whereas in the SlTPR1-overexpressers, they only appeared after 12 or 13 leaves had formed, suggesting that SlTPR1 influences flowering time. This is consistent with enhanced ethylene signalling, since it has been recently demonstrated that ethylene delays flowering in Arabidopsis by modulating the activity of DELLA proteins (Achard et al., 2007). A few attempts to knock-out SlTPR1 in planta by introducing either antisense or RNAi constructs failed to regenerate transgenic plants with altered SlTPR1 expression (data not shown), suggesting that silencing this gene might be deleterious. Taken together, our studies indicated that an appropriate level of SlTPR1 in planta is vital for development.
Some phenotypic aspects of tomato plants overexpressing SlTPR1, such as reduced stature, leaf epinasty, delayed flowering, reduced flower numbers and inflorescences, and sterility are similar to the Arabidopsis multiple loss-of-function ethylene receptor mutants (Hua and Meyerowitz, 1998). The finding that the ethylene responsive gene ChitB was more highly expressed in transgenic tomato plants is consistent with enhanced ethylene responses (Fig. 8). Altered leaf and fruit morphology, and inhibited development of the fruit abscission zone, however, are more related to auxin than ethylene responses. Auxin is known to be involved in leaf shape, vascular patterning, parthenocarpic fruit formation, and fruit morphology. Down-regulation of the auxin early responsive gene LeIAA9 and up-regulation of the small auxin up-regulated RNA gene SlSAUR1-like in SlTPR1 overexpressers (Fig. 8), together with reduced adventitious root formation of the transgenic lines in responses to auxin, indicates the involvement of SlTPR1 in cross-talk with auxin responses. LeIAA9 functions as a transcriptional repressor in auxin signalling and its expression is known to be negatively regulated by ethylene and ripening (Jones et al., 2002; Wang et al., 2005). Tomato plants with a reduced level of LeIAA9 mRNA show phenotypes similar to our SlTPR1 overexpressing lines, such as altered leaf morphology, reduced apical dominance, and the production of parthenocarpic fruits (Wang et al., 2005). The results of the present experiments are consistent with the suggestion that aspects of the phenotype of SlTPR1 overexpressing plants result from the down-regulation of LeIAA9. The degree of reduction was inversely related to the expression level of the SlTPR1 transgene (compare Figs 3, 4, and 8), and the intermediate SlTPR1 overexpressing line 3273A, which also had an intermediate level of LeIAA9 mRNA, produced parthenocarpic fruit (Figs 5, 7). The auxin-related gene SlSAUR1-like was, however, up-regulated by the overexpression of SlTPR1 in plants, an opposite pattern to LeIAA9 (Fig. 8), indicating that this gene is up-regulated by ethylene. The SAUR family is large, for example, there are 75 SAUR genes in the Arabidopsis genome, and their functions are not fully understood.
The results presented here suggest that SlTPR1 affects some auxin responses through the regulation of genes such as LeIAA9 and SlSAUR1-like. This response was quite specific since there was no effect on the expression of a number of other LeIAA genes tested (Fig. 8), but further study may reveal ethylene effects on other auxin-related gene expression. Overexpression of SlTPR1 in planta appeared to affect ethylene responses related to vegetative growth, reproductive development, and abscission, but not qualitative aspects of ripening, such as colour and texture change. However, the rate of ripening was not analysed in detail, because of the shortage of fruit (Table 2). The apparent lack of effect on fruit ripening, which is an ethylene-regulated process, might be due to the fact that SlTPR1 binds to LeETR1 and NR, whereas different ethylene receptors (LeETR4 and LeETR6) are the most important ethylene receptors in controlling tomato ripening (Kevany et al., 2007).
Some aspects of the SlTPR1 overexpression phenotypes resemble features of the epinastic (epi) mutant of tomato (Barry et al., 2001), for example, reduced plant growth, smaller cell size, and twisted epinastic leaves. Seedlings of the epi mutant also show features of a constitutive triple response in the dark in the absence of ethylene, but epi lacks the global constitutive ethylene responses seen in the ctr1 mutant. It has been proposed that the epi mutation affects a subset of ethylene responses, or acts in an independent pathway required for growth that cross-talks with the ethylene response pathway (Barry et al., 2001). It has not been possible to test whether seedlings of the overexpressing SlTPR1 show a triple response, since the plants are either sterile or produce parthenocarpic fruit. There are clear differences between epi plants and SlTPR1 overexpressers, however, since epi leads to ethylene overproduction (Fujino et al., 1998), whereas SlTPR1 overexpression does not (Table 2).
