Abstract
Quality traits such as flavour and texture are assuming a greater importance in crop breeding programmes. This study takes advantage of potato germplasm differentiated in tuber flavour and texture traits. A recently developed 44 000-element potato microarray was used to identify tuber gene expression profiles that correspond to differences in tuber flavour and texture as well as carotenoid content and dormancy characteristics. Gene expression was compared in two Solanum tuberosum group Phureja cultivars and two S. tuberosum group Tuberosum cultivars; 309 genes were significantly and consistently up-regulated in Phureja, whereas 555 genes were down-regulated. Approximately 46% of the genes in these lists can be identified from their annotation and amongst these are candidates that may underpin the Phureja/Tuberosum trait differences. For example, a clear difference in the cooked tuber volatile profile is the higher level of the sesquiterpene α-copaene in Phureja compared with Tuberosum. A sesquiterpene synthase gene was identified as being more highly expressed in Phureja tubers and its corresponding full-length cDNA was demonstrated to encode α-copaene synthase. Other potential ‘flavour genes’, identified from their differential expression profiles, include those encoding branched-chain amino acid aminotransferase and a ribonuclease suggesting a mechanism for 5′-ribonucleotide formation in potato tubers on cooking. Major differences in the expression levels of genes involved in cell wall biosynthesis (and potentially texture) were also identified, including genes encoding pectin acetylesterase, xyloglucan endotransglycosylase and pectin methylesterase. Other gene expression differences that may impact tuber carotenoid content and tuber life-cycle phenotypes are discussed.
Keywords: α-Copaene, flavour, microarray, Solanum tuberosum group Phureja, Solanum tuberosum group Tuberosum, texture, volatile
Introduction
Recent phylogenetic analysis of a wide range of wild potato species, landraces, and cultivars, indicates that cultivated potatoes have a monophyletic origin, arising from a group of extremely similar wild species classified in the Solanum brevicaule complex (Spooner et al., 2005). Thus the gene pool used to develop cultivated potatoes is considered to be fairly narrow and has not widely utilized the full diversity (and hence characteristics) of potato germplasm available. Efforts to broaden the genetic base of cultivated potatoes have been made, with some success (Bradshaw and Ramsay, 2005). Despite the purported genetic bottleneck associated with the domestication process, the eight cultivar groups recognized by Huaman and Spooner (2002) are highly variable phenotypically, with many potential uses in potato breeding programmes.
A valuable resource in this regard is S. tuberosum group Phureja, differentiated from S. tuberosum group Tuberosum on the basis of a number of important tuber quality traits such as flavour, texture, colour, and reduced tuber dormancy (De Maine et al., 1993, 1998; Dobson et al., 2004; Morris et al., 2004; Ghislain et al., 2006). Despite being able to differentiate the Phureja group of landraces based on geographical origin, this group is very similar genetically to the Tuberosum group (Spooner et al., 2007). Whatever the final taxonomic outcome, as many of the Phureja group can be clearly differentiated based on tuber quality trait parameters, they form a useful resource for the identification of key genes that underpin these traits. However, for many quality traits, the genes responsible remain to be defined, impeding molecular breeding approaches to develop improved potato germplasm.
Potato flavour is a trait of increasing importance to consumers (McGregor, 2007). However, flavour analysis is complex and likely to depend on both volatile and matrix associated compounds such as those that give rise to the umami taste (one of the five basic tastes; Morris et al., 2007). The volatiles produced by raw and cooked potatoes have been studied extensively (reviewed by Maga, 1994) and over 250 compounds have been identified in volatile fractions. Many attempts have been made to discriminate which of these components are important for potato flavour, which are specific to the method of cooking, cultivar differences, the effects of agronomic conditions, and the effects of storage (Duckham et al., 2001, 2002; Oruna-Concha et al., 2001, 2002a, b; Dobson et al., 2004). To summarize the findings of several studies on boiled potato volatiles; potentially significant compounds include those derived from lipid degradation, the Maillard reaction, and the Strecker reaction, including methional, aliphatic alcohols and aldehydes, thiols and sulphides, and methoxypyrazines (reviewed in Taylor et al., 2007). More recently, boiled tuber volatiles produced by Phureja cultivars have been compared with profiles from Tuberosum (Winfield et al., 2005; Shepherd et al., 2007). Winfield et al. (2005) compared sensory profiles of 16 Phureja clones and six Tuberosum cultivars with volatile data. Generally, the Phureja clones sampled scored considerably higher on an ‘acceptability’ scale, with four of the Tuberosums occupying the bottom five places in a ranked list of the 22 samples. The acceptability measure correlates highly with other less subjective sensory traits, such as ‘creaminess’ and ‘flavour intensity’. Principal component analysis (PCA) of volatile profiles of boiled Tuberosum and Phureja, revealed that hexanal, pentanal, pentyl-furan, and α-copaene accounted for most of the variation between the two groups (Winfield et al., 2005). Although low levels of α-copaene are seen in some Tuberosum profiles, the levels are considerably greater (up to 100-fold) in the Phureja profiles. Alpha-copaene is one of many (tens of thousands) sesquiterpene volatiles produced by plants (Glasby, 1982). Recently α-copaene has been shown to be an important aroma compound in several food plants including lettuce and carrots (Nielsen and Poll, 2007). A specific α-copaene synthase has not yet been identified from any plant species, although α-copaene is a minor part of the spectrum of sesquiterpenes produced by some sesquiterpene synthases (Chen et al., 2003). Other reported differences in the volatile profiles from cooked Phureja and Tuberosum tubers are the levels of branched chain aldehydes and esters of branched chain carboxylic acids produced both enzymatically and non-enzymatically during processing (Shepherd et al., 2007).
Texture, in common with flavour, is an important determinant of consumer preference (reviewed in Taylor et al., 2007). Although quantitative texture measurement comparisons between Phureja and Tuberosum tubers have not been published, qualitative reports indicate that Phureja accessions exhibit a boiled tuber texture described as extremely floury or crumbly (De Maine et al., 1993, 1998). In addition, many Phureja accessions produce yellow-fleshed tubers with much higher levels of carotenoids compared with those typically found in Tuberosum cultivars (Morris et al., 2004). In general, Phureja cultivars are also characterized by differences in the tuber life-cycle, for example, a much shorter dormancy period and short-day adaptations (Ghislain et al., 2006), although long-day adapted accessions have been developed for cultivation in different geographical locations (De Maine et al., 2000).
The very close genetic similarity of the two cultivar groups, yet substantial phenotypic differences that impinge on important breeding objectives, suggest relatively simple genetic architectures for the traits which differ so markedly. Thus, a comparison of gene expression profiles in representative Phureja and Tuberosum clones may reveal the identity of candidate genes that influence a wide range of traits of growing importance to consumers and producers.
In order to identify candidate genes that underpin quality trait differences between Tuberosum and Phureja tubers, transcript profiles were compared using a potato microarray, recently developed by the Potato Oligo Chip Initiative (POCI) consortium that contains gene probes based on 42 034 potato unigene sequences, using the custom Agilent platform. A detailed description of the array has recently been published (Kloosterman et al., 2008). This array has a much greater coverage of potato transcripts than was previously available and so provides an opportunity for detailed trait dissection. Using this microarray it was possible to identify consistent differences in gene expression profiles between Phureja and Tuberosum cultivars, including genes likely to impact on flavour, texture, carotenoid content, and tuber life-cycle. As an example of the utility of the approach, functional analysis of a sesquiterpene synthase gene that was consistently expressed at higher levels in Phureja tubers, demonstrated that this gene encodes an α-copaene synthase.
Materials and methods
Growth and harvesting
Potato breeding clones and cultivars examined in this study were grown in field trials during 2005 and 2006 (Dundee, UK, 56°28′27′′ N; 3°4′11′′ W) using standard agronomic practices. Tubers were sampled at four harvest periods: H1, harvested early in development when average tuber weight was 10–30 g; H2, harvested 2 weeks after H1 when average tuber weight was 40–60 g; H3, harvested 4 weeks after H1 when average tuber weight was 70–100 g and H4, harvested at maturity (following acid burn down of foliage) when average tuber weight was 125–200 g. Approximately 1 kg of average-sized tubers, from each of three independent blocks of at least five plants grown randomly throughout the field trial, were selected, pooled, and manually cut into eighths. Two opposite eighths were taken from each tuber and bulked by replicate. All tuber samples were immediately frozen in liquid nitrogen and freeze-dried. The freeze-dried samples were ground in a Retsch mill fitted with a 1 mm sieve and stored at –20 °C prior to analysis.
