Abstract
Fibroblasts located adjacent to the tumor [cancer-associated fibroblasts (CAFs)] that constitute a large proportion of the cancer-associated stroma facilitate the transformation process. In this study, we compared the biological behavior of CAFs that were isolated from a prostate tumor to their normal-associated fibroblast (NAF) counterparts. CAFs formed more colonies when seeded at low cell density, exhibited a higher proliferation rate and were less prone to contact inhibition. In contrast to the general notion that high levels of α-smooth muscle actin serve as a marker for CAFs, we found that prostate CAFs express it at a lower level compared with prostate NAFs. Microarray analysis revealed a set of 161 genes that were altered in CAFs compared with NAFs. We focused on whey acidic protein four-disulfide core domain 1 (WFDC1), a known secreted protease inhibitor, and found it to be downregulated in the CAFs. WFDC1 expression was also dramatically downregulated in highly prolific mesenchymal cells and in various cancers including fibrosarcomas and in tumors of the lung, bladder and brain. Overexpression of WFDC1 inhibited the growth rate of the fibrosarcoma HT1080 cell line. Furthermore, WFDC1 level was upregulated in senescent fibroblasts. Taken together, our data suggest an important role for WFDC1 in inhibiting proliferation of both tumors and senescent cells. Finally, we suggest that the downregulation of WFDC1 might serve as a biomarker for cellular transformation.
Introduction
Prostate cancer is one of the leading cancers diagnosed in men in the western world (1). Although the vast majority of prostate cancers stem from the epithelial compartment of the prostate, genetic and molecular studies indicate that tumorigenesis is not only solely determined by the malignant cancer cells themselves but also by the tumor stroma. Fibroblasts, that constitute the majority of the stroma, are elongated cells embedded in the connective tissue and are to a large extent responsible for its synthesis and maintenance. Fibroblasts participate in the regulation of diverse processes such as inflammation, wound healing and differentiation of their neighboring epithelial cells (2). They synthesize fibrillar extracellular matrix (ECM) vital proteins, such as type I, III and V collagens and fibronectin. In contrast, fibroblasts are also an important source of ECM-degrading proteases such as matrix metalloproteinases that stress their crucial role in maintaining the ECM homeostasis. Recent studies have established the notion that fibroblasts are prominent modifiers of cancer progression, with a specific subpopulation designated as cancer-associated fibroblasts (CAFs) that play a key role in promoting tumor initiation and progression. During prostatic carcinogenesis, periepithelial stroma undergoes progressive loss of smooth muscle accompanied by the appearance of CAFs, either through alterations in differentiation programs of smooth muscle cells or by recruitment of fibroblasts to the tumor site. CAFs isolated from malignant tissues demonstrate distinct differences from normal-associated fibroblasts (NAFs), including increased production of collagens and epithelial growth factors as well as enhanced proliferation rate (3–5). In vivo studies revealed that co-injection of CAFs derived from prostate adenocarcinoma with normal epithelia enhances the growth of the latter. Furthermore, co-injection of CAFs and immortalized epithelial cells results in tumors that surpass the control (only immortalized epithelia) tumor mass by 500-fold (6). Recent studies focused on identifying specific factors that are secreted by CAFs and potentially accelerate the transformation of neighboring epithelia. For instance, it was shown that breast cancer-derived CAFs promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion (7). Another study revealed that transforming growth factor-β signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia (8). One of the proteins that are known to be secreted from the prostate stroma and influence adjacent epithelia is the whey acidic protein four-disulfide core domain 1 (WFDC1) (9). It belongs to a family of proteins with a so-called ‘whey acidic protein’ domain that are often modulated in cancers (10). Whey acidic proteins contain at least one whey acidic protein motif consisting of ∼50 amino acids with eight highly conserved cysteine residues that form four disulfide bridges. Usually, this motif is characterized by serine protease inhibitory activity. Four members of this family were suggested as markers for various cancers: elafin, antileukoproteinase, WFDC2 and WFDC1 (10). WFDC1 was first purified from urogenital-sinus mesenchymal cell line of a rat fetus and was designated as prostate-specific 20 (ps20). When added to the media, purified ps20 inhibits the growth of PC3 human prostate metastatic cell line and enhances protein secretion by >2-fold (9). Stable ps20-expressing COS-7 cells (transformed simian fibroblasts) secrete ps20 and are growth inhibited. Additionally, COS-7 and PC3 cells are growth inhibited by bacterially produced ps20 (11). Human WFDC1 maps to chromosome 16q24, an area characterized by a frequent loss of heterozygosity in multiple cancers such as breast, prostate and liver. Watson et al. (12) integrated data from expression arrays and high-resolution comparative genomic hybridization analysis of chromosome 16q derived from 16 prostate tumors and identified six genes that were significantly downregulated compared with normal prostate tissue, among which was WFDC1. In contrast, several studies aiming at deciphering the correlation between cancer samples that harbor aberrations in chromosome 16 and WFDC1 discovered low mutation rate and no significant alteration in WFDC1 expression in various cancers (13–15). Analysis of WFDC1 expression in prostate cancer using reverse transcription–polymerase chain reaction showed that WFDC1 levels are significantly downregulated, but no mutations were detected in its coding region (16). All the above imply that WFDC1 is not a classical tumor suppressor but nonetheless can inhibit cell proliferation.
To further elucidate the effect of prostate stromal cells on the neighboring epithelia, we sought to establish an in vitro system of normal and cancer-associated fibroblasts that will permit the investigation of the phenotypic and genotypic differences between the two, with an emphasis on genes that might act in a paracrine fashion on the growth and survival of adjacent epithelia. The comparison between CAFs and NAFs demonstrated distinct phenotypes. A complementary DNA (cDNA) microarray analysis allowed us to detect specific gene signatures whose expression differed between the two cultures. Finally, we focused on WFDC1 that exhibited an altered expression pattern between NAFs and CAFs. In our experiments, we set out to assess its effect on various cell growth parameters as well as its expression pattern in various cancer tissues, cell cultures and cell lines.
Materials and methods
Cell culture
Primary human prostate fibroblasts (PFs) (designated PF179; 179, patient number) were isolated from a prostatectomy specimen, marginal and distal to the prostate tumor (CAFs and NAFs, respectively). PF179 cell cultures and WI-38 human embryonic lung fibroblasts were grown in minimum essential medium supplemented with 10% fetal calf serum, 2 mM L-glutamine, 1 mM sodium pyruvate and antibiotics. Phoenix and HT1080 cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM L-glutamine and antibiotics. Cells were maintained in a humidified incubator at 37°C and 5% CO2.