Due to its solubility in both aqueous and lipid environments, ethylene should be readily perceived by receptors residing at any cell membrane or organelle (Abeles et al., 1992). Subcellular localization studies of SlTPR1 indicated that the GFP-tagged SlTPR1 protein was localized at the plasmalemma and nuclear membrane, but was excluded from the nucleus (Fig. 10). Co-expression of SlTPR1-CFP and NR-YFP in onion epidermal cells confirmed that the two proteins co-localized at the plasmalemma and nuclear membrane (Fig. 10). The plasma and nuclear membrane localization of SlTPR1 possibly reflects its subcellular function. It is known that the human homologue of SlTPR1, TTC1, is involved in interactions with selected G-proteins and Ha-Ras (Marty et al., 2003) and that G-protein signalling occurs in the plasma membrane (Hamm, 1998). The tobacco NTHK1 ethylene receptor has also been reported to localize at the plasma membrane (Xie et al., 2003). The NR-YFP fusion protein was, however, also found at the ER (Fig. 10), which was consistent with our previous studies of the ER localization of the NR receptor (Zhong et al., 2008a), whereas SlTPR1-YFP appeared more diffuse, and did not co-localize at the ER (Fig. 10). It has also been shown that the melon ethylene receptor CmERS1 is anchored at the ER membrane, via its N-terminal transmembrane domains with its C-terminal domains facing the cytoplasm, where it could interact with proteins located in the cytoplasm (Ma et al., 2006). Initially, the Arabidopsis ETR1 receptor was reported to be at the ER membrane (Chen et al., 2002), but recently it was found predominantly to co-localize with RTE1 at the Golgi apparatus of Arabidopsis root hair cells (Dong et al., 2008). The evidence for SlTPR1 interaction with ethylene receptors in vitro (Fig. 1) and the membrane localizations suggest it would be possible for SlTPR1 and the receptors to interact in vivo, and this interaction might be significant for the ethylene receptor functions, but this remains to be tested experimentally. The variety of locations observed in different experiments suggests that individual ethylene receptors can be found in more than one place, perhaps moving as part of functional interactions; alternatively, different receptors may be located in different places. In view of the structural similarity between the different tomato ethylene receptors, it will be important to test the significance and specificity of the interaction with SlTPR1 in vivo as part of the process of testing models of SlTPR1 action. The question of which receptors bind SlTPR1 in vivo is also of critical importance, since it is beginning to become clear that some receptors are more significant than others in regulating specific subsets of ethylene responses (Kevany et al., 2007; Zhou et al., 2007). In addition, the subcellular localization should be investigated further using weak or endogenous promoters to eliminate the possibility of overexpression artefacts.
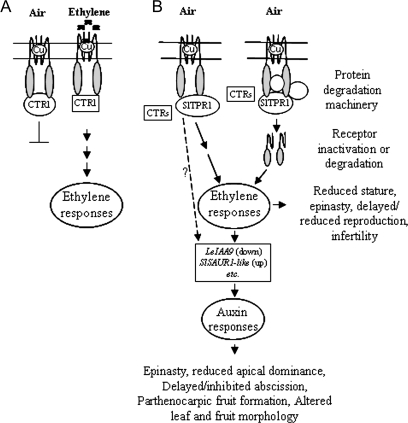
The phenotypic effects caused by overexpression of SlTPR1 in planta indicate enhanced constitutive expression of at least a subset of ethylene responses, perhaps involving a subset of ethylene receptors. There are also clear effects on auxin-related genes. There are two ways to explain these results, which are not mutually exclusive. (i) A possible mechanism for SlTPR1 in ethylene signalling is suggested from the study of TTC1 (Marty et al., 2003), which shares 50% overall similarity with SlTPR1. TTC1 interacts with Ras and competes with Raf-1 for Ras-binding (Marty et al., 2003). It is well known that the Arabidopsis ethylene receptors interact with the Raf-like protein kinase CTR1 (Clark et al., 1998; Gao et al., 2003), and the tomato ethylene receptors LeETR1 and NR also interact with several LeCTR1-like proteins (Zhong et al., 2008a; Lin et al., 2008). If SlTPR1 played a similar role to TTC1, this could explain the enhanced ethylene signalling in plants over-expressing SlTPR1. In the present ethylene signal transduction model, in the absence of ethylene, CTR1 is maintained in an active state by the receptors. This inhibits downstream components and thus ethylene response (Etheridge et al., 2005) (Fig. 11A). If SlTPR1 acts in the same fashion as the TTC1 protein to compete with LeCTRs for binding to the ethylene receptors, this would lead to CTRs remaining in a non-activated state, resulting in constitutive ethylene responses (Fig. 11B, left). (ii) Alternatively, it is also possible that association of SlTPR1 with the receptors might result in receptor inactivation or degradation, leading to enhanced ethylene sensitivity (Fig. 11B, right). It is known that reduced receptor levels can cause enhanced ethylene responses (Hua and Meyerowizt, 1998; Klee, 2002; Qu et al., 2007), and it has also been reported that the receptors are degraded in the presence of ethylene, probably through a 26S proteasome-dependent pathway (Kevany et al., 2007). Other TPR proteins, including ETO1 and CHIP (CARBOXY TERMINUS OF HSC70 INTERACTING PROTEIN)), are involved in the degradation of their interacting partners (Wang et al., 2004; McDonough and Patterson, 2003). The yeast two-hybrid screen showed that, in addition to SlTPR1, NR also interacts with several other proteins (Table 1), including INT clones 129, a zinc metalloproteinase, and 119, a ubiquitin fusion degradation protein, which could be involved in protein degradation. In this model (Fig. 11), the enhanced ethylene responses resulting from overexpression of SlTPR1 lead to altered auxin responses through an effect of LeIAA9 and SlSAUR1-like (Fig. 11B). It is unclear whether or not SlTPR1 is directly involved in auxin signalling. Further work will be required to test these hypotheses and establish the mechanism of action of SlTPR1.
