Abstract
Toll-like receptors (TLRs) may influence the development of non-Hodgkin lymphoma (NHL) given their important roles in recognizing microbial pathogens and stimulating multiple immune pathways. We conducted an investigation of TLR gene variants in a pooled analysis including three population-based case–control studies of NHL (1946 cases and 1808 controls). Thirty-six tag single-nucleotide polymorphisms (SNPs) in TLR2, TLR4 and the TLR10–TLR1–TLR6 gene cluster were genotyped. Two TLR10–TLR1–TLR6 variants in moderate linkage disequilibrium were significantly associated with NHL: rs10008492 [odds ratio for CT genotype (ORCT) 1.12, 95% confidence interval (CI) 0.97–1.30; ORTT 1.40, 95% CI 1.15–1.71; Ptrend = 0.001] and rs4833103 (ORAC 0.75, 95% CI 0.64–0.88; ORAA 0.74, 95% CI 0.62–0.90; Ptrend = 0.002; Pdominant = 0.0002). Associations with these SNPs were consistent across all the three studies and did not appreciably differ by histologic subtype. We found little evidence of association between TLR2 variation and all NHL, although the rare variant rs3804100 was significantly associated with marginal zone lymphoma (MZL), both overall (ORCT/CC 1.89, 95% CI 1.27–2.81; Pdominant = 0.002) and in two of the three studies. No associations with TLR4 variants were observed. This pooled analysis provides strong evidence that variation in the TLR10–TLR1–TLR6 region is associated with NHL risk and suggests that TLR2 variants may influence susceptibility to MZL.
Introduction
The etiology of non-Hodgkin lymphoma (NHL) is poorly understood, although various lines of evidence suggest that infections and immune dysregulation play a role. This evidence includes elevated incidence rates in immunosuppressed people and among individuals with selected autoimmune diseases, established links between specific infectious agents and rare NHL subtypes [Epstein-Barr virus and Burkitt lymphoma, human T-cell leukemia/lymphoma virus 1 and adult T-cell leukemia/lymphoma, human herpesvirus 8 and primary effusion lymphoma, Helicobacter pylori and gastric mucosa-associated lymphoid tissue (MALT) lymphoma] and associations with NHL risk for allergies and indicators of early-life exposure to infectious agents (1). In light of these findings, it is plausible that dysregulation in the recognition of and immune response to microbial pathogens may influence the risk of NHL development.
Toll-like receptors (TLRs) are a family of transmembrane receptors that play a key role in mounting an immune response against microbial pathogens. These receptors have been evolutionarily conserved to recognize specific microbial molecular components; once activated, TLRs engage a signaling cascade resulting in the stimulation of innate and adaptive immune responses targeting the invading pathogen. To date, 10 functional human TLRs have been identified (2). TLR-2 and TLR-4 are the principal receptors involved in the recognition of bacterial cell wall components; TLR-4 binds to Gram-negative bacterial lipopolysaccharides, whereas TLR-2 recognizes other cell wall molecules from a wide variety of bacteria as well as capsid ligands from several viruses (2,3). The capability of TLR-2 to recognize such a broad spectrum of ligands is the result of its ability to form heterodimer receptor complexes with TLR-1 and -6. Some endogenous molecules may also trigger TLR-mediated inflammatory responses; suspected TLR ligands include heat shock proteins, HMGB1, fibrinogen and host DNA (4).
TLRs have been hypothesized as possible contributors to lymphoma and other immunologic/inflammatory diseases given their important role in recognizing microbial ligands and stimulating multiple immune pathways (2,5,6). Single-nucleotide polymorphisms (SNPs) in TLR2 (−16933T>A; rs4696480) and TLR4 (Arg299Gly; rs4986790) have been associated with lymphoma risk in recent studies (5–7). TLR1 and TLR6, located along with TLR10 [a TLR of unknown function (8)] in a 57 kb long region on 4p14, have not been studied in detail in relation to NHL, although an uncommon TLR6 variant was significantly associated with increased risk of NHL and selected subtypes in one study (9). Variation elsewhere within the TLR10–TLR1–TLR6 gene cluster has been associated with other diseases, including asthma (10,11) and prostate cancer (12).
To further explore whether genetic variation in TLR genes influences NHL pathogenesis, we conducted a pooled investigation of tag SNPs in TLR2, TLR4 and the TLR10–TLR1–TLR6 region in three population-based case–control studies of NHL.
Materials and methods
Study population
Our study population was derived from pooling three independent population-based case–control studies, which have been described in detail previously: the National Cancer Institute (NCI)-Surveillance Epidemiology and End Results (SEER) NHL case–control study (13,14), the Connecticut NHL case–control study (15,16) and the New South Wales (NSW) NHL case–control study (17,18). Selected characteristics for each study are presented in Table I. All the three studies included first primary NHL cases only, and population controls were frequency matched to cases. The protocols for each study were approved by the following institutions: the Institutional Review Boards of the NCI and each SEER center for the NCI-SEER study; Yale University, the Connecticut Department of Public Health and the NCI for the Connecticut study and all participating institutions for the NSW study. All study participants provided informed consent.