Sarcoma samples
RNA samples were isolated from various human sarcoma specimens. Fibrosarcomas (five), liposarcomas (five), leiomyosarcoma (five), neurofibrosarcoma, dermatofibrosarcoma and rhabdomyosarcoma were kindly provided by Dr Paul Meltzer (National Institutes of Health, Bethesda).
Plasmids and constructs
The WFDC1 gene was amplified from PF179-derived random hexamer-primed cDNA with specific primers: 5′-GGGAGGAAATGCCTTTAACC-3′ and 5′-TGCTTGCCGTTGCTTTACTG-3′ using the Phusion DNA polymerase kit (Stratagene). The polymerase chain reaction product was cloned into a pGEM-T Easy vector (Promega), restricted with EcoRI (Roche) and subcloned into pBabe-Hygro retroviral vector.
Retroviral infection
See supplementary Materials and methods (available at Carcinogenesis Online) for details.
Transfections
HT1080 cells were plated at 1.2 × 105 cells per well in a six-well plate, 24 hours prior to transfection. Cells were transfected with Fugene-HD (Roche), 200 ng expression vector encoding for GFP for estimation of transfection efficiency, 1500 ng of either empty pBabe-Hygro vector as a control or pBABE-Hygro vector encoding WFDC1 and 300 ng pBlueScript plasmid (Stratagene) to complete the total DNA mass to 2 μg. Cells were then incubated for 48 h at 37°C and were either collected for RNA extraction or replated for crystal violet staining at clonal density.
Spectral karyotype analysis
Half a million cells were plated per 10 cm plate and allowed to initiate proliferation for 48 h. Then, the medium was replaced and the cells were incubated for additional 24 h. Next, the cells were incubated with colcemid (0.1 μg/ml) over night, trypsinized, lysed with hypotonic buffer and fixed in glacial acetic acid:methanol (1:4). Further analysis was performed by Dr Irit Bar-Am as described in detail in the supplementary Materials and methods (available at Carcinogenesis Online).
Isolation of total RNA and quantitative real-time polymerase chain reaction
Total RNA was isolated using the RNAeasy kit (Qiagen), according to the manufacturer's protocol. A 2 μg aliquot was reverse transcribed using MMLV-RT (Bio-RT) and random hexamer primers. Quantitative real-time polymerase chain reaction (QRT-PCR) was performed on an ABI 7300 instrument (Applied Biosystems) using Platinum SYBR Green and qPCR SuperMix (Invitrogen). Specific primers are provided in the supplementary Materials and methods (available at Carcinogenesis Online).
cDNA microarray
Total RNA was extracted using Tri-Reagent (MRC) according to the manufacturer's protocol and sent to the MicroArray department (Weizmann institute of science, Rehovot, Israel). The procedure was done using Affymetrix Human Genome U133 plus 2.0 microarray.
BrdU incorporation assay
As described previously (17).
Western blot analysis
This procedure is described in detail in the supplementary Materials and methods (available at Carcinogenesis Online).
Growth curves
Cells were grown until subconfluence density, harvested with trypsin and replated at a cell density of 2500 or 1250/cm2. Population doublings (PDLs) at each passage were calculated using the formula: PDLs = log (cell output/cell input)/log 2.
Colony-forming efficiency assay
PF179T cells were seeded in triplicates at a cell density of 20/cm2. Medium was replaced every 3–4 days. Two weeks after plating, cells were stained with crystal violet and colonies were counted. Additionally, the crystal violet was extracted with acetic acid and quantified with a spectrophotometer using a 590 nm filter.
Contact inhibition assay
PF179T cells were seeded in duplicates at a cell density of 900/cm2 plates. Medium was replaced every 3–4 days. Cells were collected and counted at each designated time point.
Results
CAFs demonstrate a higher proliferation capacity than NAFs
Primary human PFs (designated PF179) were isolated from a prostatectomy specimen, marginal and distal to the prostate tumor (CAFs and NAFs, respectively). Since CAFs isolated from malignant tissues usually demonstrate a distinct phenotype from NAFs, we compared several proliferation features between the NAFs and the CAFs.
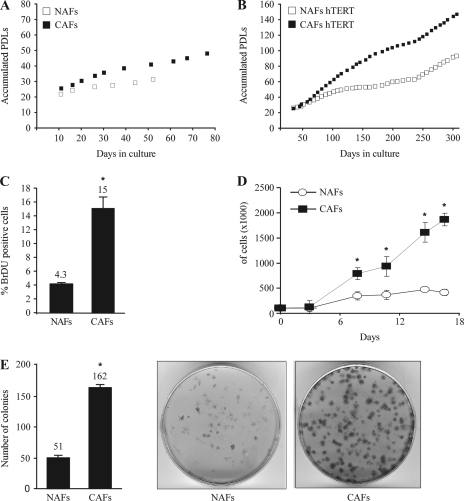
First, both cell cultures were immortalized by infection with a retroviral vector encoding for the human telomerase catalytic subunit, hTERT as reported previously (18–21). We compared the growth rate of the primary and hTERT-immortalized cell cultures by counting and replating them weekly and calculating their accumulated PDLs. As shown in Figure 1A, non-immortalized NAFs demonstrated low proliferation rate, and following 30 PDLs entered a complete growth arrest, whereas non-immortalized CAFs accumulated 45 PDLs before entering growth arrest. As expected, both immortalized NAFs and CAFs overcame growth arrest and continued to divide in culture. However, CAFs divided twice as fast as NAFs during the examined period with a P-value of 2.9 × 10−9 as calculated by Student's t-test (Figure 1B). To further substantiate this phenomenon and in order to obtain additional insights into possible changes in the cell cycle distribution, we conducted a BrdU incorporation assay that permits an accurate comparison of the proportion of cells found at the S-phase of the cell cycle. As depicted in Figure 1C and supplementary Figure 4 (available at Carcinogenesis Online), the CAFs population consisted of 15% BrdU-positive cells in contrast to only 4.3% in the NAFs population, confirming the replicative advantage of the CAFs. Next, we performed a contact inhibition assay. Normal cells demonstrate a loss of cell division due to contact with another cell when optimum size and volume of the tissue is achieved, whereas abnormal cells continue to divide. As depicted in Figure 1D, NAFs reached full confluence after 7 days in culture and maintained the same cell number throughout the duration of the experiment. CAFs, on the other hand, continued to divide regardless of cell density and after 18 days in culture reached a cell number 4-fold higher than the NAFs. Subsequently, we conducted a colony-forming efficiency assay that measures the ability of the cells to form tightly packed colonies when seeded at low cell density. As shown in Figure 1E, CAFs formed three times more colonies than the NAFs. All in all, these experiments suggest that CAFs exhibit a higher proliferative capacity as compared with NAFs.