Fig. 11.
Model of SlTPR1 action in ethylene signalling. (A) A recent model of ethylene signal transduction (Etheridge et al., 2005). (B) (i) Overexpression of SlTPR1 could result in competition for CTR1 binding to the receptors, by analogy with TTC1 competition for Raf1 binding (Marty et al., 2003), thus activating downstream ethylene responses; or (ii) SlTPR1 and other receptor-interacting proteins (perhaps including proteins encoded by INT119 and INT129, Table 1) inactivate the receptors or initiate their degradation, resulting in constitutive ethylene responses. It remains to be established which ethylene receptors interact with SlTPR1 in vivo. The enhanced ethylene responses then affect auxin responses through such as LeIAA9 and SlSAUR1-like genes. It is unclear whether or not SlTPR1 directly involved in auxin signalling (dashed arrow).
Supplementary data
The following supplementary data relating to this study are available at JXB online:
Fig. S1. Interaction assay of SlTPR1 with the ethylene receptors.
Fig. S2. Adventitious root formation on cuttings from wild type and SlTPR1 transgenic plants with and without IBA treatment.
Fig. S3. Co-localization of the SlTPR1-GFP fusion with the red-fluorescent membrane stain FM 4-64.
Table S1. Adventitious root formation on cuttings from wild type and transgenic lines treated with 0 and 1000 μg g−1 IBA.
Supplementary Material
Acknowledgments
This work was funded by a BBSRC grant 42/P09465 to DG and by the University of Nottingham.
References
- Abeles FB, Morgan PW, Saltveit ME., Jr . Ethylene in plant biology. 2nd edn. New York: Academic Press; 1992. [Google Scholar]
- Achard P, Baghour M, Chapple A, Hedden P, Van der Straeten D, Genschik P, Moritz T, Harberd NP. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proceedings of the National Academy of Sciences. USA. 2007;104:6484–6489. doi: 10.1073/pnas.0610717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams-Phillips L, Barry C, Kannanz P, Leclercq J, Bouzayen M, Giovannoni J. Evidence that CTRI-mediated ethylene signal transduction in tomato is encoded by a multigene family whose members display distinct regulatory features. Plant Molecular Biology. 2004;54:387–404. doi: 10.1023/B:PLAN.0000036371.30528.26. [DOI] [PubMed] [Google Scholar]
- Arciga L, Alexander L, Wilson I, Grierson D. Identification of a TPR protein interacting with the NR ethylene receptor in tomato. In: Vendrell M, Klee H, Pech JC, Romojaro F, editors. Biology and biotechnology of the plant hormone ethylene. III. Amsterdam: IOS Press; 2003. pp. 52–54. [Google Scholar]
- Barry CS, Blume B, Bouzayen M, Cooper W, Hamilton AJ, Grierson D. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. The Plant Journal. 1996;9:525–535. doi: 10.1046/j.1365-313x.1996.09040525.x. [DOI] [PubMed] [Google Scholar]
- Barry CS, Fox EA, Yen H, Lee S, Ying T, Grierson D, Giovannoni JJ. Analysis of the ethylene response in the epinastic mutant of tomato. Plant Physiology. 2001;127:58–66. doi: 10.1104/pp.127.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan MW. Binary Agrobacterium vector for plant transformation. Nucleic Acids Research. 1984;24:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lässle M. The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1998;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Bouchard R, Bailly A, Blakeslee JJ, et al. Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. Journal of Biological Chemistry. 2006;281:30603–30612. doi: 10.1074/jbc.M604604200. [DOI] [PubMed] [Google Scholar]
- Chang C, Shockey JA. The ethylene response pathway: signal perception to gene regulation. Current Opinion in Plant Biology. 1999;2:352–358. doi: 10.1016/s1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. Journal of Biological Chemistry. 2002;277:19861–19866. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]