Total RNA extraction from freeze-dried potato tubers
Approximately 1.5 g of freeze-dried tuber tissue was extracted with 14 ml of hot (80 °C) extraction buffer (50 mM TRIS-HCl (pH 8.0), 50 mM LiCl, 5 mM EDTA, 0.5% SDS, 50% (v/v) phenol). Sterile distilled water (10 ml) was added and the samples were vortexed for 2 min. The samples were placed on ice and 16 ml of chloroform:isoamyl alcohol (24:1 v:v) was added and vortexed as before. Following centrifugation at 4 °C at 14 000 g for 20 min, the upper aqueous layer was removed to a fresh, sterile 50 ml Sorval tube, containing an equal volume (16 ml) of 4 M LiCl. The samples were shaken and incubated overnight at –80 °C, centrifuged at 4 °C at 14 000 g for 40 min, the supernatant discarded, and the pellet resuspended in 5 ml sterile distilled water. One-tenth volume of 3 M NaOAc (pH 5.2) and 3 vols of 100% ethanol were added and the samples were incubated at –80 °C for at least 1 h.
The precipitated RNA was pelleted by centrifugation at 4 °C at 14 000 g for 40 min, washed with 10 ml of ice-cold 70% (v/v) ethanol, and centrifuged as in the previous step. The ethanol was removed and the RNA pellet allowed to air-dry prior to resuspension in 500 μl sterile distilled water. RNA samples (100 μg) were purified and genomic DNA contamination was removed using Qiagen RNeasy columns and DNase I according to the manufacturer's protocol. RNA samples were quantified using a spectrophotometer and quality tested using an RNA 6000 nano chip on an Agilent 2100 Bioanalyzer (www.chem.agilent.com). RNA samples were aliquoted in 20 μg (1 μg μl−1) batches and stored at –80 °C.
Microarray processing
Experimental design, array information, and complete datasets are available from ArrayExpress (accession number E-TABM-452 SCRI potato LD2 2006, ETABM-451 SCRI potato LD1 2005, http://www.ebi.ac.uk/microarray-as/aer/?#ae-main[0]). Total RNA was labelled by indirect incorporation of fluorescent dyes following cDNA synthesis. Reverse transcription was performed using 20 μg of total RNA in a 45 μl reaction containing 50 ng μl−1 oligo d(T)18, 0.5 mM each dATP, dCTP, dGTP, 0.2 mM dTTP, 0.3 mM aa-dUTP, 10 mM DTT, and 400 U Superscript II (Invitrogen) in 1× reaction buffer. Primers and RNA were initially heated to 70 °C for 10 min followed by cooling on ice, and the entire reaction incubated for 16 h at 42 °C. To denature the remaining RNA, 15 μl of 1 M NaOH and 15 μl of 0.5 M EDTA (pH 8.0) were added and incubated for 10 min at 65 °C. The reaction was neutralized with 15 μl of 1 M HCl. Purification of cDNA was performed using MinElute columns as recommended (Qiagen), substituting phosphate wash buffer (4.75 mM K2HPO4, 0.25 mM KH2PO4, 84% EtOH) for PB and phosphate elution buffer (3.8 mM K2HPO4, 0.2 mM KH2PO4) for EB. Cy-dye esters were added to 10 μl of cDNA in a total volume of 13 μl, containing 150 mM sodium carbonate and 1 μl of the appropriate Cy-dye (GE Healthcare) suspended in DMSO (1/10 supplied aliquot), and incubated for 1 h at room temperature in the dark. To the labelled cDNA 750 mM hydroxylamine hydrochloride was added and incubated for a further 30 min in the dark. Labelled targets for each array were combined and diluted with 24 μl sterile water and 500 μl of PB buffer (Qiagen) prior to MiniElute purification and elution with 10 μl of elution buffer. Labelling efficiency was estimated using spectrophotometry.
Labelled cDNA was made up to a volume of 200 μl with sterile water and denatured by heating at 98 °C for 3 min. The denatured cDNA was mixed with 50 μl control targets, 250 μl of 2× hybridization buffer (Agilent Technologies) and hybridization was carried out for 17 h at 60 °C as recommended. After hybridization, the arrays were washed at room temperature in the dark for 1 min each with wash solution I (6× SSPE, 0.005% N-lauroylsarcosine), followed by wash solution II (0.06× SSPE, 0.005% N-lauroylsarcosine) and dried by centrifugation.
Microarrays were scanned with an ArrayWoRx Auto scanner (Applied Precision) at suitable exposure settings for Cy3 (595 nm) and Cy5 (685 nm) at 9.75 μm resolution, generating separate TIFF images. Exposure levels were adjusted to compensate for slight variations in labelling efficiencies. Data were acquired from images using GenePix Pro software (v6.1, Molecular Devices), and median signal and background intensities were determined for the Cy3 and Cy5 channels for each spot on the microarray.
Microarray data analysis
Due to the nature of the experimental design, background-subtracted intensity values were imported into GeneSpring (v.7.3.1, Agilent Technologies) as separate Cy3 and Cy5 channels for each array. Data were normalized using default settings [(i) intensity values less than 0.01 were set to 0.01; (ii) data from each array was normalized to the 50th percentile of all measurements on the array and; (iii) the signal from each probe was subsequently normalized to the median of its value across the entire dataset)] and unreliable data with consistently low signal (<20) removed. Quality control of the datasets were performed using Principal Components Analysis (PCA) to confirm that there were no outlying replicate samples and that dye labelling had no associated bias. Data were combined from replicate samples and for both Tuberosum and Phureja accessions in a new interpretation. Two-way ANalysis Of VAriance (ANOVA, parametric test) using potato type (Phureja and Tuberosum) and harvest time as parameters was used to identify statistically significant expression profiles at a false discovery rate (P-value) of 0.05. Strict multiple testing correction (Bonferroni) was applied to ensure low false discovery rates. Filtered gene lists were clustered into two groups using the K-means algorithm with default settings (100 iterations, Pearson correlation) in GeneSpring.
Microarray genomic DNA hybridization
Genomic DNA from each of the two Tuberosum and two Phureja accessions was isolated from leaf tissue using the DNeasy Plant Mini Kit (Qiagen) as recommended and independently labelled using the Bioprime DNA Labelling System (Invitrogen). Briefly, 2 μg gDNA in 21 μl was added to 20 μl random primer reaction buffer mix and denatured at 95 °C for 5 min prior to cooling on ice. To this, 5 μl 10× dNTP mix (1.2 mM each of dATP, dGTP, dTTP; 0.6 mM dCTP; 10 mM TRIS pH 8.0; 1 mM EDTA), 3 μl of either Cy3 or Cy5 dCTP (1 mM) and 1 μl Klenow enzyme was added and incubated for 16 h at 37 °C. Labelled samples for each array were combined and unincorporated dyes removed using Qiaquick PCR purification kit (Qiagen) as recommended, eluting twice with 1× 50 μl sterile water.
Hybridizations (array 1: 333–37 Cy3 v Desiree Cy5; array 2: Maris Piper Cy3 v 333–16 Cy5), washing, scanning, and data extraction were performed using POCI microarrays as for cDNA samples. Data were imported into Genespring and default Lowess normalization applied to each array dataset.
Quantitative PCR using the universal probe library
cDNA was synthesized as described above and 25 ng was used as template for real-time PCR using the Universal Probe Library System (https://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp). Reactions were performed in 25 μl containing 1× FastStart TaqMan® Probe Master (supplemented with ROX reference dye). Gene-specific primers and probe were used at a concentration of 0.2 μM and 0.1 μM, respectively. Thermal cycling conditions were: 10 min denaturation at 95 °C followed by 40 cycles of 15 s at 94 °C, 60 s at 60 °C. The reactions were repeated in triplicate with independent cDNAs. Relative expression levels were calculated and the primers validated using the ΔΔCt method (Livak, 1997) using data obtained with the elongation factor-1α specific primers as an internal control. In the case where relative efficiencies of the target and reference amplicons were not equal, an alternative method for calculating relative quantification was used (Pfaffl, 2001). Primer and probe sequences were as follows: StPEST1_fwd, 5′-TGGGTGAGTTCGAGGGATAG-3′, StPEST1_rev, 5′-TGCATTTGCTTTGATGTCCT-3′, universal probe number 156 (5′-GCTGATGG-3′); StPAE_fwd 5′-TTGATATTACTTATCTCCAAAGTGCAG-3′, StPAE_rev, 5′-GGTGGACTTCCATCCAAACA-3′, universal probe number 17 (5′-AAGAGCTG-3′); StEF-1α_fwd, 5′-CTTGACGCTCTTGACCAGATT-3′, StEF-1α rev, 5′-GAAGACGGAGGGGTTTGTCT-3′, universal probe number 117 (5′-AGCCCAAG-3′). StSES1_fwd, 5′-AAGGAGTGAGAAAAATGCTAGGG-3′, StSES1_rev, 5′-TGAGGTCAAGTTTTTGCAATGAT-3′, universal probe number 12 (5′-CTCCTTCC-3′).