Fig. 1.
CAFs demonstrate higher proliferation capacity than NAFs. Primary (A) and hTERT-immortalized (B) NAFs and CAFs were counted and replated weekly and their accumulated PDLs were calculated as described. (C) Cells were labeled with BrdU and propidium iodide and analyzed using flow cytometry. The average proportion of S-phase (BrdU positive) cells is displayed ± standard deviation of three repeats. *Denotes a significant increase, P = 2.2 × 10−4. (D) Cells were seeded at identical confluence and were collected and counted at the indicated time points. The graph indicates the mean number of the counted cells ± standard deviation. *Denotes a significant increase, P ≤ 1 × 10−5. (E) Cells were seeded at low density in triplicates, grown for 11 days and then stained with crystal violet. Plates were scanned, and the average number of colonies was calculated. The graph indicates the mean number of colonies ± standard deviation. *Denotes a significant increase, P = 9.34 × 10−7.
Both NAFs and CAFs demonstrate genomic stability
To evaluate whether changes in the proliferation rate observed above can be attributed to gross genetic changes manifested as chromosomal aberrations, we conducted a spectral karyotyping (SKY) analysis using condensed DNA from several metaphases from each cell culture. No chromosomal aberrations were detected in none of the cell cultures examined (supplementary Figure 1 is available at Carcinogenesis Online). Based on the SKY data, we concluded that both CAFs, which lie in close proximity to the tumor and are subjected to its malignant signals, and NAFs, that are more distal to the tumor, maintained their chromosomal integrity. Thus, we hypothesize that the pronounced variations in the proliferation patterns we observed above may stem from either subtle genetic aberrations or epigenetic alterations that result in differential gene expression.
CAFs express lower levels of α-smooth muscle actin than NAFs
CAFs are often considered to be ‘activated’ fibroblasts or myofibroblasts. Dvorak (22) noted a striking resemblance between many of the signaling processes occurring in tumor progression and those involved in wound healing. An important hallmark for myofibroblasts in wound healing as well as in cancer is the expression of differentiation markers such as calponin, desmin and most commonly α-smooth muscle actin (αSMA) (23–25). However, other studies have shown downregulation of the very same markers in prostate cancer-associated stromal cells (26,27). Because of these rather contradictory data in the literature, it was important to measure the expression of αSMA as a representative marker in our cell cultures. The results presented in supplementary Figure 2A (available at Carcinogenesis Online) indicate a 10-fold decrease in the messenger RNA (mRNA) levels of αSMA in the CAFs compared with the NAFs, as measured by QRT-PCR. A comparable decrease in the protein level of αSMA was also observed using western blot (supplementary Figure 2B is available at Carcinogenesis Online). To conclude, the basal expression level of αSMA was significantly reduced in the CAFs.
NAFs and CAFs demonstrate a differential gene expression profile
Since NAFs and CAFs exhibit some differences in their biological behavior and changes in gene expression such as αSMA, we decided to adopt a genome-wide approach in order to discover specific expression signatures that might account for their distinct behavior. To that end, we performed a global expression profiling analysis using cDNA microarrays on RNA samples obtained from four distinct cell cultures: immortalized and non-immortalized NAFs and CAFs. Class comparison analyses using the BRB-ArrayTools software (28) were performed on genes that differ in their expression by at least 1.5-fold from the median expression level in at least one sample. In order to assess the effect of the hTERT introduction on the genes expression, we compared the immortalized cell cultures with the non-immortalized ones. This comparison resulted in a list of only three genes (P-value < 0.001), indicating a mild directional effect of the immortalization on the examined cell cultures. Next, we analyzed the expression profiles using the same statistical method and thresholds, for altered gene expression between the NAFs and CAFs. This analysis yielded a list of 161 gene probes that were divided into two groups: NAFs upregulated genes and CAFs upregulated genes (Tables I and II, respectively).
Table I.
NAFs upregulated genes
| Gene symbol | Fold induction | P-value | Gene symbol | Fold induction | P-value |
| SERPINB2 | 33.4 | 1.1E-04 | MME | 3.1 | 8.7E-04 |
| CD200 | 13.5 | 2.0E-04 | SAMD3 | 3.0 | 5.5E-04 |
| WFDC1 | 12.5 | 6.4E-05 | RGS7 | 3.0 | 7.6E-04 |
| JAM2 | 9.7 | 4.7E-05 | PSG1 | 3.0 | 4.8E-04 |
| AFF3 | 9.5 | 3.1E-04 | PCLO | 3.0 | 5.5E-04 |
| CORIN | 9.4 | 9.6E-06 | PRG1 | 3.0 | 6.1E-04 |
| LOC339903 | 8.8 | 7.0E-05 | PCSK7 | 2.9 | 3.6E-04 |
| OXTR | 7.6 | 4.8E-05 | SESN3 | 2.9 | 5.5E-04 |
| FAM84A/LOC400944 | 7.3 | 1.7E-04 | C21orf7 | 2.9 | 3.8E-04 |
| CCND2 | 6.6 | 5.1E-05 | FGF1 | 2.9 | 8.4E-04 |
| JPH2 | 6.4 | 2.4E-04 | DDAH1 | 2.9 | 5.7E-04 |
| Similar to yeast ARV 1 | 6.3 | 5.6E-04 | SCUBE3 | 2.9 | 5.6E-04 |
| NRXN3 | 6.3 | 5.1E-05 | SLC2A1 | 2.8 | 9.7E-04 |
| FGL2 | 6.1 | 6.3E-04 | MAMDC2 | 2.8 | 5.3E-04 |
| F2RL2 | 5.8 | 2.7E-05 | FHL1 | 2.8 | 7.1E-04 |
| SCN2A2 | 4.6 | 2.3E-04 | CXCL6 | 2.7 | 7.6E-04 |
| RASGRP2 | 4.3 | 1.0E-04 | MGC11324 | 2.5 | 9.0E-04 |
| INMT | 4.3 | 3.0E-04 | |||
| OPN3 | 4.2 | 3.0E-04 | |||
| DNAJC6 | 4.2 | 4.6E-04 | |||
| CPXM2 | 4.1 | 1.4E-04 | |||
| DMD | 3.9 | 1.2E-04 | |||
| PSG7 | 3.9 | 5.7E-04 | |||
| DKFZp761P0423 | 3.8 | 1.5E-04 | |||
| PSG4 | 3.6 | 2.5E-04 | |||
| ITGA6 | 3.4 | 7.0E-04 | |||
| PVR | 3.4 | 6.8E-04 | |||
| NEDD9 | 3.3 | 7.1E-04 | |||
| PTGDS | 3.3 | 3.8E-04 | |||
| KRTAP2-1/2-4 | 3.3 | 7.1E-04 | |||
| CLDN1 | 3.3 | 6.9E-04 | |||
| EPHB6 | 3.2 | 2.5E-04 | |||
| KIAA1913 | 3.2 | 9.7E-04 | |||
| CCND1 | 3.2 | 8.4E-04 | |||
| SMTN | 3.2 | 5.2E-04 | |||
| SYNPO2 | 3.1 | 2.4E-04 | |||
| ANXA3 | 3.1 | 2.9E-04 |
Table II.
