Abstract
Aging has been associated with damage accumulation in the genome and with increased cancer incidence. Reactive oxygen species (ROS) are produced from endogenous sources, most notably the oxidative metabolism in the mitochondria, and from exogenous sources, such as ionizing radiation. ROS attack DNA readily, generating a variety of DNA lesions, such as oxidized bases and strand breaks. If not properly removed, DNA damage can be potentially devastating to normal cell physiology, leading to mutagenesis and/or cell death, especially in the case of cytotoxic lesions that block the progression of DNA/RNA polymerases. Damage-induced mutagenesis has been linked to various malignancies. The major mechanism that cells use to repair oxidative damage lesions, such as 8-hydroxyguanine, formamidopyrimidines, and 5-hydroxyuracil, is base excision repair (BER). The BER pathway in the nucleus is well elucidated. More recently, BER was shown to also exist in the mitochondria. Here, we review the association of BER of oxidative DNA damage with aging, cancer and other diseases.
DNA damage and repair in mammalian cells
The genome of eukaryotic cells is under continuous attack from a variety of DNA-damaging agents (endogenous and exogenous), which lead to many types of DNA lesions (Table I). DNA damage that can occur include single-strand breaks (SSBs), double-strand breaks (DSBs), mismatches, chemical modifications of the bases or sugars and inter- or intrastrand cross-links (1–4). To avoid the deleterious consequences of damage accumulation, multiple DNA repair pathways have evolved, each associated with specific classes of lesions (5–8). The major source of endogenous DNA damage is reactive oxygen species (ROS) generated from normal cellular metabolism (9,10). Exogenous sources of DNA damage include environmental agents such as ultraviolet light, ionizing radiation (IR), chemicals, toxins and pollutants (11–13). Unlike proteins, lipids and RNA, DNA cannot be replaced when damaged and thus must be repaired (14–16). If the damage is not repaired, the cell may resort to induction of apoptosis or necrosis, so that the mutations are not passed on to progeny cells and do not result in disease (especially cancer). In fact, many DNA-damaging agents are used in cancer therapy to induce apoptosis of tumor cells (17–20).
Table I.
Summary of DNA lesions
| DNA damage source | Type of lesion | DNA repair pathway |
| ROS, IR, alkylating agents | Altered base, abasic site, SSBs | BER |
| Ultraviolet light, cisplatin intrastrand adducts, polyaromatic hydrocarbons | Intrastrand cross-links, bulky DNA adducts | NER |
| IR, alkylating agents, cisplatin | Interstand cross-links, DSBs | DSB repair (recombinationalrepair and end joining) |
| Replication errors | Base mismatches, insertions and deletions | Mismatch repair |
The cellular response to DNA damage is co-ordinated, in dividing cells, by cell cycle checkpoints. Cell cycle progression (proliferation) is stopped by these checkpoints to allow the DNA repair machinery time to correctly repair the damage (5). The predominant repair pathways in mammalian cells are base excision repair (BER) (Figure 1), nucleotide excision repair (NER), DSB repair and mismatch repair. BER recognizes and repairs base modifications, as well as abasic sites and DNA SSBs (8,21,22). Many, but not all, of the DNA lesions repaired by BER are products of ROS attack. Some of these oxidative lesions are formed at high rates, even in the absence of exogenous DNA-damaging agents. For example, it is estimated that 100–500 8-hydroxyguanine (8-oxoG) lesions are formed per day in a human cell (10). The formamidopyrimidine lesions 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) and 4,6-diamino-5-formamidopyrimidine are formed at similar rates as 8-oxoG after oxidative stress (23–25). The other repair pathways are also important in response to DNA damage, acting to repair a variety of lesions, several of which can also result from ROS activity. NER is the major repair pathway for removal of bulky DNA adducts, which distort the normal structure of the DNA helix. These can be formed by ultraviolet radiation (yielding adducts like thymidine dimers or 6–4 photoproducts), chemicals or ROS (26,27). Such lesions strongly disrupt transcription and replication. DSB repair is the pathway used to repair DNA sites in which both strands of the helix have been broken on the sugar phosphate backbone in relatively close proximity to each other (thus forming a DSB) (3,28). These lesions are severely genotoxic and can be caused by IR, ROS and chemicals, but also during replication as a result of replication fork arrest and collapse (7). Mismatch repair is important for repair of mismatched bases or small insertion–deletion loops that result from replication errors or polymerase slippage (29,30).
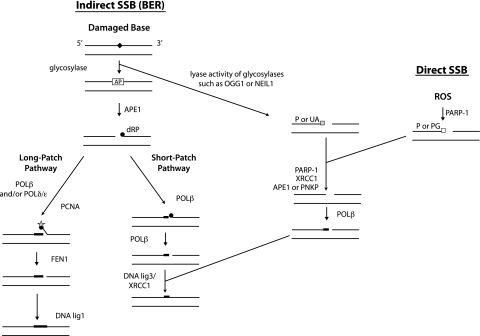
Fig. 1.
Single-strand break repair pathways. BER corrects damaged bases by first generating SSBs, through the activity of glycosylases. An alternative pathway corrects SSBs that arise directly. One of the most common sources of direct SSBs is oxidative attack by endogenous ROS. Filled diamond, damaged or non-conventional bases (e.g. uracil); AP, apurinic/apyrimidinic site; filled circles, dRP abasic residue; circle with star, abasic residue that is refractory to POLβ lyase activity; thick lines, incorporated nucleotides; P, phosphate; UA, unsaturated aldehyde; PG, 3'-phosphoglycolate.
The focus of this review is on the roles of oxidative damage and repair in carcinogenesis and the aging process. The BER pathway, the major pathway for repair of ROS-generated lesions, is described further below, as well as its importance in cancer prevention and its association with aging.
Sources and activity of ROS
The term ROS refers to a group of molecules (such as peroxides and free radicals) derived from oxygen that are highly reactive toward biomolecules. Free radicals are any atom or molecule that contains one or more unpaired electrons. They are usually more reactive than the corresponding non-radicals because they can act as oxidizing agents. ROS are constantly generated in living organisms as by-products of cellular metabolism, but can also be produced as a consequence of IR, chemotherapeutic drugs and environmental exposure to transition metals and chemical oxidants (31–34). ROS can randomly react with lipids, proteins and nucleic acids causing oxidative stress and damage in these macromolecules, leading to pathogenesis of age-related and chronic diseases including cancer (35–42). A significant portion of the extensive daily DNA damage occurring in each cell of the body is caused by ROS (43). During oxidative metabolism in mitochondria, most of the oxygen is converted to water, but ∼0.2–2% is converted to ROS (44,45); respiratory complexes I and III leak electrons directly to oxygen, producing superoxide anion (O2·−). Although relatively weak in aqueous media, O2·− can be converted to hydrogen peroxide (either spontaneously or catalyzed by superoxide dismutase), which in turn can be fully reduced to water or partially reduced to hydroxyl radical (OH·), a very powerful oxidant (46). O2·− may also react with nitric oxide to form the strong oxidant peroxynitrate (ONOO−). Despite its lesser reactivity than other ROS, hydrogen peroxide has important roles in oxidative damage and carcinogenesis since it is especially stable and diffuses easily through biological membranes, thus reaching other cellular compartments further producing cellular injury [for example, nuclear DNA (nDNA) damage], especially if it becomes converted to highly reactive OH· (47,48).
The first line of defense from the damaging effects of ROS is antioxidants, which convert the oxidants to less reactive species. Oxidative stress increases as the ratio of steady-state concentration of oxidants to antioxidant increases (49). The cellular response to oxidative damage includes DNA repair, cell cycle arrest and apoptosis (50–54). Irreversible mutations contribute to oncogenesis (55). ROS also have roles in physiological regulation of numerous cellular processes. For example, nitric oxide, a short-lived (t1/2 = up to 30 s) highly diffusible free radical generated from L-arginine (56), acts as a second messenger to regulate a variety of physiological processes such as platelet adhesion, neurotransmission and vascular permeability (57–59). Nitric oxide is not highly reactive to macromolecules but, as mentioned above, can become converted the highly reactive oxidant, ONOO− by reacting with O2·− (60).