Multiple alignment and phylogenetic analysis
Although the Pfam domain alignment database contains entries for both the stses1 domains, this was not used as the last release (22.0) was July 2007 and excluded over 100 sequences that are in Uniprot. The alignment was carried out using the CLUSTALW algorithm (default settings) as available in the CLUSTALX program. CLUSTALX warnings regarding significantly diverged sequences were investigated using the sequence annotation and by carrying out dotplot analysis against several well-aligned sequences. After this, some trimming of long sequences was carried out before realignment.
A 631 sequence alignment was used to produce a phylogenetic tree using a fast approximate Maximum Likelihood tree approach with JTT model plus gamma rate heterogeneity. Due to the large number of sequences, model selection was carried out on a subset of sequences using the BIC criterion. The value of the shape parameter was fixed at an average level of heterogeneity (α=4) which seemed reasonable based on small samples from the large alignment.
E. coli expression of sesquiterpene synthase
The expression vector pET32a (Novagen) containing the gene of interest was transformed into Origami2(DE3)pLysS competent cells (Novagen). Cultures were inoculated from a single colony in 5 ml 2× YT medium containing ampicillin (100 μg ml−1), at 37 °C and shaken at 200 rpm for 8 h. This culture was used to inoculate a 50 ml 2× YT medium containing ampicillin (100 μg ml−1), at 37 °C and shaken at 200 rpm overnight. This culture was used to inoculate a 900 ml 2× YT medium containing ampicillin (100 μg ml−1). The culture was grown for 3 h at 37 °C (OD600=0.5). IPTG (1 mM) was added and the culture was incubated for 4 h at 22 °C. The cells were harvested by centrifugation at 6000 g for 10 min and resuspended in 50 ml extraction buffer (50 mM Na2HPO4, pH 7.0, 10 mM MgCl2, 10% Nonidet NP40, 1 mg ml−1 lysozyme). The cells were lysed by three cycles of freeze/thaw and after the first cycle DNase I was added (2 U ml−1). The debris was removed by centrifugation at 20 000 g for 10 min at 4 °C. The protein of interest was purified using Novagen His-Bind Kit according to the manufacturer's protocol.
Sesquiterpene synthase assay
Purified protein (100 μl) was mixed with 5 μl 1 M DTT, 1 ml sesquiterpene assay buffer (20 mM Na2HPO4, pH 7.2, 5 mM MgCl2, 1 mM DTT, 10% glycerol) and 10 μl farnesyl pyrophosphate and incubated for 2 h at 25 °C. The reaction was extracted twice with 1.5 ml and 0.5 ml pentane then eluted through a short column of silica gel in a Pasteur pipette. The pentane extract was washed with 0.5 ml ether. The pentane/ether fraction was analysed by GC-MS on a Phenomenex ZB5 (DB-5 equivalent) column. The temperature program was 50 °C hold for 2 min then an additional 3 °C min−1 to 246 °C which was held for 4.67 min. The identities of any sesquiterpene and diterpene component, present in the samples, were confirmed on the basis of its retention index on DB-5 and its mass spectrum, using the reference library (Adams, 2001). Relative retention indices were calculated according to the method for essential oil fingerprinting recommended by the Analytical Standards Committee of the Royal Society of Chemistry (1997).
α-Copaene determination
A single tuber per replicate was transected and two ‘opposite eights’ were taken and manually diced into small segments. A randomized subsample of the segments (4–5 g) was transferred to a weighed headspace vial (20 ml) (Supelco, UK) which was sealed and reweighed prior to cooking. The vial and contents were suspended in a boiling water bath for 1 h with the bottom half of the vial containing the tuber material below the water level. Subsequently, the vial was loaded on a CombiPal autosampler (CTC Analytics, Switzerland) for automated sampling using solid phase micro extraction (SPME) and analysis by gas chromatography-mass spectrometry (GC-MS). For entrainment of headspace volatiles a carboxen/polydimethylsiloxane (PDMS) SPME fibre (23-gauge, 85 μm film; Supelco, UK) was exposed for 20 min in the headspace of the sample vial which was located in a heated incubator (50 °C). Volatiles were analysed using a Finnigan Tempus time of flight (TOF) GC-MS (Thermo Fisher, UK) fitted with a DB1701 fused silica capillary column (30 m long×0.25 mm i.d, 0.25 μm film thickness; Agilent technologies, UK). Volatiles were desorbed from the SPME fibre for 2 min at 280 °C within a programmable temperature vaporizing (PTV) injector in splitless mode. Helium was used as carrier gas at a flow rate of 1.5 ml min−1. After an initial hold for 2.0 min at 40 °C, the GC oven was programmed to increase at 10 °C min−1 up to 240 °C with a further 20 min hold at that temperature. The GC-MS interface was 250 °C and the MS was used in EI mode at 70 eV over a mass range of 25–400 amu with a source temperature of 200 °C. Data was acquired at 3 spectra s−1. The MS was tuned to give a uniform sensitivity corresponding to a signal level of approximately 350 counts for the fragment ion at m/z 501 in the mass spectrum of the tuning reference compound perfluoro-tertiary-butylamine (PFTBA) prior to measurement of α-copaene in the tuber samples. The Xcalibur software package (V. 1.4) (Thermo Fisher, UK) was used for data acquisition and analysis. Identification of α-copaene was based on comparison of mass spectral and chromatographic characteristics with those of authentic samples (Fluka, Switzerland) and with entries in a commercially available MS database (pal600K, Palisade Corporation, USA). To quantify α-copaene the selected ion chromatogram (SIC) for the major fragment ion at m/z 161 was integrated. The resulting peak areas were used directly for comparison of relative α-copaene concentrations and are expressed as area counts g−1 fresh weight of tuber and as area counts tuber−1. Values are means for two replicate samples.
Tuber cooking time
Approximately 2 cm3 of peeled tuber were cooked by steaming in a domestic steamer. The time taken for the tubers to reach an acceptable eating texture was empirically determined by two independent assessors. Tuber samples from three independent plants at stage H3 were analysed.
Results
Microarray analysis of gene expression
The strategy for this experiment was to compare tuber gene expression profiles in two Phureja clones with those from two commonly grown commercial Tuberosum cultivars, Desiree and Maris Piper. One of the chosen Phureja clones, DB333-16, contained the highest levels of α-copaene in an earlier study of cooked tuber volatiles (W Griffiths et al., unpublished data; Table 1), and shares a common parent with the other Phureja clone, Mayan Gold. Tubers were sampled at four harvest dates in 2005, giving a wide range of mean tuber sizes (detailed in the Materials and methods), so that expression profiles throughout tuber development could be compared. Following harvesting, tubers were rapidly processed by cutting into opposite eighths, an approach demonstrated to reduce the effects of any metabolite gradients (Griffiths and Dale, 2001). Two biological replicates, consisting of tubers harvested from separate plants, were analysed for each sample for the 2005 trial. A second microarray experiment was conducted in 2006, using the same site and the same germplasm, although analysis was focused on two harvest dates to give tubers equivalent to those at harvest 1 and harvest 4 from the 2005 season. In the 2006 trial, three biological replicates were analysed for each sample.
Table 1.