- Clark DG, Gubrium EK, Barrett JE, Nell TA, Klee HJ. Root formation in ethylene-insensitive plants. Plant Physiology. 1999;121:53–59. doi: 10.1104/pp.121.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proceedings of the National Academy of Sciences. USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:753–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Danhash N, Wagemakers CA, Van Kan JA, De Wit PJ. Molecular characterization of four chitinase cDNAs obtained from Cladosporium fulvum-infected tomato. Plant Molecular Biology. 1993;22:1017–1029. doi: 10.1007/BF00028974. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratrico-peptide repeats of protein phosphatase 5: implications for TPR-mediated protein–protein interactions. EMBO Journal. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Rivarola M, Resnick JS, Maggin BD, Chang C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signalling. The Plant Journal. 2008;53:275–286. doi: 10.1111/j.1365-313X.2007.03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge N, Chen YF, Schaller GE. Dissecting the ethylene pathway of Arabidopsis. Briefings in Functional Genomics and Proteomics. 2005;3:372–381. doi: 10.1093/bfgp/3.4.372. [DOI] [PubMed] [Google Scholar]
- Fujino DW, Nissen SJ, Jones AD, Burger DW, Bradford KJ. Quantification of indole-3-acetic acid in dark-grown seedlings of the diageotropica and epinastic mutants of tomato (Lycopersicon esculentum Mill.) Plant Physiology. 1988;88:780–784. doi: 10.1104/pp.88.3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. Journal of Biological Chemistry. 2003;278:34725–34732. doi: 10.1074/jbc.M305548200. [DOI] [PubMed] [Google Scholar]
- Golemis EA, Brent R. Searching for interacting proteins with the two-hybrid system, III. In: Bartel PL, Fields S, editors. The yeast two-hybrid system. New York: Oxford University Press; 1997. [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss Cross-talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. The Plant Cell. 2005;17:92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A, Barry C, Alpuche-Solis AG, Grierson D. Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. Journal of Experimental Botany. 1999;50:793–798. [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. The Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett RM, Ho C, Lin Z, Foote HCC, Fray RG, Grierson D. Antisense inhibition of the Nr gene restores normal ripening to the tomato Never-ripe mutant, consistent with the ethylene receptor-inhibition model. Plant Physiology. 2000;124:1079–1085. doi: 10.1104/pp.124.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AE, Bleecker AB. Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. The Plant Cell. 2003;15:2032–2041. doi: 10.1105/tpc.013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm HE. The many faces of G-protein signalling. Journal of Biological Chemistry. 1998;273:667–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz E. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Jones B, Frasse P, Olmos E, Zegzouti H, Li ZG, Latché A, Pech JC, Bouzayen M. Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. The Plant Journal. 2002;32:603–613. doi: 10.1046/j.1365-313x.2002.01450.x. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. Gateway vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kende H. Ethylene biosynthesis. Annual Review of Plant Physiology and Plant Molecular Biology. 1993;44:283–307. [Google Scholar]
- Kevany BM, Tieman DM, Taylor MG, Dal Cin V, Klee HJ. Ethylene receptor degradation controls the timing of ripening in tomato fruit. The Plant Journal. 2007;51:458–467. doi: 10.1111/j.1365-313X.2007.03170.x. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldman KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Klee HJ. Control of ethylene-mediated processes in tomato at the level of receptors. Journal of Experimental Botany. 2002;53:2057–2063. doi: 10.1093/jxb/erf062. Special Issue. [DOI] [PubMed] [Google Scholar]
- Lashbrook CC, Tieman DM, Klee HJ. Differential regulation of the tomato ETR gene family throughout plant development. The Plant Journal. 1998;15:243–252. doi: 10.1046/j.1365-313x.1998.00202.x. [DOI] [PubMed] [Google Scholar]
- Lin Z, Alexander L, Hackett R, Grierson D. LeCTR2, a CTR1-like protein kinase from tomato, plays a role in ethylene signalling, development and defence. The Plant Journal. 2008;54:1083–1093. doi: 10.1111/j.1365-313X.2008.03481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln JE, Corders S, Read E, Fisher RL. Regulation of gene expression by ethylene during Lycopersicon esculentum (tomato) fruit development. Proceedings of the National Academy of Sciences. USA. 1987;84:2793–2797. doi: 10.1073/pnas.84.9.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Cui ML, Sun HJ, Takada K, Mori H, Kamada H, Ezura H. Subcellular localization and membrane topology of the melon ethylene receptor CmERS1. Plant Physiology. 2006;141:587–597. doi: 10.1104/pp.106.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty C, Browning DD, Ye RD. Identification of tetratricopeptide repeat 1 as an adaptor protein that interacts with heterotrimeric G proteins and the small GTPase Ras. Molecular and Cell Biology. 2003;23:3847–3858. doi: 10.1128/MCB.23.11.3847-3858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress and Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton S, Fray RG, Brown S, Grierson D. Ethylene receptor expression is regulated during fruit ripening, flower senescence and abscission. Plant Molecular Biology. 1996;31:1227–1231. doi: 10.1007/BF00040839. [DOI] [PubMed] [Google Scholar]