CAFs upregulated genes
| Gene symbol | Fold induction | P-value | Gene symbol | Fold induction | P-value |
| IL13Rα2 | 42.7 | 6.0E-07 | ZFPM2 | 4.1 | 4.5E-04 |
| SPON2 | 35.8 | 1.6E-06 | IGFBP3 | 4.1 | 9.3E-04 |
| TNXB | 11.8 | 3.9E-04 | SLC19A2 | 4.0 | 4.2E-04 |
| NFIB | 9.8 | 3.7E-05 | MFAP5 | 4.0 | 5.9E-04 |
| GLDN | 9.8 | 1.5E-05 | ID4 | 4.0 | 4.9E-04 |
| PCOLCE2 | 9.5 | 4.8E-06 | PCTK2 | 3.8 | 3.7E-04 |
| ASB5 | 8.8 | 5.7E-05 | STEAP1 | 3.8 | 3.6E-04 |
| LIMS3 | 8.5 | 4.5E-06 | SLC27A6 | 3.7 | 4.9E-04 |
| GATA3 | 8.4 | 6.2E-05 | CA12 | 3.7 | 5.1E-04 |
| FAM38B | 7.3 | 2.4E-04 | EGR3 | 3.7 | 1.5E-04 |
| SGCG | 6.8 | 3.9E-04 | DOK5 | 3.5 | 4.2E-04 |
| MOCOS | 6.6 | 1.2E-05 | L1MS1 | 3.4 | 4.7E-04 |
| GADD45B | 6.3 | 5.6E-04 | SOCS3 | 3.4 | 4.0E-04 |
| RCN3 | 6.2 | 3.3E-04 | KLF9 | 3.4 | 3.6E-04 |
| ID2 /// ID2B | 6.2 | 4.0E-05 | ALDH1A3 | 3.4 | 2.8E-04 |
| KLF10 | 6.0 | 9.1E-04 | FGF9 | 3.3 | 4.8E-04 |
| ACVR2A | 6.0 | 1.4E-05 | ANKRD10 | 3.3 | 2.0E-04 |
| PEAR1 | 5.9 | 1.8E-05 | IRF2BP2 | 3.3 | 5.2E-04 |
| RFX3 | 5.6 | 6.2E-05 | CCDC8 | 3.1 | 2.9E-04 |
| VIT | 5.6 | 9.7E-05 | EYA1 | 3.1 | 2.8E-04 |
| FAM49A | 5.5 | 9.8E-04 | ARIH1 | 3.0 | 4.0E-04 |
| COL14A1 | 5.3 | 5.0E-04 | MYLIP | 3.0 | 9.6E-04 |
| ZNF521 | 5.3 | 1.1E-04 | FNBP1L | 3.0 | 6.7E-04 |
| LOC200230 | 5.1 | 1.5E-04 | CLDN11 | 2.9 | 8.0E-04 |
| PDE3B | 5.1 | 3.6E-05 | GPC6 | 2.9 | 8.3E-04 |
| EBF | 5.0 | 9.5E-04 | NFIA | 2.9 | 3.7E-04 |
| AQP3 | 4.9 | 3.2E-05 | PDE4DIP | 2.8 | 4.2E-04 |
| MBP | 4.8 | 4.6E-04 | KCNK1 | 2.8 | 8.4E-04 |
| FRAS1 | 4.7 | 2.2E-04 | B4GALT1 | 2.8 | 7.0E-04 |
| INSIG1 | 4.6 | 1.5E-04 | RBMS1 | 2.8 | 7.9E-04 |
| CLEC2B | 4.6 | 9.8E-04 | MEIS1 | 2.7 | 6.1E-04 |
| SIPA1L2 | 4.5 | 1.5E-04 | PDE4D | 2.7 | 9.7E-04 |
| IL7 | 4.5 | 4.0E-04 | SH3D19 | 2.7 | 7.8E-04 |
| MN1 | 4.4 | 5.2E-05 | SPAG9 | 2.6 | 8.4E-04 |
| FLRT2 | 4.4 | 6.1E-04 | CXCL12 | 2.5 | 9.0E-04 |
| PBEF1 | 4.4 | 2.3E-04 | NFKBIZ | 2.5 | 9.6E-04 |
| CRABP2 | 4.2 | 6.0E-04 |
In concordance with the notion that the stromal cells affect their surroundings via the ECM, we used the ‘DAVID Bioinformatics’ software (29) to identify genes that are related to the extracellular region. After filtration of the microarray data into a list consisting of the 2000 most differentially expressed genes, we observed a significant enrichment of extracellular space-related genes (9.32%, P-value 1.6 × 10−5). These genes were then clustered using the Cluster 3.0 software (30) according to their expression pattern (supplementary Figure 3A is available at Carcinogenesis Online). To validate the microarray data, we conducted QRT-PCR analyses for a representative number of candidate genes. We have chosen to focus on three genes from each group that were both ECM related and reported previously in the literature as differentially expressed in human cancers. From the CAFs upregulated gene list, we examined interleukin (IL) 13 receptor α2, Spondin2 and IL7. IL13 receptor α2 is a subunit of the IL13 receptor complex that was shown to be highly expressed in ovarian cancer specimens when compared to normal ovary samples (31). Spondin2 is a cell adhesion protein that was identified as a candidate marker of prostate cancer (32). IL7 is a cytokine important for B and T cell development that can be produced by some human tumor cells and is involved in tumor development and progression (reviewed in ref. 33). From the NAFs upregulated gene list, we examined Periostin, matrix metalloproteinase3 and WFDC1. Periostin induces cell attachment and spreading and plays a role in cell adhesion. Its mRNA expression was shown to be markedly downregulated in a variety of human cancer cell lines and in human lung and bladder tumors (34,35). Matrix metalloproteinase3 is an ECM-degrading proteinase that on the one hand might prompt normal mammary epithelial cells to disaggregate and undergo epithelial to mesenchymal transition and become invasive (36) and on the other hand is secreted from senescent fibroblasts (37). WFDC1 is a secreted growth inhibitor that is known to be downregulated in the stroma of prostate cancer (16). As depicted in supplementary Figure 3B (available at Carcinogenesis Online), we found that the expression levels of the various genes examined by QRT-PCR agreed with the expression profile seen by the cDNA microarray analysis.