Relative concentrations of α-copaene and cooking time of tubers from Solanum tuberosum group Phureja cultivar Mayan Gold and line 333-16 and Solanum tuberosum group Tuberosum cultivars Maris Piper and Desiree
| Cultivar/line | Mean tuber weight | α-Copaene contenta | Cooking time |
| (g) | (Area counts g−1 FW tuber−1) | (min) | |
| Mayan Gold | 142.7±39.3 | (86.00±21.21)×103 | 9–10 |
| 333-16 | 72.0±2.1 | (245.70±39.67)×103 | 9–10 |
| Maris Piper | 167.6±63.4 | (1.19±0.19)×102 | 19–20 |
| Desiree | 117.2±6.6 | (1.17±0.06)×102 | 19–20 |
Expressed as area counts from integrated selected ion chromatogram (SIC) for the fragment ion at m/z 161. Results are means of two replicates.
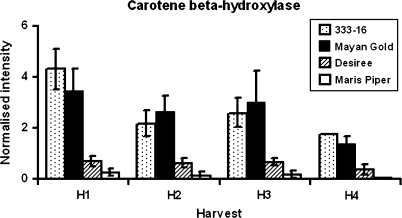
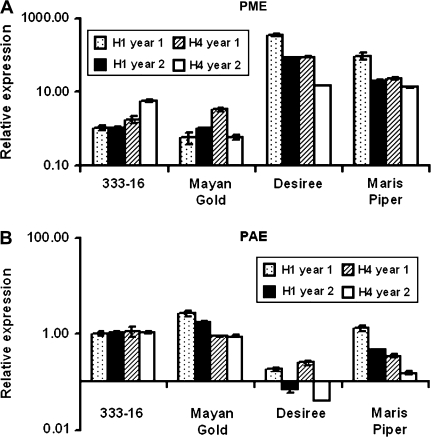
Microarray data were analysed using standard statistical approaches in GeneSpring software (Materials and methods). Only gene expression profiles that were consistently different in both Phureja clones compared with both Tuberosum clones were considered likely to underpin the traits of interest. In addition, the Phureja flavour, texture, and colour traits are consistent across seasons (De Maine et al., 2000; Griffiths et al., 2007) and so gene expression profiles from two growing seasons were compared to provide further focus on the traits of interest, and to reduce the background variation in expression levels unrelated to these traits. To illustrate this point, using ANOVA (see Materials and methods), 2075 genes were shown to be significantly up- or down-regulated in both Phureja lines compared with both Tuberosum lines in 2005. A lower number of genes (1386) fulfilled this criterion in the 2006 trial (analysed independently from the previous season's data), but this could be attributed to fewer experimental factors (time points) being applied and the higher level of replication. However, by comparing these gene lists, the number of common members was further reduced to 864. This subset of genes (consistently up- or down-regulated in both Phureja cultivars compared with both Tuberosum cultivars in the two growing seasons) was of particular interest. Using K-means clustering (see Materials and methods) this group consists of 309 genes that are significantly and consistently up-regulated (see Supplementary Table S1a at JXB online) and 555 genes down-regulated (see Supplementary Table S1b at JXB online) in Phureja compared with Tuberosum. The entire dataset has been deposited into ArrayExpress (accession numbers E-TABM-452 SCRI potato LD2 2006, ETABM-451 SCRI potato LD1 2005, http://www.ebi.ac.uk/microarray-as/aer/?#ae-main[0]). Approximately 46% of the genes in these lists can be identified from their annotation and amongst these are several obvious candidates that may help to explain the Phureja/Tuberosum trait differences. Examples include carotene β-hydroxylase (MICRO.7880.C2_1119), involved in carotenoid biosynthesis that is up-regulated in the Phureja tubers (Fig. 1); several genes involved in cell wall metabolism were also differentially expressed – a pectin acetylesterase (MICRO.4427.C3_1465) and xyloglucan endotransglycosylase (MICRO.4152.C1_825) were generally expressed at higher levels in the Phureja tubers, whereas a pectin methylesterase gene (MICRO.4403.C1_728) was expressed at lower levels (Fig. 2). In general, expression patterns for this set of genes were consistent in the two years of the trial although the high level of PAE expression in H1, year 1 for Maris Piper was an exception (Fig. 2).
Fig. 1.
Carotene β-hydroxylase gene expression profile in Phureja (333-16, Mayan Gold) and Tuberosum (Desiree, Maris Piper) cultivars during tuberization (harvest stages H1 to H4: see Materials and methods for details) as determined by microarray analysis. Values are the means of two biological replicates (apart from the H4 sample where only 1 reliable datapoint was obtained) and error bars represent standard error of the mean.
Fig. 2.
(A) Expression profiles of pectin methylesterase (PME) and (B) pectin acetylesterase (PAE) in Phureja (333-16, Mayan Gold) and Tuberosum (Desiree, Maris Piper) cultivars during tuberization (harvest stages H1 and H4: see Materials and methods for details) as determined by quantitative RT-PCR. Year 1 is 2005, year 2 is 2006. Values are the means of three replicates and error bars represent the standard error of the mean. For each gene, values are normalized to the expression level in 333-16, H1 year 1.
Isolation and characterization of an α-copaene synthase gene
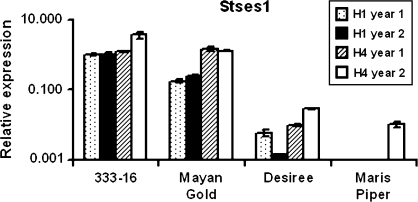
Only two probes on the array, annotated as representing terpene synthase genes, hybridized more strongly to Phureja tuber cDNA than that from Tuberosum. The ESTs corresponding to the probes for these sequences are very similar showing greater than 92% identity over a 740 bp overlap. Many of the nucleotide differences between the two ESTs correspond to frameshifts in the translated sequence and it is likely that both cDNAs represent the same gene. The 60-mer probe for the MICRO.8755.C4_692 sequence contained only two mismatches with the corresponding MICRO.8755.C3_977 sequence. The probe sequence for the MICRO.8755.C3_977 sequence is towards the 3′ of the cDNA for which the corresponding sequence of the MICRO.8755.C4_692 EST is not available. Translation of the EST sequences also demonstrated that the 3′ coding sequence was not present in either of these EST sequences. In order to obtain a full-length clone, the EST sequences were compared to the DFCI potato gene index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=potato) sequences using the BLASTN algorithm (Altschul et al., 1997) and the best hit gave a tentative consensus sequence (TC141591) with 99% identity to MICRO.8755.C3_977 and 92% identity to MICRO.8755.C4_692. Clones of two of the ESTs that constituted TC141591 were obtained from the Arizona Institute of Genomics (http://genome.arizona.edu) and fully sequenced (GenBank numbers BQ121961 and BG590598). The full coding sequence was then amplified by PCR from cDNA from Phureja 333-16 tubers. The translated coding sequence of the Phureja cDNA (stses1) was greater than 95% identical to that derived from the microarray EST sequences, with obvious sequence errors in the EST sequences where frameshifts occurred. RT-PCR analysis confirmed the microarray expression result in tuber samples and demonstrated that expression of this gene in Phureja was higher than in Tuberosum (Fig. 3).
Fig. 3.
Sesquiterpene synthase (stses1) gene expression profile in Phureja (333-16, Mayan Gold) and Tuberosum (Desiree, Maris Piper) cultivars during tuberization (harvest stages H1 and H4: see Materials and methods for details) as determined by quantitative RT-PCR analysis. Year 1 is 2005, year 2 is 2006. Values are the means of three replicates and error bars represent the standard error of the mean.
Multiple alignment and phylogenetic analysis
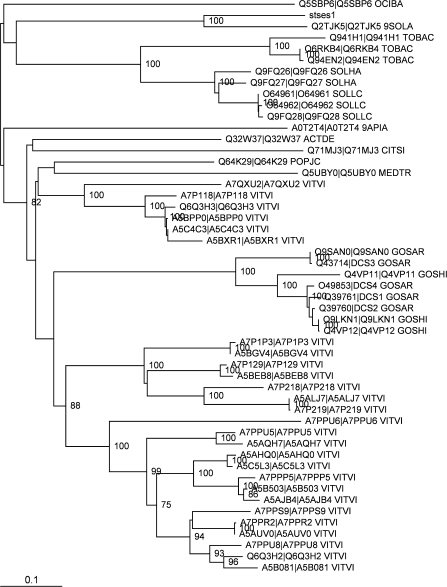
Phylogenetic analysis of the terpene synthase gene (termed stses1) was carried out to determine which class of terpene synthase the gene was likely to encode (Fig. 4). The gene clustered with one other gene, annotated as a sesquiterpene synthase gene (Uniprot entry Q2TJK5) from leaf tissue of Fabiana imbricata, an evergreen shrub common in Chile and Argentina and used for traditional medicine (http://www.ncbi.nlm.nih.gov/pubmed/16372369). There is little information about the Q2TJK5 entry (submitted 1 Jan 2006) except for evidence regarding protein presence at the transcript level (the associated EMBL accession AY860847 is a mRNA entry). This cluster was significantly significant (bootstrap support 100%) and joins the other clusters deep in the tree, (at a distance of 0.61 amino acid replacements per position), suggesting that stses1 and Q2TJK5 form a distinct, uncharacterized class of sesquiterpene synthase. The two sequences are not highly similar and have diverged by 0.33 amino acid replacements per position (73% identity) whereas sequences outside the cluster only have 52% identity or less. With only two sequences in a cluster that is diverged from the other cluster, it is difficult to describe the particular sequence characteristics of this new cluster.