- Qu X, Hall BP, Gao ZY, Schaller GE. A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biology. 2007;7:1–15. doi: 10.1186/1471-2229-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Wen CK, Shockey JA, Chang C. REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proceedings of the National Academy of Sciences. USA. 2006;103:7917–7922. doi: 10.1073/pnas.0602239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A, Schapire AL, Bressan RA, Harfouche AL, Hasegawa PM, Valpuesta V, Botella MA. The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiology. 2006;142:1113–1126. doi: 10.1104/pp.106.085191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DJ. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Smith CJS, Slater A, Grierson D. Rapid appearance of an mRNA correlated with ethylene synthesis encoding a protein of molecular weight 35000. Planta. 1986;168:94–100. doi: 10.1007/BF00407014. [DOI] [PubMed] [Google Scholar]
- Smith CJS, Watson CF, Bird CR, Ray J, Schuch W, Grierson D. Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Molecular and General Genetics. 1990;224:477–481. doi: 10.1007/BF00262443. [DOI] [PubMed] [Google Scholar]
- Tieman DM, Taylor MG, Ciardi JA, Klee HJ. The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proceedings of the National Academy of Sciences, USA. 2000;97:5663–5668. doi: 10.1073/pnas.090550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D, Klee HJ. Differential expression of two novel members of the tomato ethylene-receptor family. Plant Physiology. 1999;120:165–172. doi: 10.1104/pp.120.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TS, Swain SM, Olszewski NE. Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiology. 2001;126:1250–1258. doi: 10.1104/pp.126.3.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni JJ. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech JC, Bouzayen M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. The Plant Cell. 2005;17:2676–2692. doi: 10.1105/tpc.105.033415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Yoshida H, Lurin C, Ecker JR. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature. 2004;428:945–950. doi: 10.1038/nature02516. [DOI] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. The Plant Cell. 2002:S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by Neverripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Xie C, Zhang JS, Zhou HL, Li J, Zhang ZG, Wang DW, Chen SY. Serine/threonine kinase activity in the putative histidine kinase-like ethylene receptor NTHK1 from tobacco. The Plant Journal. 2003;33:385–393. doi: 10.1046/j.1365-313x.2003.01631.x. [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffmann NE. Ethylene biosynthesis and its regulation in higher plants. Annual Review of Plant Physiology. 1984;35:155–189. [Google Scholar]
- Zarembinski TI, Theologis A. Ethylene biosynthesis and action: a case of conservation. Plant Molecular Biology. 1994;26:1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]
- Zhong S, Lin Z, Fray R, Grierson D. Improved plant transformation vectors for fluorescent protein tagging. Transgenic Research. 2008b;17:985–987. doi: 10.1007/s11248-008-9199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Lin Z, Grierson D. Tomato ethylene receptor interaction: visualization of NEVER-RIPE interaction with multiple CTRs at the endoplasmic reticulum. Journal of Experimental Botany. 2008a;59:965–972. doi: 10.1093/jxb/ern021. [DOI] [PubMed] [Google Scholar]
- Zhou D, Kalaitzis P, Matoo AK, Tucker ML. The mRNA for an ETR1 homologue in tomato is constitutively expressed in vegetative and reproductive tissues. Plant Molecular Biology. 1996;30:1331–1338. doi: 10.1007/BF00019564. [DOI] [PubMed] [Google Scholar]
- Zhou X, Liu Q, Xie F, Wen CK. RTE1 is a Golgi associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiology. 2007;145:75–86. doi: 10.1104/pp.107.104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.