A significant consequence of oxidative stress is DNA lesions, which result in genomic instability and various pathologies (22,61,62). ROS generate a variety of DNA lesions, including oxidized DNA bases, abasic sites, SSBs and DSBs (63). There are >100 types of oxidative base modifications in mammalian DNA (64,65). Studies have suggested that mitochondrial DNA (mtDNA) may accumulate more oxidative DNA damage than nDNA (66,67). This has been ascribed to localization of the mtDNA near the inner mitochondrial membrane where oxidants are formed, as well as lack of histones and diminished DNA repair activities (68). DNA bases are sensitive to ROS oxidation, particularly guanine due to its low redox potential (69). So, not surprisingly, 8-oxoG is one of the most abundant and well-characterized DNA lesions generated by ROS (reviewed in ref. 70). It has been estimated that ∼180 guanines are oxidized to 8-oxoG per mammalian cell per day (10). 8-oxoG is a highly mutagenic miscoding lesion that can lead to G:C to T:A transversion mutations (71).
Mechanisms of the BER response to oxidative DNA damage in the nucleus
The BER pathway is the primary mechanism for repair of oxidative base lesions, such as 8-oxoG and formamidopyrimidines (4,6-diamino-5-formamidopyrimidine, FapyG). Other base modifications repaired by BER include uracil and 3-methyladenine, resulting from cytosine deamination and alkylating agents, respectively (72,73). BER is a critical process for genomic maintenance, as highlighted by the severe phenotypes seen in animals deficient in BER function, including cancer, premature aging and metabolic defects (74,75). BER has alternative pathways depending on the damage and the responsive enzymes, as described below and illustrated in Figure 1. During short-patch BER, only one new nucleotide is incorporated in the place of the damaged base; during long-patch BER, from two to six new nucleotides are incorporated.
The initial step in BER uses DNA glycosylases, which cleave the N-glycosyl bond between the sugar and the base, thus releasing the damaged base to form an abasic site, also termed apurinic/apyrimidinic (AP) site. Abasic sites can also arise through spontaneous depurination and depyrimidination (76,77). The efficient repair of abasic sites is critical because they are highly mutagenic (78,79). There are several different glycosylases specific for certain lesions. Individual glycosylases may recognize more than one type of damage, and each specific modification may be recognized by more than one type of glycosylase, giving a degree of redundancy in the process. For example, OGG1 has strong specificity to 8-oxoG and FapyG lesions, whereas NEIL1 efficiently removes FapyG and 4,6-diamino-5-formamidopyrimidine (80,81). The BER target lesions (glycosylase-specific damage products) are classified as follows: oxidized/reduced bases (such as 8-oxoG or formamidopyrimidines), alkylated bases (typically methylated), deaminated bases (such as uracil) or base mismatches (7).
DNA glycosylases are classified as mono- or bifunctional, depending on their reaction mechanisms. Monofunctional glycosylases (having only glycosylase activity; such as UNG) require another enzyme (predominantly APE1) for the incision of the resulting abasic sugar residue [leaving behind a 5′-deoxyribose phosphate (dRP)]. If the glycosylase is bifunctional (having both glycosylase and AP lyase activity; such as OGG1 or NEIL1), then both base excision and an incision 3′ to the AP site can occur (82), resulting in a SSB that harbors a 3′-α,β-unsaturated aldehyde (for example from OGG1 activity) or 3′-phosphate (for example from NEIL1 activity). SSBs can also arise directly by ROS-induced disintegration of oxidized deoxyribose, generating a 3′-phosphoglycolate terminus. The SSB requires removal of the altered 3′-terminal groups prior to polymerization and/or ligation. The enzymes APE1 (82–84) and polynucleotide kinase (85,86) have the ability to remove 3′-obstructive termini, particularly unsaturated aldehyde and phosphate groups, respectively. APE1 also has the ability to remove 3′-phosphoglycolate groups generated during direct SSBs Accessory proteins such as poly(ADP-ribose)polymerase-1 (1,87) and XRCC1 (1,88) aid in this process by forming complexes with repair proteins at the SSBs and helping with repair protein recruitment and chromatin modification. After removal of obstructive termini or APE incision 5′ to the AP site, replacement of the excised nucleotide is performed by polymerase (POLβ) (89). This enzyme also removes the dRP group left behind by APE1 incision (Figure 1, short-patch pathway). Often if the 5′ terminal is refractory to this POLβ AP lyase activity, strand displacement synthesis is required by way of incorporation of multiple nucleotides (Figure 1, long-patch pathway); in this case, several enzymes (proliferating cell nuclear antigen, FEN1 and POLβ and/or POLδ/ε) act together to remove the blocking terminus. The final step of BER entails ligation of the remaining nick, by either LIG1 alone or LIG3–XRCC1 complex (90).
There is evidence of co-operation between the enzymes at successive steps in BER. For example, studies indicate that, in short-patch BER, a specific interaction of APE1 and POLβ occurs that assembles the polymerase onto the AP site in DNA, serving to accelerates the excision of 5′-terminal dRP by POLβ (89,91). Besides the core enzymes that participate in BER, several other protein factors have been identified to modulate BER activity. Such auxiliary proteins interact with the essential BER proteins and/or the DNA to enhance the enzyme activity or efficiency of the reactions (92).
BER in the mitochondria
Although much attention has been paid to the effects of BER on nDNA, mitochondria harbor a critical component of the total cellular DNA and should therefore not be overlooked. Recent work underscores the differences in the manner that oxidative DNA damage is handled in the mitochondria as compared with the nucleus (93). Moreover, mitochondrial and nuclear mutations have been linked to aging and several neurodegenerative diseases (94–100). Therefore, it is of great interest to determine similarities and differences between nuclear and mitochondrial repair mechanisms.
Within each mammalian cell, there are hundreds to thousands of mitochondria, each containing two to five copies of circular supercoiled DNA. The mtDNA encodes components of the electron transport chain including 13 structural genes, 22 transfer RNAs and 2 ribosomal RNAs. Although limited information is available about the molecular organization of DNA in mammalian mitochondria, it is known that mtDNA are not protected by histones but instead associated with the inner membrane, near the ROS-generating electron transport chain (101). This localization makes mtDNA more prone to damage than nDNA. It has been observed that mtDNA contain a higher steady-state level of oxidative damage compared with nDNA (64,102). Although it was originally suggested that mitochondria had no DNA repair (103), later findings demonstrated that mammalian mitochondria efficiently remove DNA damage repaired by BER (104–106).
Several BER proteins have been identified in the mitochondria and these are generally either identical to nDNA repair proteins or isoforms of nuclear proteins arising from differential splicing or alternative transcription initiation sites (107,108). Nuclear and mitochondrial adenine-DNA-glycosylases are generated by alternatively spliced forms of the human MYH gene (109). In addition, the human NTH1 protein (glycosylase that is involved in repair of oxidized pyrimidines) has a putative mitochondrial targeting sequence and has been found both in nuclear and mitochondrial extracts (110). The human OGG1 gene produces two major distinct transcripts via alternative splicing: α-OGG1 and β-OGG1. The α-OGG1 protein localizes mainly in nuclei, whereas β-OGG1 appears to be localized exclusively in mitochondria. The β-OGG1 isoform does not have any measurable glycosylase activity; it lacks the C-terminal α-O helix present in α-OGG1 that is essential for 8-oxoG DNA glycosylase activity (111). Mitochondrial and nuclear forms of uracil DNA glycosylase (UDG, also termed UNG) are encoded by alternative splicing and transcription from different positions in the UNG gene (112). The APE1 protein exerts repair activity in both nuclei and mitochondria (113), and recent evidence suggests that the mitochondrial form is an N-terminal truncation of the full-length APE1 (114). In vitro studies have demonstrated that POLγ, the only DNA polymerase present in mitochondria (115,116), has both polymerase and dRP lyase activity and is therefore the major enzyme for the repair synthesis step in BER (117). Very recently, three studies have reported the presence of long-patch BER in mitochondria (118–120). While these studies all find long-patch repair, two of them (119,120) also find the presence of FEN1 in the mitochondrial extracts from human cells, whereas the third (118) detects flap endonuclease activity, but suggests it is not from FEN1.