Due to its ECM localization and relevance to prostate cancer, we decided to focus our study on WFDC1.
WFDC1 expression level is repressed in highly prolific cell cultures and in tumors
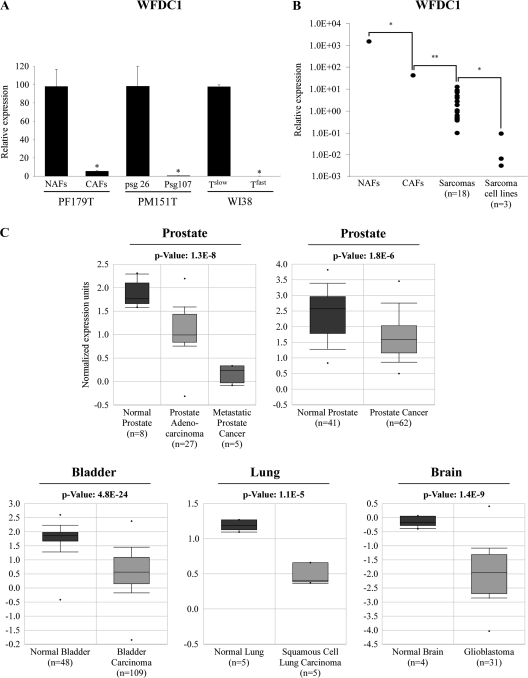
Since WFDC1 level was significantly decreased in CAFs (Table II, supplementary Figure 3 is available at Carcinogenesis Online), which is in agreement with previously published studies showing a decrease of its level in prostate reactive stroma (16), we decided to further characterize the expression pattern of WFDC1. First, we examined the generality of our observation. To that end, we utilized two systems that were previously established in our lab by immortalization of human primary mesenchymal cells. In both systems, following a prolonged culturing period, cells spontaneously acquired an accelerated growth rate and lost the expression of key tumor suppressors (38,39). The first system was the WI-38 lung fibroblasts consisting of early slow-growing passages (Tslow) and fast-growing passages (Tfast). The second system was the early (passage 27) and late (passage 107) passages of PM151T prostate-derived immortalized smooth muscle cells. As expected, a marked reduction of WFDC1 mRNA level in the late passage-derived samples was observed in both systems (Figure 2A). These observations prompted us to measure the level of WFDC1 in several cell lines of mesenchymal origin as well as in human sarcomas. We found that WFDC1 is expressed at low levels in human sarcomas (n = 18) and at very low to undetectable levels in the transformed mesenchymal cell lines U2OS, HT1080 and SAOS, compared with the PFs (Figure 2B). To further confirm the hypothesis that WFDC1 repression is a frequent event during malignant transformation, we searched the ‘Oncomine’ database (40) for clinical tumor sample studies with significant differential expression of WFDC1 compared to their corresponding normal tissues. We found that WFDC1 was significantly decreased in prostate cancers compared with normal prostate tissues. Metastatic prostate cancers exhibited even lower levels of WFDC1 expression than prostate adenocarcinomas (41,42). WFDC1 downregulation was also observed in other cancer types, namely, bladder, lung and brain (43–45) (Figure 2C).
Fig. 2.
WFDC1 expression level is repressed in highly prolific cell cultures and in tumors. (A) Basal expression levels of WFDC1 in various hTERT-immortalized mesenchymal cells (PF179T NAFs, PM151T psg26 and WI-38 Tslow) and their highly prolific counterparts (PF179T CAFs, PM151T psg107 and WI-38 Tfast) were determined by QRT-PCR. The results are presented as a mean ± SD of two duplicate runs from a representative experiment. *Denotes a significant decrease of WFDC1, P ≤ 0.02. (B) RNA was produced from PF179T cell cultures, 18 sarcoma specimens (as described in Materials and Methods) and 3 fibrosarcoma cell lines: SAOS, U2OS and HT1080. QRT-PCR analysis was conducted for WFDC1 expression. The results are displayed on a logarithmic scale. *Denotes a significant decrease, P < 0.03. **P < 1.2 × 10−16. (C) The Oncomine database was queried for studies in which WFDC1 expression significantly differs between cancer and normal samples as described in Material and Methods. The P-value designates the probability that the observed difference in WFDC1 expression between normal and cancer samples is due to random chance. The bars represent 25–75% interval of normalized WFDC1 expression levels in each group. The deviation lines represent 10–90% percentiles and dots represent samples with minimum and maximum expression level in a given group.
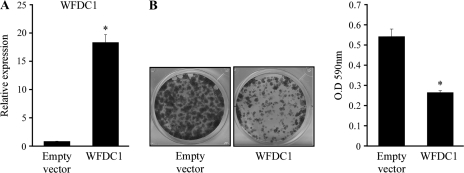
Owing to the correlation between the reduction in WFDC1 mRNA and cell proliferation rate, we sought to assess the growth-inhibiting effect of WFDC1 in HT1080 cells, a human fibrosarcoma cell line, in which WFDC1 is not expressed. To that end, we transfected HT1080 with a retroviral vector containing either the WFDC1 coding sequence or with an empty vector as a control and examined their colony formation efficiency. We found that overexpression of WFDC1 exerted an inhibitory effect on the colony formation efficiency of HT1080 cells (Figure 3). This result is in agreement with the notion that WFDC1 expression does not only correlate with low proliferation rate but may also directly inhibit proliferation when introduced exogenously. Moreover, WFDC1 inhibitory function might explain the selective pressure that underlies its downregulation in malignant tissues.