Fig. 4.
Phylogenetic tree showing the relationships among stses1 and the top 49 hits from the BLAST search. Bootstrap support values greater than 70% are shown. The scale bar represents a genetic distance of 0.1 amino acid replacements per position.
Phenotypic comparison of Phureja and Tuberosum tubers
Relative concentrations of α-copaene g−1 tuber fresh weight were 2000-fold and 700-fold greater, respectively, in Phureja line 333-16 and cultivar Mayan Gold than in the Tuberosum cultivars Maris Piper and Desiree (Table 1). As an indication of textural differences between the tuber types, the time taken to steam tuber samples to an acceptable eating texture was empirically determined. The Phureja tubers reached an acceptable texture in 9–10 min whereas, for the Tuberosum cultivars, cooking time was 19–20 min (Table 1).
Validation of gene expression patterns
Even though Phureja and Tuberosum lines are believed to be very similar genetically (Spooner et al., 2005), it could not be dismissed that at least part of the differential hybridization could be due to nucleotide polymorphism within the probe binding sites. The long probe length of Agilent microarrays (60-mer) compared to other platforms, means they are less sensitive to polymorphism, with effects of SNPs (Single Nucleotide Polymorphisms) only apparent at very specific base positions and at lower intensity levels (Hughes et al., 2001). In order to test whether the expected low levels of polymorphism would impact on hybridization levels, genomic DNA from each of the four accessions used in this study were hybridized to POCI arrays. When data from each potato type were combined and visualized on a scatter plot (see Supplementary Fig. S2 at JXB online), extremely high levels of correlation (R2=0.981) were observed indicating that Agilent arrays are suitable for comparisons between genotypes used in this study. Ratio analysis of gDNA hybridization for the four genes described in detail (carotene β-hydroxylase, PAE, PME, and stses1) indicate no significant variation from 1 (Tuberosum:Phureja range 0.9–1.0).
As a further control, independent qRT-PCR analysis was carried out for three of the genes of interest. For each, similar patterns of expression were observed whether using the microarray or qRT-PCR analysis. As the qRT-PCR assay was designed to a region outwith the microarray 60-mer probe sequence, the similarity of patterns would suggest that expression differences were unlikely to be due to polymorphisms affecting one or other of the assays.
Identification of stses1 as an α-copaene synthase
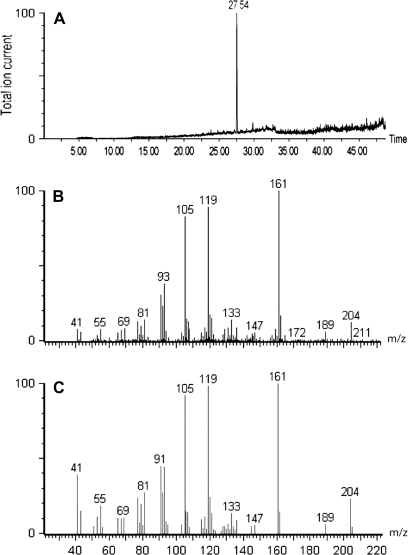
In Arabidopsis, 40 terpenoid synthase genes have been identified by genome sequence analysis (Aubourg et al., 2002). The function of many terpenoid synthases has been deduced by expression in E. coli, and analysis by GC-MS of the reaction products when fed with the appropriate prenyl diphosphate precursor (Facchini and Chappell, 1992). Although it seemed likely that stses1 did encode protein with sesquiterpene synthase activity, to determine the nature of the sesquiterpene products produced, functional analysis was required. Therefore, the stses1 cDNA was expressed as a thioredoxin fusion protein in E. coli. Previous work has demonstrated that for other, closely related, sesquiterpene synthases, the thioredoxin moiety at the N-terminus has little effect on either the reaction kinetics or spectrum of reaction products obtained (Prosser et al., 2004). The stses1 open reading frame was cloned into the vector pET32b and transformed into the expression host E. coli Origami2(DE3)pLysS. Soluble protein was extracted from cultures induced for 3 h with IPTG and partially purified by Ni-chelate chromatography. The products from assays using the affinity-purified protein were assayed by GC-MS. There was only one peak in the total ion GC-MS trace (Fig. 5A) and mass spectrum and retention time confirmed that this was α-copaene (Fig. 5B, C).
Fig. 5.
GC-MS analysis of the products produced by the putative sesquiterpene synthase. (A) Total ion current trace. (B) Mass spectra of peak at retention time 27.54 min. (C) Adam's library spectrum for α-copaene.
Developmental gene expression
The design of the microarray experiment and the subsequent statistical analysis (two-way ANOVA) used in this study also allows identification of developmentally regulated gene expression. Over 900 genes (see Supplementary Table S2 at JXB online) were differentially regulated in a temporal manner across both seasons and these include many genes previously reported to be important factors in tuber development (Kloosterman et al., 2005) representing a rich resource for identification of previously uncharacterized candidates, although this is outwith the scope of this current report.
Other genes of interest
A number of other genes implicated in additional trait differences between Phureja and Tuberosum can be identified solely on the basis of the annotation information available (Table 2; see Supplementary Fig. S1 at JXB online). These include additional genes potentially involved in flavour and texture as well as genes related to dormancy and tuberization, hormone metabolism, and carotenoid biosynthesis.
Table 2.
Genes potentially involved in tuber quality traits identified by microarray analysis as being differentially expressed between Phureja and Tuberosum
| Gene description | POCI array IDa | Top hit accession number | Process | Higher in |
| Branched chain amino acid aminotransferase | MICRO.2772.C2_1399 | AAF07191 | Flavour | Phureja |
| Sesquiterpene synthase | MICRO.8755.C3_977 | AAX40666 | Flavour | Phureja |
| Glutamate ammonia ligase | MICRO.3959.C1_623 | NP_190886 | Flavour (glutamate biosynthesis) | Phureja |
| Glutamine synthetase I | STMDI41TV_515 | CAB63844 | Flavour (glutamate biosynthesis) | Phureja |
| Ribonuclease | MICRO.5716.C1_596 | AAD50436 | Flavour (nucleotide formation?) | Phureja |
| Carotene β-hydroxylase | MICRO.7880.C2_1119 | ABI23730 | Flesh colour | Phureja |
| Pectin acetylesterase | MICRO.4427.C3_1465 | CAA67728 | Texture (cell wall biosynthesis) | Phureja |
| Xyloglucan endotransglycosylase | MICRO.4152.C1_825 | AAG00902 | Texture (cell wall biosynthesis) | Phureja |
| NAD-dependent epimerase | bf_arrayxxx_0046b02.t7m.scf_638 | ABE78360 | Texture (cell wall biosynthesis) | Phureja |
| Nucleotide-rhamnose synthase | MICRO.444.C1_634 | NP_564806 | Texture (cell wall biosynthesis) | Phureja |
| Chitinase | MICRO.15095.C1_874 | CAA54374 | Tuber life cycle | Phureja |
| FRIGIDA | MICRO.1851.C1_1 | CAM06912 | Tuber life cycle | Phureja |
| β-amylase | MICRO.13823.C1_1872 | AAK84008 | Starch structure | Phureja |
| GABA transaminase subunit 3 | MICRO.15425.C2_1257 | AAO92257 | Flavour (glutamate biosynthesis) | Tuberosum |
| Cystathione γ synthase I | MICRO.1118.C2_1798 | AAF74981 | Flavour (methionine biosynthesis) | Tuberosum |
| Pectin methylesterase | MICRO.4403.C1_728 | AAF23891 | Texture (cell wall biosynthesis) | Tuberosum |
| ent-Kaurene oxidase | MICRO.10720.C2_566 | AAO85520 | Tuber life cycle | Tuberosum |
| Dimethylallyl transferase | MICRO.2151.C3_724 | CAA59170 | Tuber life cycle | Tuberosum |
POCI, potato oligo chip initiative; ID, identification.