Association of oxidative DNA damage and BER with cancer and aging
ROS-induced DNA damage is believed to contribute to carcinogenesis, aging and neurodegeneration. Oxidative damage to DNA may lead to mutations that activate oncogenes or inactivate tumor suppressor genes (32,121). Many DNA repair pathways, as well as other cellular stress response pathways, such as cell cycle arrest and apoptosis, determine the probability of genetic alterations turning into neoplastic events. Specific DNA lesions have been strongly implicated in tumorigenesis. For example, high levels of 8-oxoG and other oxidative lesions have been found in urine and tumor tissue DNA from patients with a variety of malignancies (39,122–126). In addition, antioxidant and repair enzymes activities may be altered in cancers (126–130). Thus, one can envision that BER could be of critical importance in cancer prevention.
Many experimental approaches have linked ROS-generated oxidative DNA damage and the associated avoidance and repair processes to carcinogenesis. Early studies showed that exposure of mouse fibroblasts to ROS can lead to cellular transformation (131). However, ROS have a wide range of potential cellular effects, depending on the dose and cellular environment. For example, ROS can also induce cellular senescence or apoptosis, which are considered anticancer mechanisms since they inhibit cell proliferation or kill the cell. A more definitive approach to understanding the role of ROS in cancer is to increase ROS levels by inhibiting antioxidant defenses. Mice deficient (by gene knockout) in the antioxidant superoxide dismutase enzyme can have dramatic phenotypes ranging from increased rates of cancer to death (soon after birth) (130,132). In addition, mice deficient in the antioxidant catalase and simultaneously restricted in nutritional antioxidant vitamin E had an elevated incidence of mammary tumors (128).
Carcinoma cells are characterized by persistent oxidative stress and high levels of ROS. Suggested contributions to this include an altered metabolic pathway involving nicotinamide adenine dinucleotide phosphate oxidase, tumor-associated macrophages delivering sublethal oxidative stress and an inadequate tumor vascular network (133,134). For example, human tumor cells in vitro have a much enhanced production of hydrogen peroxide relative to non-transformed cell lines (135). Also, high levels of 8-oxoG have been found in human renal carcinomas (136) and in breast carcinoma cells (134) relative to respective normal tissue. Reports also indicate that cancer cells have a suppressed antioxidant system. For example, catalase gene expression has been shown to be lowered in human hepatoma cells (137). Moreover, tumor cells are typically low in both manganese and copper–zinc superoxide dismutase (138). The effect of persistent oxidative stress on cancer cells is to further promote cancer growth and metastasis of the cell clones. One way it does this is through induction of DNA damage that may lead to further mutation. It may also activate growth-promoting transcription factors or modulate genes important in apoptosis or proliferation (39,139). In addition, it may damage certain protease inhibitors, thus allowing aberrant action of proteases involved in tumor invasion and metastasis (elastin, plasminogen activator and plasmin) (135). The chronic inflammation associated with carcinomas also stimulates large production of ROS, thus causing additional genetic instability. The high oxidative stress in tumor cells creates selective pressure for cells having enhanced growth, invasion and metastasis characteristics (36,134).
Aging is the highest risk factor for cancer (140). Considerable evidence suggests that oxidative stress has a role in the organismal aging process. The free radical theory of aging proposes that aging may be in part due to free radical-dependent cellular damage accumulation. The premise in this theory is that free radicals (through continuous attack of protein, lipids and DNA) are involved in producing the aging changes associated with the environment, disease and the intrinsic aging process (40,141,142). Many of the studies on oxidative damage and the aging process have been done on the short-lived invertebrates Drosophila melanogaster and Caenorhabditis elegans or on the mammalian model systems rats and mice (40). In the case of Drosophila and C.elegans, results strongly suggest that oxidative stress and antioxidant defenses play a critical role in life span. In rats and mice, caloric restriction (with associated reduced free radical production) significantly increases the average life span and slows age-related decline. There are several lines of evidence linking aging and genome maintenance pathways. Accelerated aging is observed in mice defective in DNA repair pathways, such as NER and DSB repair, telomere maintenance and mitochondrial genome replication (143). In the case of BER, the use of genetic defects to link BER to aging has been limited. Most BER proteins are necessary for embryonic development; so mice with designed knockout of key BER proteins (exceptions are the functionally redundant DNA glycosylases) are embryonic lethal. Defects in glycosylases and more mild defects in other BER genes (such as point mutations or haploinsufficiency) are generally connected to elevated cancer susceptibility (described in the next section) but have yielded less insight into the relationship between BER and aging. Recently, it was demonstrated that the mice deficient in the mammalian Sir2 homolog, SIRT6, a chromatin-associated protein required for normal BER maintenance of genomic integrity, display age-related degenerative phenotypes (74). Multiple factors can contribute to aging and life span, such as telomere shortening, hormone levels and multiple targets of ROS, making it difficult to establish a direct role of the BER enzymes in counteracting aging. In this respect, yeast is a simpler, more tractable, system since it is without hormones and does not inactivate telomerase. Studies in yeast have shown that defects in several BER enzymes shorten chronological life span (144).
Mitochondria are the major physiological site for the generation of ROS and play a key role in mediating apoptosis. Thus, the mitochondria's activity in producing oxidative stress (and associated damage to mtDNA) and in cellular turnover is thought to significantly contribute to human aging, cancer and neurodegeneration. The mitochondrial theory of aging proposes that mtDNA mutations caused by ROS accumulate in the cell, leading to damaged respiratory chain proteins, thereby generating more ROS, which in turn causes higher mutation rates (145). There are a variety of organisms with shortened or enhanced life span and associated alterations in mitochondrial metabolism or ROS generation (145). Moreover, accumulation of point mutations and deletions of mtDNA in a variety of tissues with age have been reported for humans (146), monkeys (147) and mice (148). Also, there are many reports indicating an association between mitochondrial metabolic/molecular alterations and a variety of human cancers (149). A recent study found that mtDNA mutations can contribute to tumor cell metastasis (150). While mitochondria accumulate a great amount of genetic damage, the existence of multiple copies of the mitochondrial genome per cell may render these organelles relatively tolerant to high levels of damaged DNA, through complementation by the remaining mitochondrial genomes. Mitochondrial BER has been shown to be important in disease and aging. For instance, dysfunction of the mitochondrial-specific BER enzyme POLγ has been associated with mtDNA mutation disorders, such as progressive external ophthalmoplegia, parkinsonism and Alpers syndrome (151). Also, mice lacking the DNA repair proofreading function of POLγ show signs of premature aging and accumulated mutations in their mtDNA (100). In addition, expression of β-OGG1 and incision of 8-oxoG were both reduced in mitochondrial extracts from the prostate cancer cell lines PC-3 and DU-145 (126). Moreover, defective BER of 8-oxoG in mitochondria of MCF-7 and MDA-MB-468 breast cancer cell lines has been reported (152). The many distinct alterations in mtDNA structure and function in cancer cells, relative to normal cells, make mtDNA an attractive molecular marker for early detection of cancer. In fact, mutant mtDNA is readily detectable in urine, blood and saliva samples from cancer patients (153).
The above-mentioned studies provide evidence that cellular responses to ROS modulate carcinogenesis and aging. Since BER is the main pathway for repair of oxidative lesions, investigations into relating specific BER enzymes to disease is justified. Some of these studies have been discussed above with respect to cancer and aging, including the role of mtDNA repair. More detailed clinical aspects are discussed below.
Clinical manifestations of alterations of BER pathway genes
The importance of the proper functioning of the BER pathway to sustain life is illustrated in the phenotypes seen in mouse knockout strains. Gross BER defects appear to be incompatible with life. Mouse knockouts of genes coding for core BER proteins, including XRCC1 (154), POLβ (155,156), APE/HAP1 (157), FEN1 (158) and DNA ligase I (159), are embryonic lethal. Other steps of the BER pathway may, however, be protected by redundancy. Whereas MYH and OGG1 knockout mice show very little phenotype, MYH/OGG1 double mutant mice show high susceptibility to tumor formation (160).