Fig. 3.
Overexpression of WFDC1 in the HT1080 cell line inhibits its proliferation rate. Cells were transiently transfected with either a retroviral vector containing the WFDC1 coding sequence or an empty vector. (A) Cells were tested for WFDC1 expression using QRT-PCR. The results are presented as a mean ± SD of two duplicate runs from a representative experiment. *Denotes a significant increase in WFDC1 expression compared with the control, P ≤ 4.6 × 10−4. (B) Cells were seeded at a low density, incubated for 4 days and dyed with crystal violet. The crystal violet was removed with acetic acid and quantified with a spectrophotometer using a 590 nm filter (right hand graph). The graph indicates the mean number of three repeats ± standard deviation. *Denotes a significant decrease in OD 590 nm compared with the control, P ≤ 1.1 × 10−4.
WFDC1 is upregulated during cellular senescence
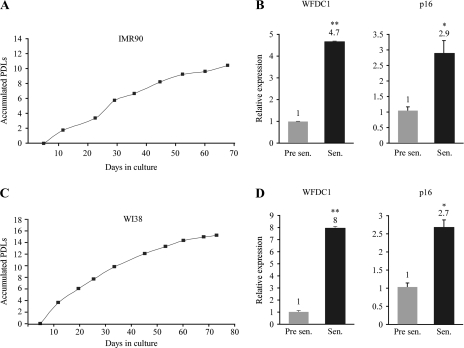
Cellular senescence was initially described by Hayflick (46), as he pointed out that normal human fibroblasts undergo robust cell division in tissue culture, followed by a gradual decrease in PDL rate over time, until they cease to divide altogether. Nonetheless, these cells remain viable and metabolically active despite their inability to proliferate even in the presence of nutrients and growth factors. Other than growth arrest, there are no widely specific markers for cellular senescence. Senescence-associated β-galactosidase activity is detected by histochemical staining in most senescent cells; however, it is also induced in dense cell populations and prolonged confluence in culture. In addition, p16, a known regulator of senescence, which is expressed by many but not all senescent cells, is often used as a marker as well (47). Since WFDC1 expression level was high in slowly dividing cells, we set to evaluate its levels in cells undergoing senescence. To that purpose, we used two normal human primary lung fibroblasts cell types: WI38 and IMR90. IMR90 cells were growth arrested after 12 PDLs, whereas WI38 ceased to proliferate after 14 PDLs (Figure 4A and C). RNA was produced from pre-senescent and senescent cells and the expression levels of p16 and WFDC1 were measured by QRT-PCR. As expected, p16 was elevated upon senescence (Figure 4B and D). WFDC1 mRNA level was increased as well, implying yet again on its production by cells with reduced proliferation rate, and suggesting that WFDC1 level might be considered as a potential marker for senescent cells.
Fig. 4.
IMR90 and WI38 cells were grown until the onset of replicative senescence. The left panels show their accumulated PDLs along time (A and C). WFDC1 and p16 expression levels were evaluated by QRT-PCR in young, pre-senescent cells (Pre sen.) and in cells that entered replicative senescence (Sen.) (B and D). The results are presented as a mean ± SD of two duplicate runs from a representative experiment. *Denotes a significant increase in WFDC1, P ≤ 0.02, **P ≤ 7.7 × 10−5.
Discussion
It is well accepted that malignant transformation is greatly dependent on the tumor stroma. CAFs, that constitute a large proportion of the cancer-associated stroma, play a key role in malignant transformation mainly via the activation of ECM-related genes (2). The aim of our study was to further characterize the biological behavior of prostate-derived CAFs and uncover their possible altered gene expression patterns.
The prostate CAFs studied here exhibited a higher proliferation rate than their NAFs counterparts, formed more colonies when seeded at low cell density and were less prone to contact inhibition. SKY analysis suggested that both CAFs and NAFs maintain their chromosomal integrity and ploidy. Therefore, the observed variations in the proliferation patterns were probably due to subtle genetic or epigenetic changes that are non-detectable by SKY analysis. These results are in agreement with other studies that showed enhanced production of collagens, epithelial growth factors and rapid proliferation rate of CAFs (3–5). Higher proliferation rate possibly grants the CAFs a long-term advantage over other cell types in the stroma compartment such as NAFs and smooth muscle cells and enables them to become the predominant cell type in the vicinity of the tumor. Furthermore, factors secreted from the CAFs might accelerate tumor progression (48). Thus, it might be of great importance for the tumor cells to favor CAFs growth in the tumor surroundings via reciprocal interactions. Elucidation of the cellular pathways that govern CAFs proliferation might serve as a platform to target their growth and that of their adjacent tumor cells.
Due to similarities between CAFs and myofibroblasts that take part in the wound healing process, CAFs are often considered to be activated fibroblasts or myofibroblasts (22). For example, both are recruited by platelet-derived growth factor and activated by transforming growth factor-β (48). Despite these similarities, these processes might last months and years in the context of cancer, in contrast to a transient period in the wound healing process. An accepted hallmark for myofibroblasts in wound healing as well as in cancer is the expression of differentiation markers such as calponin, desmin and most commonly α-SMA (23–25). In contrast, Tuxhorn et al. (26) observed high expression of αSMA in 95% of the normal prostate stroma compared with only 37% of the reactive stroma. This study consisted of 84 normal prostate stroma samples that were compared with 89 samples taken from stromal cells adjacent to a Gleason 3 tumor. A similar pattern of expression was observed also with the differentiation marker calponin. In addition, a study aimed at examining reactive stroma as a predictor of biochemical-free recurrence in prostate cancer, that was performed on a cohort of 545 patients, showed that reduced desmin and αSMA are hallmarks of cancer-associated reactive stroma, relative to normal fibromuscular stroma (27). We examined the basal level expression of αSMA in the NAFs and CAFs and found a 10-fold decrease in the expression of αSMA in the CAFs. Prostate carcinoma cells, especially those that have progressed into a higher level of malignancy, secrete transforming growth factor-β and perhaps promote the myofibroblasts phenotype in adjacent stroma. The expression level of differentiation markers in the stroma might be dependent on the malignant stage of the adjacent epithelial cells and may account for the contradictive reports of the differentiation marker levels in CAFs. Owing to the increasing controversy that exists in the literature, and in light of our own findings, our conclusion is that αSMA cannot serve as a definitive general marker of CAFs.