Table I.
Selected characteristics of the NCI-SEER, Connecticut and NSW NHL case–control studies
| NCI-SEER | Connecticut | NSW | |
| Location | Residents of the Iowa, Detroit, Los Angeles and Seattle SEER registries | Residents of the Connecticut SEER registry | Residents of the Australian State of NSW or the Australian Capital Territory via the NSW Central Cancer Registry |
| Time period | 1998–2000 | 1996–2000 | 2000–2001 |
| Age range (years) | 20–74 | 21–84 | 20–74 |
| Eligibility criteria | Excluded known HIV-positive individuals | Excluded males | Excluded known HIV-positive individuals and organ transplant recipients |
| Histopathologic classification system | ICD-O-2 | REAL | WHO + ICD-O-3 |
| Control selection | <65 years: random digit dialing | <65 years: random digit dialing | Electoral rolls |
| ≥65 years: medicare files | ≥65 years: medicare files | ||
| Matching variables | Age (5 year groups), sex, race and SEER area | Age (5 year groups) | Age (5 year groups), sex, state or territory |
| Study population (participation)a | Cases: n = 1321 (76%) | Cases: n = 601 (72%) | Cases: n = 694 (85%) |
| Controls: n = 1057 (52%) | Controls: n = 717 (<65 years: 69%; ≥65 years: 47%) | Controls: n = 694 (61%) | |
| Risk factor information | Self-administered questionnaire, in-person interview | Self-administered questionnaire, in-person interview | Self-administered questionnaire, telephone interview |
| DNA source | Venous blood or mouthwash buccal cell sample | Venous blood or mouthwash buccal cell sample | Venous blood sample |
| Restrictions for this analysis | Excluded participants who provided a buccal cell sample | Included participants of European or Asian ethnicity (97% of participants) | |
| Genotyped for this analysisb | Cases: n = 1001 | Cases: n = 436 | Cases: n = 524 |
| Controls: n = 834 | Controls: n = 517 | Controls: n = 474 |
HIV, human immunodeficiency virus; ICD-O-2/3, International Classification of Diseases for Oncology, second/third revision; REAL, Revised European American Lymphoma system; WHO, World Health Organization.
Participation was defined as the percentage interviewed among those approached.
Study participants who did not provide a biologic specimen, did not have sufficient material for DNA extraction or sufficient DNA for genotyping or whose genotyped sex was discordant from the questionnaire data were excluded from this analysis. As described in Materials and Methods, the final analytic population further excluded participants with a low sample completion rate (NCI-SEER: 11 cases, 6 controls; Connecticut: 2 controls; NSW: 4 cases, 9 controls).
NHL pathology classification
All cases were histologically confirmed by the local diagnosing pathologist in the NCI-SEER study and by central review by two independent pathologists in the Connecticut study. In the NSW study, all cases were histologically confirmed by the local diagnostic pathologist; a confirmatory central pathology slide review was performed for cases of <90% certainty when they had consented to access to their pathological specimens (19). In the present analyses, we grouped cases into NHL subtypes according to the World Health Organization classification (20) using the International Lymphoma Epidemiology Consortium (InterLymph) guidelines (21). Subtype-specific analyses were done for the four most common NHL subtypes: diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), marginal zone lymphoma (MZL) and chronic lymphocytic leukemia (CLL)/small lymphocytic lymphoma (SLL). Our studies primarily included SLL rather than CLL cases because these diseases were not considered the same entity until the World Health Organization classification was introduced in 2001 (20).
Biological samples and DNA extraction
Study participants who did not provide a biologic specimen, did not have sufficient material for DNA extraction or sufficient DNA for genotyping or whose genotyped sex was discordant from the questionnaire data were excluded from this analysis. For the NCI-SEER study, DNA was extracted from blood clots or buffy coats (BBI Biotech, Gaithersburg, MD) using Puregene Autopure DNA extraction kits (Gentra Systems, Minneapolis, MN) and from buccal cell samples by phenol–chloroform extraction methods (22). Genotype frequencies for individuals who provided blood compared with buccal cells were equivalent (23). For the Connecticut study, DNA was extracted from the blood samples using phenol–chloroform extraction methods; subjects providing buccal cells only were excluded from this analysis (22). For the NSW study, DNA was extracted from buffy coats using Qiagen QIAamp® DNA Blood Midi Kits by laboratory staff at the Viral Epidemiology Section, SAIC-Frederick, NCI-Frederick.