There are many human hereditary diseases associated with defective cellular responses to DNA damage. Principal defective responses include NER (examples include Xeroderma pigmentosum, Cockayne syndrome and Trichothiodystrophy), DNA strand break repair (Ataxia telangiectasia and Nijmegen breakage syndrome), helicase defects (Bloom, Werner and Rothmund-Thomson syndromes), mismatch repair (hereditary non-polyposis colon cancer), chromosomal stability (Fanconi anemia) and class switch recombination (Hyper-IGM syndrome). Cancer susceptibility is a common theme among individuals with DNA repair gene mutations. DNA damage compounded with defects in DNA repair can lead to genomic instability and consequential tumorigenesis. Autosomal recessive familial cancer-prone syndromes such as Ataxia telangiectasia, Bloom syndrome, Fanconi's anemia, Rothmund-Thomson syndrome, Werner's syndrome and Xeroderma pigmentosum are examples of how defects in DNA repair can lead to genomic instability.
Genetic diseases caused by mutations in BER pathway genes are less common than those caused by other DNA repair pathway genes. Interestingly, however, 30% of all human tumors examined have variant POLβ proteins, with approximately half having a single amino acid change (161). An early link between an inherited defect in BER and cancer was reported in 2002, when a family with a phenotype similar to familial adenomatous polyposis was shown to have mutations in the gene encoding the human MutY homolog (162). It appears that the MutY homolog mutations found caused a reduced capacity to initiate the repair of 8-oxoG•A mismatches, which probably led to an increased number of G–T transversions in the adenomatous polyposis coli gene that resulted in the inactivation of the adenomatous polyposis coli protein.
Recessive mutations of UNG in three patients have been found to be one cause of the primary immune deficiency disorder Hyper-IGM syndrome (163). In a pathway initiated by activation-induced cytidine deaminase, UNG is required to trigger somatic hypermutation and class switch recombination of antibody genes, which are involved in the generation of high-affinity antibodies. In these patients, recessive mutations were found in the catalytic domain shared by the mitochondrial and nuclear isoforms of UNG. B cells from these patients had a severe impairment in class switch recombination at a DNA precleavage step and the pattern of somatic hypermutation was also found to be abnormal. These patients have not, as yet, developed cancer, though 28% of older (>18 month) UNG knockout mice developed lymphomas compared with only 1.3% of control animals (164).
Two different miscoding mutations on separate alleles of LIG1 were found in a patient with lymphoma, growth retardation, sun sensitivity and immunodeficiency (165). A fibroblast strain derived from this patient showed retarded joining of Okazaki fragments, indicative of a defect in lagging strand DNA synthesis, and increased sensitivity to several DNA-damaging agents (166).
Mutations in DNA repair genes are also linked to neurodegeneration. Examples include Ataxia telangiectasia (ATM), Xeroderma pigmentosum (XPA-G), Cockayne syndrome (CSA and B) and Werner syndrome (WS) (167). In addition, Trichothiodystrophy, Bloom syndrome and Rothmund–Thomson syndrome all have mental retardation as part of their syndromes (167). It has been hypothesized that neurons from Alzheimer's disease patients may have a genetic defect in BER (168). In fact, increased 8-oxoG levels and mitochondrial deletions, as well as deficient BER, have been found in Alzheimer's disease patients (98,169,170). Because neuronal cells have high metabolic rates, it has been proposed that BER may play a more important role in repairing DNA damage in neurons (both in the nucleus and mitochondria) than in other cell types (171). Oxidative stress plays a significant role in neuronal cell death associated with a variety of neurodegenerative disease. Elevated levels of oxidative damage are seen in the cortical region of brains that have Lewy bodies and associated dementia (41) and in the chronic active plaques of patients with multiple sclerosis (172). Moreover, the mitochondrial isoform of OGG1 is elevated in the substantia nigra of Parkinson's patients (173); and APE1 expression and activity are decreased in amyotrophic lateral sclerosis (ALS) patients with sporadic ALS (174). Missense mutations in APE1 were also found in eight of 11 patients with sporadic and familial ALS (175). However, one report indicates that APE1 was increased in ALS patients (176).
More subtle associations between cancer and other BER genes are being explored through single-nucleotide polymorphism (SNP) association studies. Individuals with certain polymorphisms of BER genes may have a greater susceptibility to cancer after being exposed to environmental toxins. Polymorphisms may help explain why individuals with equal exposure to certain environmental carcinogens, such as cigarette smoke, may be at different risks for the development of cancer. The majority of lung cancers, for instance, are associated with smoking but only a small minority of smokers will develop the disease.
As with many clinical association studies involving SNPs, epidemiological findings for BER SNPs have been inconsistent. Several genes have been sufficiently investigated to where meta-analysis of prior studies of BER gene associations with cancer cohorts has been performed, though inconsistencies still exist. A meta-analysis of seven studies (>3000 cases and 3000 controls) by Hung et al. (177) revealed a 24% increased risk of lung cancer among subjects homozygous for the variant OGG1 Ser326Cys polymorphism (326Cys/Cys) [odds ratio (OR) = 1.24, 95% confidence interval (CI) = 1.01–1.53]. Experimental evidence has shown that this isoform exhibits decreased DNA glycosylase activity in the repair of 8-oxoG (178). In addition, subjects with the 326Cys/Cys genotype have higher levels of oxidized guanine in lymphocyte DNA following ex vivo treatment with sodium dichromate (179). However, when Kiyohara et al. (180) performed a meta-analysis, using an additional four studies to the original seven studies by Hung et al. (177), there was no longer an association between the OGG1 Ser326Cys polymorphism and lung cancer risk. The presence of heterogeneity in association studies may compromise the interpretation of meta-analysis and make conclusions difficult. Heterogeneity may result from variables such as ethnicity and study design. In the case of the OGG1 Ser326Cys polymorphism, further studies are warranted, with attention toward reducing heterogeneity.
Other BER genes have also been studied with meta-analysis, especially XRCC1. Polymorphisms of XRCC1 have been extensively evaluated in cancer association studies, in part because of their relative high frequency in the population. The three most frequently analyzed polymorphisms of XRCCI are Arg194Trp, Arg399Gln and Arg280His. In a meta-analysis of 16 studies of tobacco-related cancers, using ∼5000 cases and 6000 controls, individuals heterozygous for XRCC1 Arg194Trp were found to have a protective effect (OR = 0.86, 95% CI = 0.77–0.95) (177). This was in agreement with a large meta-analysis by Hu et al. (181) (38 case–control studies, ∼12 000 cancer cases and >14 000 controls), which also found a decreased cancer risk (OR = 0.89, 95% CI = 0.81–0.98) with the variant genotypes (Trp/Trp + Arg/Trp) compared with the wild-type Arg/Arg genotype. A mutagen sensitivity assay using bleomycin, which showed individuals with the wild-type allele had more chromosomal breaks per cell (182), also supports the findings of the meta-analyses. However, meta-analyses of studies examining specific cancers and the Arg194Trp polymorphism have not shown significant associations. When lung cancer risk alone was analyzed in two meta-analyses, one based on eight studies in nine different ethnic populations (180) and another based on five studies that also looked at aerodigestive tract cancers (including oral cavity and pharynx) (177), no significant associations were found. There was also no association between Arg194Trp and breast cancer risk in a meta-analysis of 11 studies (∼6000 cases and ∼6000 controls) of Caucasian and Asian populations (183).
The XRCC1 Arg399Gln polymorphism, on the other hand, has shown associations with specific cancers, especially in Asian populations, but not overall cancer risk. A meta-analysis of 17 studies analyzing this polymorphism and lung cancer risk (>7000 cases and ∼9000 controls) showed that individuals with the 399Gln/Gln genotype had an increased risk of lung cancer among Asians (OR = 1.34, 95% CI = 1.16–1.54) but not among Caucasians (180). In another meta-analysis of four studies (∼1600 cases and ∼1600 controls), Zhang et al. (183) found an association in Asian populations between the Arg399Gln polymorphism and breast cancer (OR = 1.6, 95% CI = 1.1–2.3). An accompanying meta-analysis by Zhang et al. (183) of eight studies (∼8000 cases and ∼8000 controls) did not find the same association in the Caucasian population. In terms of overall cancer risk, Hu et al. (181) did not find any association between the XRCC1 Arg399Gln polymorphism and a general risk of cancer.