Discussion
Previous reports have demonstrated that S. tuberosum group Phureja is differentiated from S. tuberosum group Tuberosum on the basis of a number of important tuber quality traits such as flavour, texture, colour, and reduced tuber dormancy (De Maine et al., 1993, 1998; Dobson et al., 2004; Morris et al., 2004, 2007; Ghislain et al., 2006). This study provides a unique and comprehensive examination of the differences in gene expression that exists between these groups. The POCI 44 000 feature microarray is the best microarray platform currently available to analyse global gene expression in potato (described in Kloosterman et al., 2008) representing 42 034 unigene sequences, thereby enabling a much more complete analysis of gene expression than has hitherto been achievable. Most potato microarray experiments performed to date have used the widely accessible spotted cDNA array produced by The Institute for Genomic Research (TIGR) which contains around 12 000 cDNA clones (http://www.tigr.org/tdb/potato/microarray_desc.shtml). It has been estimated that 35 000 genes are expressed in tomato (Van der Hoeven et al., 2002) and a similar number are probably expressed in potato. Therefore, the TIGR array probably only contains c. 35% of the transcriptome, which is a serious limitation for global expression studies. In addition, with spotted cDNA arrays it is inherently difficult to achieve a high degree of discrimination between similar sequences such as members of multigene families. Approximately 50% of the sequences on the POCI microarray were not represented on the TIGR array (Kloosterman et al., 2008).
Previous phytochemical analysis has identified clear differences in the volatile profile produced by cooked Phureja compared with Tuberosum (Winfield et al., 2005; Shepherd et al., 2007) and the aim of the current study is to identify candidate genes for such differences. The use of the POCI microarray to assist cloning of the α-copaene synthase gene highlights the utility of this approach. The sesquiterpene synthase identified appears to produce α-copaene as its sole reaction product from farnesyl diphosphate. The much higher expression level of this particular sesquiterpene synthase gene in Phureja tubers corresponds with the much higher level of α-copaene in the Phureja boiled tuber volatiles. Thus it would appear that this gene underpins the difference. As phylogenetic analysis indicates that this gene is somewhat separated from other sesquiterpene synthases, it will be of interest to see if this gene clusters with other α-copaene synthases that may be important for flavour in other vegetables. A recent report establishes that α-copaene has high odour impact in lettuce and carrots (Nielsen and Poll, 2007). The relative importance of volatile compounds compared with non-volatile taste compounds associated with the food matrix, is not clear for potato. Recently, levels of umami compounds have been shown to be elevated in Phureja tubers and that this increase correlates positively with sensory evaluation data (Morris et al., 2007). However, the large differences in Phureja and Tuberosum volatiles may also contribute to the differences in sensory perception. With the identification of the potato α-copaene synthase gene, it is now possible to manipulate the tuber volatile profile through transgenic modification providing a means of testing the importance of this class of volatile to sensory evaluation.
Esterified branched-chain carboxylic acids such as 2-methylpropionic acid methyl ester and 2-methylbutanoic acid methyl ester are other volatiles produced in much higher levels from cooked Phureja tubers than from Tuberosum (Shepherd et al., 2007). These esters are derived from branched chain amino acids such as valine, leucine, and isoleucine in a reaction pathway that is thought to involve branched chain amino acid aminotransferase (BCAAT; Beck, 2005). It is reasonable to speculate that this elevated level is due to the higher expression level of the BCAAT in Phureja than in Tuberosum tubers (Table 1) and again suggests that a transgenic route in which the gene is over-expressed or silenced may resolve the issue of its contribution to overall volatile level and flavour. Another amino acid-derived volatile that has been implicated in baked potato flavour is methional and its amount has previously been increased in potato tubers by elevating the amount of its precursor methionine by metabolic engineering (Di et al., 2003). Interestingly, cystathione γ-synthase I, the gene over-expressed by Di et al. to increase tuber methional content, was expressed at a higher level in Tuberosum. However, methional level was not significantly different between boiled Phureja and Tuberosum tubers (data not shown).
Glutamate is likely to be a determinant of flavour in potato tubers due to its contribution to the umami taste (Morris et al., 2007). Genes encoding glutamate ammonia ligase and glutamine synthetase I (enzymes involved in glutamate biosynthesis) were expressed at higher levels in Phureja cultivars. Conversely, another gene involved in glutamate biosynthesis, γ-aminobutyric acid transaminase, was expressed at a higher level in Tuberosum cultivars. Further investigation of gene expression profiles of these and other genes involved in tuber glutamate biosynthesis may reveal those that account for higher levels of glutamate in Phureja tubers. In addition, it is still not clear to what extent the import of amino acids from the leaves has on the tuber amino acid content (Fischer et al., 1998).
Other non-volatile compounds that are important in potato flavour include the 5′-nucleotides adenosine-5′-monophosphate (5′-AMP), inosine-5′-monophosphate (5′-IMP), and guanosine-5′-monophosphate (5′-GMP). The levels of 5′-AMP and 5′-GMP are consistently and significantly higher in Phureja tubers compared with Tuberosum (Morris et al., 2007). Previously it has been suggested that 5′-nucleotides accumulate due to the action of nucleases during cooking processes, particularly due to RNA degradation (Buri and Solms, 1971). Despite the proposed involvement of RNases, no gross differences in RNase, or phosphohydrolytic enzyme, activities could be detected in extracts from Tuberosum and Phureja (Morris et al., 2007). Our current study has identified a ribonuclease (MICRO.5716.C1_596; Table 2) that is more highly expressed in Phureja than Tuberosum that may yet provide a mechanism for 5′-ribonucleotide formation in potato tubers on cooking.
Based on differential expression patterns between Phureja and Tuberosum tubers several genes involved in cell wall synthesis are candidates for those genes that account for tuber texture differences. The most clear-cut examples include genes encoding pectin acetylesterase (MICRO.4427.C3_1465), xyloglucan endotransglycosylase (MICRO.4152.C1_825), and pectin methylesterase (MICRO.4403.C1_728). Orthologous genes have been shown to impact on the texture of fruit from many species (reviewed in Fischer and Bennet, 1991), however a role in potato tuber texture has not previously been demonstrated. Indeed in the study of Pilling et al. (2000), it was not possible to detect significant transcription of PEST1 (equivalent to MICRO.4403.C1_728) in the tuber. This is clearly different from the results presented here, possibly due to the more sensitive microarray and PCR-based detection methods used in this study. As pectin is a major component of the cell wall and the middle lamella, its structure is likely to be an important factor in texture in potato tubers as well as other plant tissues (Fischer and Bennet, 1991). The pectin molecule consists of a backbone of α−(1–4)-D-galacturonic acid and contains some regions with alternating L-rhamnose and D-galacturonic acid (Voragen et al., 2001). Twenty to eighty per cent of the rhamnose units in the backbone are branched, with side chains composed by galactose, arabinose, and small amounts of fucose and mannose. Some of the galacturonic acids in the pectin are methyl esterified or acetylated. Non-methylated D-galacturonic acid sequences are sites for cross-linking polymeric chains through the site-specific interaction with Ca2+ (Pilnik and Voragen, 1991). The content of unesterified uronic acid residues can be increased by the action of pectin methylesterase. The binding of pectin chains has a co-operative nature and an increase in Ca2+ binding efficiency contributes to higher tissue firmness (Andersson et al., 1994). The much higher (c. 10-fold) expression level of the PEST1 gene in Tuberosum tubers may underpin a higher degree of Ca2+ cross-linking and contribute to the firmer texture compared with Phureja tubers.
Other expression differences that may be implicated in tuber texture include NAD-dependent epimerase (bf_arrayxxx_0046b02.t7m.scf_638) and nucleotide-rhamnose synthase (MICRO.444.C1_634), potentially involved in cell wall biosynthesis. Textural differences may also arise from differences in starch structure (reviewed in Taylor et al., 2007) and so the higher expression level of a β-amylase gene (MICRO.13823.C1_1872) may also be significant.