Several studies utilized genome-wide expression profiling in order to characterize the altered gene expression in the prostate reactive stroma, either on a large cohort of patients specimens or in a exogenously modified stroma (49–51). We report here for the first time on a cDNA microarray analysis that compared normal and cancer-associated PFs from the same patient without exogenous intervention. This approach was chosen in order to identify specific gene signatures that might be altered between closely related cells. Bioinformatical analysis revealed a list of 161 differentially expressed gene transcripts. Among them, 96 were upregulated in the CAFs compared with 65 in the NAFs. We also witnessed an enrichment in extracellular-related genes among the differentially expressed transcripts, which is in agreement with the notion that fibroblasts are able to influence their microenvironment via the secretion of proteins such as collagens, cytokines and growth factors (48). Although the exact function of some of these factors and their contribution to malignancy are not fully understood, their deregulation is consistent in various cancers. We identified some novel genes whose expression differed between NAFs and CAFs that were both ECM related and previously reported to exhibit a deregulated expression in cancer. Future studies might utilize the altered gene signatures of the CAFs to identify possible specific markers of the reactive stroma.
In our present study, we focused on the altered expression of WFDC1 that we found to be downregulated in the CAFs. WFDC1 is a member of a family composed primarily of secreted serine protease inhibitors, that was first reported to be secreted from rat mesenchymal cells and to act as a growth inhibitor of the PC3 prostatic cell line in vitro (9). However, WFDC1 might exert its effect in different manners. In a study conducted by McAlhany et al. (52), WFDC1 was shown to promote endothelial cell motility but did not have an effect on endothelial cell proliferation. Furthermore, in vivo experiments in mice revealed that WFDC1 causes an elevation in microvascular density of implanted tumors and an increase in their volume, suggesting that WFDC1 might facilitate angiogenesis. All in all, these data suggest that WFDC1 is a multifunctional protein and that its end point effect depends on the specificity of the cell or its context. Although WFDC1 is localized in the human genome to chromosome 16q24, an area of frequent loss of heterozygosity in multiple cancers (11), and despite its downregulation in prostate cancer (16), accumulated evidence support the notion that WFDC1 is not a classical tumor suppressor gene. In our study, we showed that WFDC1 expression was dramatically downregulated during prolonged culturing in two mesenchymal cellular systems and in CAFs. We also discovered that WFDC1 is significantly downregulated in clinical tumors of mesenchymal origin as well as in several carcinomas including prostate, bladder, lung and brain and that prostate metastases exhibit even lower level of WFDC1 expression than primary prostate adenocarcinoma. Overexpression of WFDC1 exerted an inhibitory effect on the growth rate of HT1080 cells. Furthermore, we report here for the first time that WFDC1 mRNA level is upregulated in fibroblasts undergoing senescence. In conclusion, WFDC1 function is related to the inhibition of cell growth. Although its role as a tumor suppressor is not established, its absence might serve as a biomarker for cellular transformation in a tissue/cell-specific manner.
Supplementary material
Supplementary Materials and methods and Figures 1–4 can be found at http://carcin.oxfordjournals.org/
Funding
Center of Excellence grant from Flight Attendant Medical Research Institute, FP6 grant Prima project 504587; Yad Abraham Center for Cancer Diagnosis and Therapy.
Supplementary Material
Acknowledgments
This publication reflects the authors' views and not necessarily those of the European Community. The European Community is not liable for any use that may be made of the information contained herein. V.R. is the incumbent of the Norman and Helen Asher Professorial Chair Cancer Research at the Weizmann institute.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CAFs
cancer-associated fibroblasts
- cDNA
complementary DNA
- ECM
extracellular matrix
- IL
interleukin
- mRNA
messenger RNA
- NAF
normal-associated fibroblast
- PDL
population doubling
- PF
prostate fibroblast
- ps20
prostate-specific 20
- QRT-PCR
quantitative real-time polymerase chain reaction
- SKY
spectral karyotyping
- αSMA
α-smooth muscle actin
- WFDC1
whey acidic protein four-disulfide core domain 1
References
- 1.Jemal A, et al. Cancer statistics, 2005. CA Cancer J. Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, et al. Fibroblasts in cancer. Nat. Rev. Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 3.Bauer EA, et al. Enhanced collagenase production by fibroblasts derived from human basal cell carcinomas. Cancer Res. 1979;39:4594–4599. [PubMed] [Google Scholar]
- 4.Rasmussen AA, et al. Paracrine/autocrine regulation of breast cancer by the insulin-like growth factors. Breast Cancer Res. Treat. 1998;47:219–233. doi: 10.1023/a:1005903000777. [DOI] [PubMed] [Google Scholar]
- 5.van den Hooff A. Stromal involvement in malignant growth. Adv. Cancer Res. 1988;50:159–196. doi: 10.1016/s0065-230x(08)60437-6. [DOI] [PubMed] [Google Scholar]
- 6.Olumi AF, et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orimo A, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 8.Bhowmick NA, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 9.Rowley DR, et al. Purification of a novel protein (ps20) from urogenital sinus mesenchymal cells with growth inhibitory properties in vitro. J. Biol. Chem. 1995;270:22058–22065. doi: 10.1074/jbc.270.37.22058. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard D, et al. Proteins with whey-acidic-protein motifs and cancer. Lancet Oncol. 2006;7:167–174. doi: 10.1016/S1470-2045(06)70579-4. [DOI] [PubMed] [Google Scholar]
- 11.Larsen M, et al. Molecular cloning and expression of ps20 growth inhibitor. A novel WAP-type “four-disulfide core” domain protein expressed in smooth muscle. J. Biol. Chem. 1998;273:4574–4584. doi: 10.1074/jbc.273.8.4574. [DOI] [PubMed] [Google Scholar]