In meta-analyses examining smoking associations with the XRCC1 Arg399Gln polymorphism, Hung et al. (177) showed that individuals homozygous for the variant polymorphism (Gln/Gln) appeared to be at increased risk of tobacco-related cancers among light smokers (six combined studies, OR = 1.38, 95% CI = 0.99–1.94) but decreased risk among heavy smokers (seven combined studies, OR = 0.71, 95% CI = 0.51–0.99). According to the authors of this study, the discrepancy may have resulted from different mechanisms affecting cells exposed to light versus heavy tobacco smoke. Hung et al. (177) note that the XRCC1 399Gln allele has been shown to be associated with higher mutagen sensitivity (182) and higher levels of DNA adducts (184) that may explain the increased risk of tobacco-related cancers among light smokers. Possible explanations for the decreased risk among heavy smokers include enhanced apoptosis at the time of cell division from heavy tobacco smoking or induction of DNA repair capacity in response to DNA damage.
The XRCC1 Arg280His polymorphism has had conflicting results by meta-analysis. In two separate meta-analyses (177,180) studies, there was no association between cancer risk and the XRCC1 Arg280His polymorphism. However, the meta-analysis of 38 case–control studies by Hu et al. (181) did find an overall increase risk of cancer (OR = 1.19, 95% CI = 1.00–1.42) for the variant genotypes (His/His + Arg/His) of the Arg280His polymorphism compared with the wild-type homozygote.
One other meta-analysis has been reported for a BER gene. A meta-analysis of the APE1 polymorphism Asp148Glu (166) combining ∼1400 cases and 1700 controls from four studies showed no association with lung/upper aerodigestive tract cancer risk.
Many single cohort or larger consortium-based SNP association studies without further meta-analysis have been performed looking for associations of various BER pathway genes with overall cancer risk or for specific cancers. Studies have also been performed looking at polymorphisms with longevity (and specifically for BER, no association was seen in a study of XRCCI polymorphisms) (185). As with polymorphism studies in general, repeated analysis in different populations will be necessary to draw reliable conclusions. Future meta-analyses will be needed as studies become available.
Conclusions
There is extensive evidence indicating that ROS damage to nDNA, mtDNA, proteins and lipids is associated with disease and aging. DNA damage accumulation leads to genomic instability resulting in cancer and age-associated disorders. Many of the studies revealing these connections involve modifying genes in model organisms (especially mice) or treating cultured mouse or human cells with oxidants, antioxidants or agents that alter specific processes (especially DNA repair pathways). It is important, however, to keep in mind that associations found in mouse models may not hold true to disease associations in humans since the damage response pathways may differ between species. Moreover, conclusions based on cultured human cells may not reflect the processes or responses in vivo. Direct relationships between BER and disease manifestations have been difficult to establish, probably due to the critical role of BER in maintaining genomic integrity, as demonstrated by the fact that full knockouts of all the core BER proteins (APE1, POLβ, FEN1 and XRCC1) result in embryonic lethality. More connections have been made between impaired NER and human disease. However, work with BER SNPs and meta-analysis has provided extensive circumstantial evidence for a role of BER SNPs in human disease manifestations. Since it is now evident that there is effective BER in mammalian mitochondria, the crucial importance of BER in mtDNA integrity and disease manifestations can no longer be overlooked.
Funding
National Institute on Aging/National Institutes of Health Intramural Research Program.
Acknowledgments
We would like to thank Dr Anne-Cecile Bayne and Maria Aamann for proofreading this manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- AP
apurinic/apyrimidinic
- BER
base excision repair
- CI
confidence interval
- dRP
5′-deoxyribose phosphate
- DSB
double-strand break
- FapyG
2,6-diamino-4-hydroxy-5-formamidopyrimidine
- IR
ionizing radiation
- mtDNA
mitochondrial DNA
- nDNA
nuclear DNA
- NER
nucleotide excision repair
- OR
odds ratio
- 8-oxoG
8-hydroxyguanine
- POL
polymerase
- ROS
reactive oxygen species
- SNP
single-nucleotide polymorphism
- SSB
single-strand break
References
- 1.Caldecott KW. Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair (Amst) 2007;6:443–453. doi: 10.1016/j.dnarep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 2.de BJ, et al. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 3.Helleday T, et al. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel TA, et al. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 5.Houtgraaf JH, et al. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc. Revasc. Med. 2006;7:165–172. doi: 10.1016/j.carrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Larsen NB, et al. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion. 2005;5:89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Sancar A, et al. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DM, III, et al. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic. Biol. Med. 2002;32:804–812. doi: 10.1016/s0891-5849(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 11.Fry RC, et al. Genome-wide responses to DNA-damaging agents. Annu. Rev. Microbiol. 2005;59:357–377. doi: 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- 12.Norbury CJ, et al. Cellular responses to DNA damage. Annu. Rev. Pharmacol. Toxicol. 2001;41:367–401. doi: 10.1146/annurev.pharmtox.41.1.367. [DOI] [PubMed] [Google Scholar]
- 13.Spry M, et al. DNA repair pathways and hereditary cancer susceptibility syndromes. Front. Biosci. 2007;12:4191–4207. doi: 10.2741/2380. [DOI] [PubMed] [Google Scholar]
- 14.Ganong BR. Roles of lipid turnover in transmembrane signal transduction. Am. J. Med. Sci. 1991;302:304–312. [PubMed] [Google Scholar]
- 15.Meyer S, et al. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 16.Shringarpure R, et al. Protein turnover by the proteasome in aging and disease. Free Radic. Biol. Med. 2002;32:1084–1089. doi: 10.1016/s0891-5849(02)00824-9. [DOI] [PubMed] [Google Scholar]
- 17.Altucci L, et al. Retinoids and TRAIL: two cooperating actors to fight against cancer. Vitam. Horm. 2004;67:319–345. doi: 10.1016/S0083-6729(04)67017-8. [DOI] [PubMed] [Google Scholar]
- 18.Crighton D, et al. Splicing DNA-damage responses to tumour cell death. Biochim. Biophys. Acta. 2004;1705:3–15. doi: 10.1016/j.bbcan.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Sedletska Y, et al. Cisplatin is a DNA-damaging antitumour compound triggering multifactorial biochemical responses in cancer cells: importance of apoptotic pathways. Curr. Med. Chem. Anticancer Agents. 2005;5:251–265. doi: 10.2174/1568011053765967. [DOI] [PubMed] [Google Scholar]
- 20.Sun SY, et al. Apoptosis as a novel target for cancer chemoprevention. J. Natl Cancer Inst. 2004;96:662–672. doi: 10.1093/jnci/djh123. [DOI] [PubMed] [Google Scholar]
- 21.Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat. Res. 2005;591:45–59. doi: 10.1016/j.mrfmmm.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 22.Seeberg E, et al. The base excision repair pathway. Trends Biochem. Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 23.Dizdaroglu M, et al. Modification of DNA bases in chromatin of intact target human cells by activated human polymorphonuclear leukocytes. Cancer Res. 1993;53:1269–1272. [PubMed] [Google Scholar]
- 24.Kasprzak KS, et al. Oxidative DNA base damage and its repair in kidneys and livers of nickel(II)-treated male F344 rats. Carcinogenesis. 1997;18:271–277. doi: 10.1093/carcin/18.2.271. [DOI] [PubMed] [Google Scholar]
- 25.Mori T, et al. DNA base damage generated in vivo in hepatic chromatin of mice upon whole body gamma-irradiation. Int. J. Radiat. Biol. 1993;64:645–650. doi: 10.1080/09553009314551881. [DOI] [PubMed] [Google Scholar]
- 26.Costa RM, et al. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85:1083–1099. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 27.Leibeling D, et al. Nucleotide excision repair and cancer. J. Mol. Histol. 2006;37:225–238. doi: 10.1007/s10735-006-9041-x. [DOI] [PubMed] [Google Scholar]