As well as flavour and texture, Phureja tubers have a much shorter dormancy period than Tuberosum (De Maine et al., 2000) and so some gene expression differences may reflect the changes in tuber life-cycle. For example, for both harvest years, although the expression of PME is substantially lower in Phureja than in Tuberosum tubers, there is a consistent difference in expression pattern between H1 and H4 (Fig. 2). In Tuberosum, higher expression is seen at H1 compared with H4, whereas for Phureja the opposite was observed. It is intriguing that a FRIGIDA-like gene (MICRO.1851.C1_1) is up-regulated in Phureja tubers (see Supplementary Fig. S1 and Supplementary Table S1A at JXB online). FRIGIDA is a gene that controls flowering time in Arabidopsis (Johansen et al., 2000). However, it is becoming well known that there are commonalities between the flowering time and tuberization time response (Rodriguez-Falcon et al., 2006) and so the up-regulation of FRIGIDA in Phureja may also be implicated in tuber life-cycle differences. Gibberellins and cytokinins are also implicated in the control of the tuber life-cycle (Fernie and Willmitzer, 2001) and so differences in the expression levels of gibberellin and cytokinin biosynthetic genes are of potential importance. In this study the expression level of ent-kaurene oxidase (MICRO.10720.C2_566) is lower in Phureja tubers as is the expression of dimethylallyl transferase (MICRO.2151.C3_724), an enzyme involved in cytokinin metabolism (Kakimoto, 2001; see Supplementary Fig. 1 and Supplementary Table S1B at JXB online).
The higher level of tuber carotenoid content in many Phureja accessions is well documented (Morris et al., 2004). It has recently been demonstrated that an allele of the carotene β-hydroxylase (bch) gene co-segregates with yellow tuber flesh in the progeny of a white-flesh×yellow-flesh cross, making bch an excellent candidate for the classical Y locus required for yellow tuber flesh (Brown et al., 2006). Interestingly, the only known carotenoid biosynthetic gene that was significantly up-regulated was that encoding carotene β-hydroxylase, consistent with its identification as the Y locus. A mechanistic understanding of how enhanced carotene β-hydroxylase expression could lead to enhanced tuber carotenoid content is currently lacking, however, and it is clear that other genes are also important in the control tuber carotenoid content (Brown et al., 2006; Giuliano et al., 2008).
In conclusion, the comparison of gene expression profiles of potato germplasm differentiated in quality traits presented here has resulted in the identification of candidate genes involved in a wide range of processes that warrant further investigation. In addition to using transgenic models, it will also be of interest to use a genetic approach to identify key regulatory genes involved in potato tuber quality. Mapping populations (including a Phureja×Tuberosum cross) that may help in the identification of quantitative trait loci (QTL) associated with potato flavour, texture, and dormancy have been generated (G Bryan et al., unpublished data). Co-localization of trait QTL (flavour, texture, colour, and dormancy) with the map locations of candidate genes will help to identify the key genes that contribute to the trait. This approach will become more feasible with the progression of the potato genome sequencing project by the Genomics Sequencing Consortium (http://www.potatogenome.net) established with the objective of elucidating the complete sequence of the potato genome by 2009 and the development of high throughput SNP-based mapping technologies. Mapping genes of unknown function (54%) of the significant gene expression differences may enable the identification of those genes worthy of further investigation.
Supplementary data
Further supplementary data can be found at JXB online.
Fig. S1. Gene expression profiles of other genes possibly affecting tuber quality traits in Phureja (333-16, Mayan Gold) and Tuberosum (Desiree, Maris Piper) cultivars during tuber development (harvest stages H1 to H4; see Materials and methods for details) as determined by microarray analysis.
Fig. S2. A scatter plot of mean probe intensities of gDNA hybridizations to POCI microarray.
Table S1. Genes differentially expressed between tubers of group Phureja and Tuberosum based on ANOVA analysis: (a) probes with significantly higher levels of expression in Phureja; (b) probes with significantly higher levels of expression in Tuberosum; (c) a summary of the microarray data for the sets of genes in (a) and (b) from the 2006 trial.
Table S2. Genes differentially expressed in tubers between harvest time points based on ANOVA analysis.
Supplementary Material
Acknowledgments
This work was funded by the Scottish Government Rural and Environmental Research and Analysis Directorate and EU-SOL project number PL 016214. We are grateful to Nathan Hawkins, Rothamsted Research, for carrying out the GC-MS analysis, to Heather Ross, SCRI, for determining tuber cooking times and to Clare Booth, SCRI, for carrying out the gDNA hybridizations.
References
- Adams RP. Identification of essential oils components by gas chromatography/quadrupole mass spectroscopy. Illinois, USA: Allured Publishing Corporation; 2001. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Analytical Standards Committee of the Royal Society of Chemistry. Application of gas–liquid chromatography to the analysis of essential oils, XVII. Fingerprinting of essential oils by temperature-programmed gas–liquid chromatography using capillary columns with non-polar stationary phases. Analyst. 1997;122:1167–1174. [PubMed] [Google Scholar]
- Andersson A, Gekas V, Lind I, Oliveira F. Effect of preheating on potato texture. Critical Reviews in Food Science and Nutrition. 1994;34:229–251. doi: 10.1080/10408399409527662. [DOI] [PubMed] [Google Scholar]
- Aubourg S, Lecharny A, Bohlmann J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Molecular Genetics and Genomics. 2002;267:730–745. doi: 10.1007/s00438-002-0709-y. [DOI] [PubMed] [Google Scholar]
- Beck HC. Branched-chain fatty acid biosynthesis in a branched-chain amino acid aminotransferase mutant of Staphylococcus carnosus. FEMS Microbiology Letters. 2005;243:37–44. doi: 10.1016/j.femsle.2004.11.041. [DOI] [PubMed] [Google Scholar]
- Bradshaw JE, Ramsay G. Utilization of the Commonwealth potato collection in potato breeding. Euphytica. 2005;146:9–19. [Google Scholar]
- Brown CR, Kim TS, Ganga Z, Haynes K, De Jong D, Jahn M, Paran I, De Jong W. Segregation of total carotenoid in high level potato germplasm and its relationship to beta-carotene hydroxylase polymorphism. American Journal of Potato Research. 2006;83:365–372. [Google Scholar]
- Buri R, Solms J. Ribonucleic acid: a flavor precursor in potatoes. Naturwissenschaften. 1971;58:56. doi: 10.1007/BF00620813. [DOI] [PubMed] [Google Scholar]
- Chen F, Tholl D, D'Auria JC, Farooq A, Pichersky E, Gershenzon J. Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. The Plant Cell. 2003;15:481–494. doi: 10.1105/tpc.007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maine M, Carrol CP, Torrance CJW. Culinary quality of tubers derived from Solanum phureja and Solanum tuberosum×Solanum phureja hybrids. Journal of Agricultural Science. 1993;120:213–217. [Google Scholar]
- De Maine M, Lees A, Bradshaw J. Soft-rot resistance combined with other tuber characters in long day-adapted Solanum phureja. Potato Research. 1998;41:69–82. [Google Scholar]
- De Maine M, Lees AK, Muir DD, Bradshaw JE, Mackay GR. Long-day-adapted Phureja as a resource for potato breeding and genetical research. In: Khurana SMP, Shekhawat GS, Singh BP, Pandey SK, editors. Potato: global research and development. Vol. 1. Shimla, India: Indian Potato Association; 2000. pp. 134–137. [Google Scholar]
- Di R, Kim J, Martin MN, Leustek T, Jhoo J, Ho CT, Tumer NE. Enhancement of the primary flavor compound methional in potato by increasing the level of soluble methionine. Journal of Agricultural and Food Chemistry. 2003;51:5695–5702. doi: 10.1021/jf030148c. [DOI] [PubMed] [Google Scholar]