- 12.Watson JE, et al. Integration of high-resolution array comparative genomic hybridization analysis of chromosome 16q with expression array data refines common regions of loss at 16q23-qter and identifies underlying candidate tumor suppressor genes in prostate cancer. Oncogene. 2004;23:3487–3494. doi: 10.1038/sj.onc.1207474. [DOI] [PubMed] [Google Scholar]
- 13.Gratias S, et al. Allelic loss in a minimal region on chromosome 16q24 is associated with vitreous seeding of retinoblastoma. Cancer Res. 2007;67:408–416. doi: 10.1158/0008-5472.CAN-06-1317. [DOI] [PubMed] [Google Scholar]
- 14.Saffroy R, et al. Analysis of alterations of WFDC1, a new putative tumour suppressor gene, in hepatocellular carcinoma. Eur. J. Hum. Genet. 2002;10:239–244. doi: 10.1038/sj.ejhg.5200795. [DOI] [PubMed] [Google Scholar]
- 15.Kurose K, et al. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat. Genet. 2002;32:355–357. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- 16.Watson JE, et al. Molecular analysis of WFDC1/ps20 gene in prostate cancer. Prostate. 2004;61:192–199. doi: 10.1002/pros.20100. [DOI] [PubMed] [Google Scholar]
- 17.Milyavsky M, et al. Prolonged culture of telomerase-immortalized human fibroblasts leads to a premalignant phenotype. Cancer Res. 2003;63:7147–7157. [PubMed] [Google Scholar]
- 18.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 19.Vaziri H, et al. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, et al. Myc activates telomerase. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, et al. Human endothelial cell life extension by telomerase expression. J. Biol. Chem. 1999;274:26141–26148. doi: 10.1074/jbc.274.37.26141. [DOI] [PubMed] [Google Scholar]
- 22.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 23.Gabbiani G. The myofibroblast in wound healing and fibrocontractive diseases. J. Pathol. 2003;200:500–503. doi: 10.1002/path.1427. [DOI] [PubMed] [Google Scholar]
- 24.Tsukada T, et al. HHF35, a muscle actin-specific monoclonal antibody. II. Reactivity in normal, reactive, and neoplastic human tissues. Am. J. Pathol. 1987;127:389–402. [PMC free article] [PubMed] [Google Scholar]
- 25.Ronnov-Jessen L, et al. Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiol. Rev. 1996;76:69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- 26.Tuxhorn JA, et al. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin. Cancer Res. 2002;8:2912–2923. [PubMed] [Google Scholar]
- 27.Ayala G, et al. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin. Cancer Res. 2003;9:4792–4801. [PubMed] [Google Scholar]
- 28.BRB Array Tool Version 3.5.0 Beta 2. (2006) http://linus.nci.nih.gov/BRB-ArrayTools.html. [Google Scholar]
- 29.Dennis G, Jr, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 30.Cluster Software 3.0. http://bonsai.ims.u-tokyo.ac.jp. [Google Scholar]
- 31.Kioi M, et al. Interleukin-13 receptor alpha2 chain: a potential biomarker and molecular target for ovarian cancer therapy. Cancer. 2006;107:1407–1418. doi: 10.1002/cncr.22134. [DOI] [PubMed] [Google Scholar]
- 32.Edwards S, et al. Expression analysis onto microarrays of randomly selected cDNA clones highlights HOXB13 as a marker of human prostate cancer. Br. J. Cancer. 2005;92:376–381. doi: 10.1038/sj.bjc.6602261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Rawi MA, et al. Interleukin-7 (IL-7) and IL-7 receptor (IL-7R) signalling complex in human solid tumours. Histol. Histopathol. 2003;18:911–923. doi: 10.14670/HH-18.911. [DOI] [PubMed] [Google Scholar]
- 34.Kim CJ, et al. Periostin is down-regulated in high grade human bladder cancers and suppresses in vitro cell invasiveness and in vivo metastasis of cancer cells. Int. J. Cancer. 2005;117:51–58. doi: 10.1002/ijc.21120. [DOI] [PubMed] [Google Scholar]
- 35.Yoshioka N, et al. Suppression of anchorage-independent growth of human cancer cell lines by the TRIF52/periostin/OSF-2 gene. Exp. Cell Res. 2002;279:91–99. doi: 10.1006/excr.2002.5590. [DOI] [PubMed] [Google Scholar]
- 36.Lochter A, et al. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J. Cell Biol. 1997;139:1861–1872. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parrinello S, et al. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J. Cell Sci. 2005;118:485–496. doi: 10.1242/jcs.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kogan I, et al. hTERT-immortalized prostate epithelial and stromal-derived cells: an authentic in vitro model for differentiation and carcinogenesis. Cancer Res. 2006;66:3531–3540. doi: 10.1158/0008-5472.CAN-05-2183. [DOI] [PubMed] [Google Scholar]
- 39.Milyavsky M, et al. Transcriptional programs following genetic alterations in p53, INK4A, and H-Ras genes along defined stages of malignant transformation. Cancer Res. 2005;65:4530–4543. doi: 10.1158/0008-5472.CAN-04-3880. [DOI] [PubMed] [Google Scholar]
- 40.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapointe J, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl Acad. Sci. USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanaja DK, et al. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–3882. [PubMed] [Google Scholar]
- 43.Wachi S, et al. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez-Carbayo M, et al. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J. Clin. Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 45.Bredel M, et al. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–8689. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 46.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 47.Campisi J, et al. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 48.Weinberg RA. The Biology of Cancer. 2007. [Google Scholar]
- 49.Gerdes MJ, et al. Localization of transforming growth factor-beta1 and type II receptor in developing normal human prostate and carcinoma tissues. J. Histochem. Cytochem. 1998;46:379–388. doi: 10.1177/002215549804600312. [DOI] [PubMed] [Google Scholar]
- 50.Eastham JA, et al. Transforming growth factor-beta 1: comparative immunohistochemical localization in human primary and metastatic prostate cancer. Lab. Invest. 1995;73:628–635. [PubMed] [Google Scholar]
- 51.Steiner MS, et al. Transforming growth factor-beta 1 overproduction in prostate cancer: effects on growth in vivo and in vitro. Mol. Endocrinol. 1992;6:15–25. doi: 10.1210/mend.6.1.1738367. [DOI] [PubMed] [Google Scholar]
- 52.McAlhany SJ, et al. Promotion of angiogenesis by ps20 in the differential reactive stroma prostate cancer xenograft model. Cancer Res. 2003;63:5859–5865. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.