- 28.Cahill D, et al. Mechanisms of eukaryotic DNA double strand break repair. Front. Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 29.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schofield MJ, et al. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 31.Ercal N, et al. Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr. Top. Med. Chem. 2001;1:529–539. doi: 10.2174/1568026013394831. [DOI] [PubMed] [Google Scholar]
- 32.Klaunig JE, et al. The role of oxidative stress in carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 33.Kovacic P, et al. Mechanisms of anti-cancer agents: emphasis on oxidative stress and electron transfer. Curr. Pharm. Des. 2000;6:277–309. doi: 10.2174/1381612003401046. [DOI] [PubMed] [Google Scholar]
- 34.Wink DA, et al. Chemical biology of nitric oxide: regulation and protective and toxic mechanisms. Curr. Top. Cell. Regul. 1996;34:159–187. doi: 10.1016/s0070-2137(96)80006-9. [DOI] [PubMed] [Google Scholar]
- 35.Chakravarti B, et al. Oxidative modification of proteins: age-related changes. Gerontology. 2007;53:128–139. doi: 10.1159/000097865. [DOI] [PubMed] [Google Scholar]
- 36.Cooke MS, et al. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 37.Evans MD, et al. Oxidative DNA damage and disease: induction, repair and significance. Mutat. Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Filipcik P, et al. The role of oxidative stress in the pathogenesis of Alzheimer's disease. Bratisl. Lek. Listy. 2006;107:384–394. [PubMed] [Google Scholar]
- 39.Karihtala P, et al. Reactive oxygen species and antioxidant mechanisms in human tissues and their relation to malignancies. APMIS. 2007;115:81–103. doi: 10.1111/j.1600-0463.2007.apm_514.x. [DOI] [PubMed] [Google Scholar]
- 40.Kregel KC, et al. An integrated view of oxidative stress in aging: basic mechanisms, functional effects, and pathological considerations. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;292:R18–R36. doi: 10.1152/ajpregu.00327.2006. [DOI] [PubMed] [Google Scholar]
- 41.Lyras L, et al. Oxidative damage to proteins, lipids, and DNA in cortical brain regions from patients with dementia with Lewy bodies. J. Neurochem. 1998;71:302–312. doi: 10.1046/j.1471-4159.1998.71010302.x. [DOI] [PubMed] [Google Scholar]
- 42.Thomson A, et al. Oxidative stress and antioxidants in intestinal disease. Dig. Dis. 1998;16:152–158. doi: 10.1159/000016859. [DOI] [PubMed] [Google Scholar]
- 43.Lindahl T, et al. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 44.Chance B, et al. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 45.Hansford RG, et al. Dependence of H2O2 formation by rat heart mitochondria on substrate availability and donor age. J. Bioenerg. Biomembr. 1997;29:89–95. doi: 10.1023/a:1022420007908. [DOI] [PubMed] [Google Scholar]
- 46.Turrens JF. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boveris A, et al. Mitochondrial metabolic states regulate nitric oxide and hydrogen peroxide diffusion to the cytosol. Biochim. Biophys. Acta. 2006;1757:535–542. doi: 10.1016/j.bbabio.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 48.Ray G, et al. Oxidants, antioxidants and carcinogenesis. Indian J. Exp. Biol. 2002;40:1213–1232. [PubMed] [Google Scholar]
- 49.Sies H. Oxidative stress: oxidants and antioxidants. Exp. Physiol. 1997;82:291–295. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- 50.Halliwell B. Reactive oxygen species and the central nervous system. J. Neurochem. 1992;59:1609–1623. doi: 10.1111/j.1471-4159.1992.tb10990.x. [DOI] [PubMed] [Google Scholar]
- 51.Koppal T, et al. Peroxynitrite-induced alterations in synaptosomal membrane proteins: insight into oxidative stress in Alzheimer's disease. J. Neurochem. 1999;72:310–317. doi: 10.1046/j.1471-4159.1999.0720310.x. [DOI] [PubMed] [Google Scholar]
- 52.Markesbery WR, et al. Oxidative alterations in Alzheimer's disease. Brain Pathol. 1999;9:133–146. doi: 10.1111/j.1750-3639.1999.tb00215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz JB, et al. Potassium deprivation-induced apoptosis of cerebellar granule neurons: a sequential requirement for new mRNA and protein synthesis, ICE-like protease activity, and reactive oxygen species. J. Neurosci. 1996;16:4696–4706. doi: 10.1523/JNEUROSCI.16-15-04696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.See V, et al. Oxidative stress induces neuronal death by recruiting a protease and phosphatase-gated mechanism. J. Biol. Chem. 2001;276:35049–35059. doi: 10.1074/jbc.M104988200. [DOI] [PubMed] [Google Scholar]
- 55.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 56.Nathan C, et al. Regulation of biosynthesis of nitric oxide. J. Biol. Chem. 1994;269:13725–13728. [PubMed] [Google Scholar]
- 57.Davis KL, et al. Novel effects of nitric oxide. Annu. Rev. Pharmacol. Toxicol. 2001;41:203–236. doi: 10.1146/annurev.pharmtox.41.1.203. [DOI] [PubMed] [Google Scholar]
- 58.Moncada S, et al. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 1991;43:109–142. [PubMed] [Google Scholar]
- 59.Palmer RM, et al. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature. 1987;327:524–526. doi: 10.1038/327524a0. [DOI] [PubMed] [Google Scholar]
- 60.Huie RE, et al. The reaction of no with superoxide. Free Radic. Res. Commun. 1993;18:195–199. doi: 10.3109/10715769309145868. [DOI] [PubMed] [Google Scholar]
- 61.Lundberg R, et al. Deficient DNA end joining activity in extracts from fanconi anemia fibroblasts. J. Biol. Chem. 2001;276:9543–9549. doi: 10.1074/jbc.M008634200. [DOI] [PubMed] [Google Scholar]
- 62.Wilson DM, et al. Base excision repair and the central nervous system. Neuroscience. 2007;145:1187–1200. doi: 10.1016/j.neuroscience.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 63.Krokan HE, et al. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Croteau DL, et al. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J. Biol. Chem. 1997;272:25409–25412. doi: 10.1074/jbc.272.41.25409. [DOI] [PubMed] [Google Scholar]
- 65.Dizdaroglu M. Chemical determination of free radical-induced damage to DNA. Free Radic. Biol. Med. 1991;10:225–242. doi: 10.1016/0891-5849(91)90080-m. [DOI] [PubMed] [Google Scholar]
- 66.Wallace DC, et al. Sequence analysis of cDNAs for the human and bovine ATP synthase beta subunit: mitochondrial DNA genes sustain seventeen times more mutations. Curr. Genet. 1987;12:81–90. doi: 10.1007/BF00434661. [DOI] [PubMed] [Google Scholar]
- 67.Yakes FM, et al. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl Acad. Sci. USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mecocci P, et al. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann. Neurol. 1993;34:609–616. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 69.Neeley WL, et al. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 70.Dizdaroglu M, et al. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic. Biol. Med. 2002;32:1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 71.Grollman AP, et al. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 72.Krokan HE, et al. Uracil in DNA—occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 73.Sedgwick B, et al. Repair of alkylated DNA: recent advances. DNA Repair (Amst) 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 74.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 75.Vartanian V, et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl Acad. Sci. USA. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCullough AK, et al. Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 77.Mol CD, et al. DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu. Rev. Biophys. Biomol. Struct. 1999;28:101–128. doi: 10.1146/annurev.biophys.28.1.101. [DOI] [PubMed] [Google Scholar]
- 78.Lindahl T. Repair of intrinsic DNA lesions. Mutat. Res. 1990;238:305–311. doi: 10.1016/0165-1110(90)90022-4. [DOI] [PubMed] [Google Scholar]
- 79.Loeb LA. Apurinic sites as mutagenic intermediates. Cell. 1985;40:483–484. doi: 10.1016/0092-8674(85)90191-6. [DOI] [PubMed] [Google Scholar]
- 80.Morland I, et al. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parsons JL, et al. NEIL1 excises 3′ end proximal oxidative DNA lesions resistant to cleavage by NTH1 and OGG1. Nucleic Acids Res. 2005;33:4849–4856. doi: 10.1093/nar/gki816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hazra TK, et al. Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:193–205. doi: 10.1016/s0079-6603(01)68100-5. [DOI] [PubMed] [Google Scholar]