- Dobson G, Griffiths DW, Davies HV, McNicol JW. Comparison of fatty acid and polar lipid contents of tubers from two potato species, Solanum tuberosum and Solanum phureja. Journal of Agricultural and Food Chemistry. 2004;52:6306–6314. doi: 10.1021/jf049692r. [DOI] [PubMed] [Google Scholar]
- Duckham SC, Dodson AT, Bakker J, Ames JM. Volatile components of baked potato flesh. A comparison of eleven potato cultivars. Nahrung/Food. 2001;45:317–323. doi: 10.1002/1521-3803(20011001)45:5<317::AID-FOOD317>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Duckham SC, Dodson AT, Bakker J, Ames JM. Effect of cultivar and storage time on the volatile flavor components of baked potato. Journal of Agricultural and Food Chemistry. 2002;50:5640–5648. doi: 10.1021/jf011326+. [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Chappell J. Gene family for an elicitor-induced sesquiterpene cyclase in tobacco. Proceedings of the National Academy of Sciences, USA. 1992;89:11088–11092. doi: 10.1073/pnas.89.22.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Willmitzer L. Molecular and biochemical triggers of potato tuber development. Plant Physiology. 2001;127:1459–1465. [PMC free article] [PubMed] [Google Scholar]
- Fischer RL, Bennett AB. Role of cell wall hydrolases in fruit ripening. Annual Review of Plant Physiology and Plant Molecular Biology. 1991;42:675–703. [Google Scholar]
- Fischer WN, Andre B, Rentsch D, Krolkiewicz S, Tegeder M, Breitkreuz K, Frommer WB. Amino acid transport in plants. Trends in Plant Science. 1998;3:188–195. [Google Scholar]
- Ghislain M, Andrade D, Rodriguez F, Hijmans R, Spooner D. Genetic analysis of the cultivated potato Solanum tuberosum L. Phureja group using RAPDs and nuclear SSRs. Theoretical and Applied Genetics. 2006;113:1515–1527. doi: 10.1007/s00122-006-0399-7. [DOI] [PubMed] [Google Scholar]
- Giuliano G, Tavazza R, Diretto G, Beyer P, Taylor M. Metabolic engineering of carotenoid biosynthesis in higher plants. Trends in Biotechnology. 2008;26:139–145. doi: 10.1016/j.tibtech.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Glasby JS. Encyclopedia of terpenoids. Chichester: Wiley; 1982. [Google Scholar]
- Griffiths DW, Dale MFB. Effect of light exposure on the glycoalkaloid content of Solanum phureja tubers. Journal of Agricultural and Food Chemistry. 2001;49:5223–5227. doi: 10.1021/jf010656r. [DOI] [PubMed] [Google Scholar]
- Griffiths DW, Dale MFB, Morris WL, Ramsay G. Effect of season and post-harvest storage on the carotenoid content of Solanum phureja potato tubers. Journal of Agricultural and Food Chemistry. 2007;55:379–385. doi: 10.1021/jf0620822. [DOI] [PubMed] [Google Scholar]
- Huaman Z, Spooner DM. Reclassification of landrace populations of cultivated potatoes (Solanum sect. Petota) American Journal of Botany. 2002;89:947–965. doi: 10.3732/ajb.89.6.947. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Mao M, Jones AR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nature Biotechnology. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant and Cell Physiology. 2001;42:677–685. doi: 10.1093/pcp/pce112. [DOI] [PubMed] [Google Scholar]
- Kloosterman B, Vorst O, Hall RD, Visser RGF, Bachem CW. Tuber on a chip: differential gene expression during potato tuber development. Plant Biotechnology Journal. 2005;3:505–519. doi: 10.1111/j.1467-7652.2005.00141.x. [DOI] [PubMed] [Google Scholar]
- Kloosterman B, De Koeyer D, Griffiths R, et al. The potato transcriptome: a new look at transcriptional changes during tuber development using the POCI array. Comparative and Functional Genomics. 2008 doi: 10.1007/s10142-008-0083-x. (in press) [DOI] [PubMed] [Google Scholar]
- Livak KJ. 1997. [Google Scholar]
- Maga JA. Potato flavor. Food Reviews International. 1994;10:1–48. [Google Scholar]
- McGregor I. The fresh potato market. In: Vreugdenhil D, editor. Potato biology and biotechnology: advances and perspectives. Elsevier; 2007. pp. 3–26. [Google Scholar]
- Morris WL, Ducreux L, Griffiths DW, Stewart D, Davies HV, Taylor MA. Carotenogenesis during tuber development and storage in potato. Journal of Experimental Botany. 2004;55:975–982. doi: 10.1093/jxb/erh121. [DOI] [PubMed] [Google Scholar]
- Morris WL, Ross HA, Ducreux LJM, Bradshaw JE, Bryan GJ, Taylor MA. Umami compounds are a determinant of the flavor of potato (Solanum tuberosum L.) Journal of Agricultural and Food Chemistry. 2007;55:9627–9633. doi: 10.1021/jf0717900. [DOI] [PubMed] [Google Scholar]
- Nielsen GS, Poll L. Determination of odour active aroma compounds in a mixed product of fresh-cut iceberg lettuce, carrot and green bell pepper. Developments in Food Science. 2007;43:517–520. [Google Scholar]
- Oruna-Concha MJ, Duckham SC, Ames JM. Comparison of volatile compounds isolated from the skin and flesh of four potato cultivars after baking. Journal of Agricultural and Food Chemistry. 2001;49:2414–2421. doi: 10.1021/jf0012345. [DOI] [PubMed] [Google Scholar]
- Oruna-Concha MJ, Bakker J, Ames JM. Comparison of the volatile components of two cultivars of potato cooked by boiling, conventional baking and microwave baking. Journal of the Science of Food and Agriculture. 2002a;82:1080–1087. [Google Scholar]
- Oruna-Concha MJ, Bakker J, Ames JM. Comparison of the volatile components of eight cultivars of potato after microwave baking. Lebensmittel-Wissenschaft und-Technologie. 2002b;35:80–86. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling J, Willmitzer L, Fisahn J. Expression of a Petunia inflata pectin methyl esterase in Solanum tuberosum L. enhances stem elongation and modifies cation distribution. Planta. 2000;210:391–399. doi: 10.1007/PL00008147. [DOI] [PubMed] [Google Scholar]
- Pilnick W, Voragen AGJ. The significance of endogenous and exogenous pectin enzymes in fruit and vegetable processing. In: Fox PE, editor. Food enzymology. London: Elsevier Applied Science; 1991. pp. 303–336. [Google Scholar]
- Prosser I, Altug IG, Phillips AL, Konig WA, Bouwmeester HJ, Beale MH. Enantiospecific (+)- and (–)-germacrene D synthases, cloned from goldenrod, reveal a functionally active variant of the universal isoprenoid-biosynthesis aspartate-rich motif. Archives of Biochemistry and Biophysics. 2004;432:136–144. doi: 10.1016/j.abb.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Falcon M, Bou J, Prat S. Seasonal control of tuberization in potato: conserved elements with the flowering response. Annual Review of Plant Biology. 2006;57:151–180. doi: 10.1146/annurev.arplant.57.032905.105224. [DOI] [PubMed] [Google Scholar]
- Shepherd T, Dobson G, Marshall R, Verrall S, Conner S, Griffiths D, Stewart D, Davies H. Profiling of metabolites and volatile flavour compounds from Solanum species using gas chromatograph-mass spectrometry. In: Nikolau B, Wurtele E, editors. Concepts in plant metabolomics. Springer; 2007. pp. 208–219. [Google Scholar]
- Spooner DM, McLean K, Ramsay G, Waugh R, Bryan GJ. A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proceedings of the National Academy of Sciences, USA. 2005;102:14694–14699. doi: 10.1073/pnas.0507400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner DM, Nunez J, Trujillo G, del Rosario Herrera M, Guzman F, Ghislain M. Extensive simple sequence repeat genotyping of potato landraces supports a major re-evaluation of their gene pool structure and classification. Proceedings of the National Academy of Sciences, USA. 2007;104:19398–19403. doi: 10.1073/pnas.0709796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MA, McDougall GJ, Stewart D. Potato flavour and texture. In: Vreugdenhil D, editor. Potato biology and biotechnology: advances and perspectives. Elsevier; 2007. pp. 525–540. [Google Scholar]
- Van der Hoeven R, Ronning C, Giovannoni J, Martin G, Tanksley S. Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. The Plant Cell. 2002;14:1441–1456. doi: 10.1105/tpc.010478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voragen F, Beldman G, Schols H. Chemistry and enzymology of pectins. In: McCleary BV, Prosky L, editors. Advanced dietary fibre technology. Oxford: Blackwell Science; 2001. pp. 379–398. [Google Scholar]
- Winfield M, Lloyd D, Griffiths DW, Bradshaw JE, Muir D, Nevison IM, Bryan GJ. Assessing organoleptic attributes of Solanum tuberosum and S. phureja potatoes. Aspects of Applied Biology. 2005;76:127–135. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.