- 83.Demple B, et al. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amst) 2005;4:1442–1449. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 84.Wilson DM, III, et al. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat. Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 85.Rasouli-Nia A, et al. Stable down-regulation of human polynucleotide kinase enhances spontaneous mutation frequency and sensitizes cells to genotoxic agents. Proc. Natl Acad. Sci. USA. 2004;101:6905–6910. doi: 10.1073/pnas.0400099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wiederhold L, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 87.D'Amours D, et al. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 88.Brem R, et al. XRCC1 is required for DNA single-strand break repair in human cells. Nucleic Acids Res. 2005;33:2512–2520. doi: 10.1093/nar/gki543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilson SH. Mammalian base excision repair and DNA polymerase beta. Mutat. Res. 1998;407:203–215. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 90.Tomkinson AE, et al. Completion of base excision repair by mammalian DNA ligases. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:151–164. doi: 10.1016/s0079-6603(01)68097-8. [DOI] [PubMed] [Google Scholar]
- 91.Bennett RA, et al. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl Acad. Sci. USA. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fan J, et al. Protein-protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic. Biol. Med. 2005;38:1121–1138. doi: 10.1016/j.freeradbiomed.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 93.Oka S, et al. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J. 2008;27:421–432. doi: 10.1038/sj.emboj.7601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Polyak K, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat. Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 95.Shadel GS, et al. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 96.Akbari M, et al. Cytotoxicity and mutagenicity of endogenous DNA base lesions as potential cause of human aging. Mech. Ageing Dev. 2008;129:353–365. doi: 10.1016/j.mad.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 97.Altieri F, et al. DNA damage and repair: from molecular mechanisms to health implications. Antioxid. Redox Signal. 2008;10:891–937. doi: 10.1089/ars.2007.1830. [DOI] [PubMed] [Google Scholar]
- 98.Chang SW, et al. The frequency of point mutations in mitochondrial DNA is elevated in the Alzheimer's brain. Biochem. Biophys. Res. Commun. 2000;273:203–208. doi: 10.1006/bbrc.2000.2885. [DOI] [PubMed] [Google Scholar]
- 99.Forman MS, et al. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat. Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 100.Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 101.Stuart JA, et al. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res. 2005;33:3722–3732. doi: 10.1093/nar/gki683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brown WM, et al. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J. Mol. Evol. 1982;18:225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- 103.Clayton DA, et al. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. PNAS. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.LeDoux SP, et al. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 105.Pettepher CC, et al. Repair of alkali-labile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J. Biol. Chem. 1991;266:3113–3117. [PubMed] [Google Scholar]
- 106.Taffe BG, et al. Gene-specific nuclear and mitochondrial repair of formamidopyrimidine DNA glycosylase-sensitive sites in Chinese hamster ovary cells. Mutat. Res. 1996;364:183–192. doi: 10.1016/s0921-8777(96)00031-6. [DOI] [PubMed] [Google Scholar]
- 107.Nakabeppu Y. Regulation of intracellular localization of human MTH1, OGG1, and MYH proteins for repair of oxidative DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:75–94. doi: 10.1016/s0079-6603(01)68091-7. [DOI] [PubMed] [Google Scholar]
- 108.Pinz KG, et al. Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol. Cell. Biol. 1998;18:1257–1265. doi: 10.1128/mcb.18.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ohtsubo T, et al. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takao M, et al. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998;26:2917–2922. doi: 10.1093/nar/26.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hashiguchi K, et al. The C-terminal alphaO helix of human Ogg1 is essential for 8-oxoguanine DNA glycosylase activity: the mitochondrial beta-Ogg1 lacks this domain and does not have glycosylase activity. Nucleic Acids Res. 2004;32:5596–5608. doi: 10.1093/nar/gkh863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nilsen H, et al. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tell G, et al. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid. Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 114.Chattopadhyay R, et al. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006;34:2067–2076. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Copeland WC, et al. DNA polymerase gamma in mitochondrial DNA replication and repair. ScientificWorldJournal. 2003;3:34–44. doi: 10.1100/tsw.2003.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu. Rev. Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 117.Longley MJ, et al. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc. Natl Acad. Sci. USA. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Akbari M, et al. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 119.Liu P, et al. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell. Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Szczesny B, et al. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dreher D, et al. Role of oxygen free radicals in cancer development. Eur. J. Cancer. 1996;32A:30–38. doi: 10.1016/0959-8049(95)00531-5. [DOI] [PubMed] [Google Scholar]
- 122.Cooke MS, et al. Progress in the analysis of urinary oxidative DNA damage. Free Radic. Biol. Med. 2002;33:1601–1614. doi: 10.1016/s0891-5849(02)01146-2. [DOI] [PubMed] [Google Scholar]
- 123.Cooke MS, et al. Evidence for attenuated cellular 8-oxo-7,8-dihydro-2′-deoxyguanosine removal in cancer patients. Biol. Chem. 2006;387:393–400. doi: 10.1515/BC.2006.053. [DOI] [PubMed] [Google Scholar]
- 124.Jungst C, et al. Oxidative damage is increased in human liver tissue adjacent to hepatocellular carcinoma 2. Hepatology. 2004;39:1663–1672. doi: 10.1002/hep.20241. [DOI] [PubMed] [Google Scholar]
- 125.Lu J, et al. [Study of 8-OH-dG and its correlation with several cancer related gene in lung cancer tissues] Wei Sheng Yan Jiu. 2003;32:304–307. [PubMed] [Google Scholar]
- 126.Trzeciak AR, et al. Cellular repair of oxidatively induced DNA base lesions is defective in prostate cancer cell lines, PC-3 and DU-145. Carcinogenesis. 2004;25:1359–1370. doi: 10.1093/carcin/bgh144. [DOI] [PubMed] [Google Scholar]
- 127.Cullen JJ, et al. Expression of antioxidant enzymes in diseases of the human pancreas: another link between chronic pancreatitis and pancreatic cancer. Pancreas. 2003;26:23–27. doi: 10.1097/00006676-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 128.Ishii K, et al. Prevention of mammary tumorigenesis in acatalasemic mice by vitamin E supplementation. Jpn. J. Cancer Res. 1996;87:680–684. doi: 10.1111/j.1349-7006.1996.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim IS. Down-regulation of human FEN-1 gene expression during differentiation of promyelocytic leukemia cells. Exp. Mol. Med. 1998;30:252–256. doi: 10.1038/emm.1998.37. [DOI] [PubMed] [Google Scholar]
- 130.Li Y, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat. Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 131.Zimmerman R, et al. Active oxygen acts as a promoter of transformation in mouse embryo C3H/10T1/2/C18 fibroblasts. Proc. Natl Acad. Sci. USA. 1984;81:2085–2087. doi: 10.1073/pnas.81.7.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Elchuri S, et al. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 133.Brown NS, et al. Hypoxia and oxidative stress in breast cancer. Oxidative stress: its effects on the growth, metastatic potential and response to therapy of breast cancer. Breast Cancer Res. 2001;3:323–327. doi: 10.1186/bcr315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Toyokuni S, et al. Persistent oxidative stress in cancer. FEBS Lett. 1995;358:1–3. doi: 10.1016/0014-5793(94)01368-b. [DOI] [PubMed] [Google Scholar]
- 135.Szatrowski TP, et al. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51:794–798. [PubMed] [Google Scholar]
- 136.Okamoto K, et al. Formation of 8-hydroxy-2′-deoxyguanosine and 4-hydroxy-2-nonenal-modified proteins in human renal-cell carcinoma. Int. J. Cancer. 1994;58:825–829. doi: 10.1002/ijc.2910580613. [DOI] [PubMed] [Google Scholar]
- 137.Sato K, et al. Negative regulation of catalase gene expression in hepatoma cells. Mol. Cell. Biol. 1992;12:2525–2533. doi: 10.1128/mcb.12.6.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sun Y. Free radicals, antioxidant enzymes, and carcinogenesis. Free Radic. Biol. Med. 1990;8:583–599. doi: 10.1016/0891-5849(90)90156-d. [DOI] [PubMed] [Google Scholar]
- 139.Amstad P, et al. Oxidants stress induces the proto-oncogenes, C-fos and C-myc in mouse epidermal cells. Bull. Cancer. 1990;77:501–502. [PubMed] [Google Scholar]
- 140.Marusyk A, et al. Declining cellular fitness with age promotes cancer initiation by selecting for adaptive oncogenic mutations. Biochim. Biophys. Acta. 2008;1785:1–11. doi: 10.1016/j.bbcan.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Harman D. Free radicals in aging. Mol. Cell. Biochem. 1988;84:155–161. doi: 10.1007/BF00421050. [DOI] [PubMed] [Google Scholar]
- 142.Harman D. Free radical theory of aging: history. EXS. 1992;62:1–10. doi: 10.1007/978-3-0348-7460-1_1. [DOI] [PubMed] [Google Scholar]
- 143.Lombard DB, et al. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 144.Maclean MJ, et al. Base excision repair activities required for yeast to attain a full chronological life span. Aging Cell. 2003;2:93–104. doi: 10.1046/j.1474-9728.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- 145.Balaban RS, et al. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 146.Corral-Debrinski M, et al. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat. Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 147.Schwarze SR, et al. High levels of mitochondrial DNA deletions in skeletal muscle of old rhesus monkeys. Mech. Ageing Dev. 1995;83:91–101. doi: 10.1016/0047-6374(95)01611-3. [DOI] [PubMed] [Google Scholar]
- 148.Khaidakov M, et al. Accumulation of point mutations in mitochondrial DNA of aging mice. Mutat. Res. 2003;526:1–7. doi: 10.1016/s0027-5107(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 149.Modica-Napolitano JS, et al. Mitochondria and human cancer. Curr. Mol. Med. 2007;7:121–131. doi: 10.2174/156652407779940495. [DOI] [PubMed] [Google Scholar]
- 150.Ishikawa K, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 151.Graziewicz MA, et al. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem. Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 152.Mambo E, et al. Defective repair of 8-hydroxyguanine in mitochondria of MCF-7 and MDA-MB-468 human breast cancer cell lines. Cancer Res. 2002;62:1349–1355. [PubMed] [Google Scholar]
- 153.Fliss MS, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 154.Tebbs RS, et al. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 155.Cabelof DC, et al. Base excision repair deficiency caused by polymerase beta haploinsufficiency: accelerated DNA damage and increased mutational response to carcinogens. Cancer Res. 2003;63:5799–5807. [PubMed] [Google Scholar]
- 156.Sobol RW, et al. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 157.Xanthoudakis S, et al. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Larsen E, et al. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell. Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bentley D, et al. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat. Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- 160.Xie Y, et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 161.Starcevic D, et al. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 162.Al-Tassan N, et al. Inherited variants of MYH associated with somatic G:C–>T:A mutations in colorectal tumors. Nat. Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 163.Imai K, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 164.Nilsen H, et al. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 2003;22:5381–5386. doi: 10.1038/sj.onc.1206860. [DOI] [PubMed] [Google Scholar]
- 165.Webster AD, et al. Growth retardation and immunodeficiency in a patient with mutations in the DNA ligase I gene. Lancet. 1992;339:1508–1509. doi: 10.1016/0140-6736(92)91266-b. [DOI] [PubMed] [Google Scholar]
- 166.Barnes DE, et al. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69:495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 167.Rolig RL, et al. Linking DNA damage and neurodegeneration. Trends Neurosci. 2000;23:417–424. doi: 10.1016/s0166-2236(00)01625-8. [DOI] [PubMed] [Google Scholar]
- 168.Boerrigter ME, et al. DNA repair and Alzheimer's disease. J. Gerontol. 1992;47:B177–B184. doi: 10.1093/geronj/47.6.b177. [DOI] [PubMed] [Google Scholar]
- 169.Lezza AM, et al. Mitochondrial DNA 4977 bp deletion and OH8dG levels correlate in the brain of aged subjects but not Alzheimer's disease patients. FASEB J. 1999;13:1083–1088. doi: 10.1096/fasebj.13.9.1083. [DOI] [PubMed] [Google Scholar]
- 170.Weissman L, et al. Defective DNA base excision repair in brain from individuals with Alzheimer's disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007;35:5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fishel ML, et al. DNA repair in neurons: so if they don't divide what's to repair? Mutat. Res. 2007;614:24–36. doi: 10.1016/j.mrfmmm.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 172.Lu F, et al. Oxidative damage to mitochondrial DNA and activity of mitochondrial enzymes in chronic active lesions of multiple sclerosis. J. Neurol. Sci. 2000;177:95–103. doi: 10.1016/s0022-510x(00)00343-9. [DOI] [PubMed] [Google Scholar]
- 173.Fukae J, et al. Expression of 8-oxoguanine DNA glycosylase (OGG1) in Parkinson's disease and related neurodegenerative disorders. Acta Neuropathol. (Berl) 2005;109:256–262. doi: 10.1007/s00401-004-0937-9. [DOI] [PubMed] [Google Scholar]
- 174.Kisby GE, et al. Evidence of reduced DNA repair in amyotrophic lateral sclerosis brain tissue. Neuroreport. 1997;8:1337–1340. doi: 10.1097/00001756-199704140-00004. [DOI] [PubMed] [Google Scholar]
- 175.Olkowski ZL. Mutant AP endonuclease in patients with amyotrophic lateral sclerosis. Neuroreport. 1998;9:239–242. doi: 10.1097/00001756-199801260-00012. [DOI] [PubMed] [Google Scholar]
- 176.Shaikh AY, et al. DNA base-excision repair enzyme apurinic/apyrimidinic endonuclease/redox factor-1 is increased and competent in the brain and spinal cord of individuals with amyotrophic lateral sclerosis. Neuromolecular Med. 2002;2:47–60. doi: 10.1007/s12017-002-0038-7. [DOI] [PubMed] [Google Scholar]
- 177.Hung RJ, et al. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am. J. Epidemiol. 2005;162:925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 178.Kohno T, et al. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene. 1998;16:3219–3225. doi: 10.1038/sj.onc.1201872. [DOI] [PubMed] [Google Scholar]
- 179.Lee AJ, et al. Interindividual variability in response to sodium dichromate-induced oxidative DNA damage: role of the Ser326Cys polymorphism in the DNA-repair protein of 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA glycosylase 1. Cancer Epidemiol. Biomarkers Prev. 2005;14:497–505. doi: 10.1158/1055-9965.EPI-04-0295. [DOI] [PubMed] [Google Scholar]
- 180.Kiyohara C, et al. Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: a meta-analysis. Lung Cancer. 2006;54:267–283. doi: 10.1016/j.lungcan.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 181.Hu Z, et al. XRCC1 polymorphisms and cancer risk: a meta-analysis of 38 case-control studies. Cancer Epidemiol. Biomarkers Prev. 2005;14:1810–1818. doi: 10.1158/1055-9965.EPI-04-0793. [DOI] [PubMed] [Google Scholar]
- 182.Wang Y, et al. From genotype to phenotype: correlating XRCC1 polymorphisms with mutagen sensitivity. DNA Repair (Amst) 2003;2:901–908. doi: 10.1016/s1568-7864(03)00085-5. [DOI] [PubMed] [Google Scholar]
- 183.Zhang Y, et al. Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiol. Biomarkers Prev. 2006;15:353–358. doi: 10.1158/1055-9965.EPI-05-0653. [DOI] [PubMed] [Google Scholar]
- 184.Matullo G, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 185.Wilding CS, et al. Genotype profiles of loci encoding DNA repair enzymes in newborn and elderly populations: no evidence of association with longevity. Biogerontology. 2006;7:35–41. doi: 10.1007/s10522-005-6042-1. [DOI] [PubMed] [Google Scholar]