Genotyping
Thirty-six tag SNPs for TLR10–TLR1–TLR6 (n = 11), TLR2 (n = 11) and TLR4 (n = 14) were selected from the designable set of common SNPs (minor allele frequency ≥ 0.05) genotyped in the Caucasian (CEU) population sample of the HapMap Project (Data Release 20/Phase II, NCBI B35 assembly, dbSNP b125). Tag SNP selection was done using the software application TagZilla (v1.1; http://tagzilla.nci.nih.gov/), which employs the pairwise binning algorithm of Carlson et al. (24). For each gene, SNPs within the region 20 kb 5′ of the ATG-translation initiation codon and 10 kb 3′ of the end of the last exon were binned using a binning threshold of r2 >0.80. When there were multiple transcripts available for genes, the primary transcript was assessed. Genotyping was performed at the National Cancer Institute Core Genotyping Facility (Gaithersburg, MD) using the Illumina GoldenGate platform, with the 36 tag SNPs included in an oligo pool assay (OPA) investigating genetic variants from different candidate pathways. Data for an additional non-synonymous TLR4 SNP (D299G; rs4986790) that had been previously genotyped by TaqMan assay in the NCI-SEER (14) and Connecticut studies were also included in the pooled analysis, increasing the total number of investigated SNPs to 37. Gene coverage (the number of genotyped SNPs divided by the number of bins from the designable set of SNPs in HapMap Build 20 CEU samples) was calculated to be 61, 69 and 94% for TLR10–TLR1-TLR6, TLR2 and TLR4, respectively.
All investigated SNPs in TLR1, TLR2, TLR4, TLR6 and TLR10 had genotype concordance rates and assay completion rates ≥99%. For each SNP, an exact test of Hardy–Weinberg proportions was performed among Caucasian controls. One TLR2 SNP deviated significantly from Hardy–Weinberg proportions (rs4696187, P = 0.02).
We excluded samples with OPA-wide sample completion rates <90% (NCI-SEER: 11 cases, 6 controls; Connecticut: 0 cases, 2 controls; NSW: 4 cases, 9 controls). The final analytic population from pooling the three studies consisted of 1946 cases and 1808 controls (NCI-SEER, 990/828; Connecticut, 436/515; NSW, 520/465).
Statistical methods
Statistical analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC) unless otherwise specified. Odds ratios (ORs) and 95% confidence intervals (CIs) estimating the relative risk of NHL in relation to SNP genotype were calculated using unconditional logistic regression adjusting for age, race/ethnicity, sex and study center. Analyses restricted to non-Hispanic Caucasians were also performed. Tests for trend (i.e. assuming a codominant genetic model) were performed by modeling the number of rare alleles (0, 1 or 2) as a continuous variable. In general, P-values for SNPs reported in the Results are from the trend test. For rs4833103, which showed consistent evidence of association under a dominant genetic model throughout our analyses, both trend test P-values and P-values assuming a dominant model are reported.
Gene-level statistical tests of association of TLR10–TLR1–TLR6, TLR2 and TLR4 were conducted using two approaches. First, we performed the ‘MinP’ test, which assesses the true statistical significance of the smallest P-trend within each gene region by permutation-based resampling methods (10 000 permutations) that automatically adjust for the number of tag SNPs tested within that gene and the underlying linkage disequilibrium (LD) pattern (25,26). These analyses were performed using the MATLAB Statistics Toolbox™ 6.2 (The Mathworks, Natick, MA). Second, we conducted a likelihood ratio test comparing the log likelihood for a model containing main-effect parameters (assuming a codominant model) for all non-correlated SNPs (r2 < 0.80 among controls) within a gene region to the log likelihood for a model with these parameters removed (27).
Two types of haplotype analyses were conducted among non-Hispanic Caucasian subjects only. We evaluated risk associated with haplotypes defined by SNPs within a sliding window of three loci across a gene region (Haplo.Stats, haplo.score.slide, version 1.3.1; http://cran.r-project.org/web/packages/haplo.stats/index.html). A global score statistic was used to summarize the evidence of association of disease with the haplotypes for each window. We also conducted analyses of haplotypes inferred from all tag SNPs across the gene region (Haplo.Stats, haplo.glm, minimum haplotype frequency 1%). These region-wide haplotype analyses were performed after visualizing the haplotype structures across TLR10–TLR1–TLR6, TLR2 and TLR4 and noting the absence of separate haplotype blocks within each region (Haploview version 4.0; http://www.broad.mit.edu/mpg/haploview/). For both approaches, codominant haplotype effects were modeled with adjustment for age, sex and study center.
In addition to analyses of all NHL, we investigated associations with specific histologic subtypes (FL, DLBCL, MZL and CLL/SLL). Polytomous regression modeling was used to simultaneously calculate subtype-specific SNP effects and to construct tests of heterogeneity in codominant SNP effects across subtypes. Subtype-specific gene-level tests and haplotype analyses were performed using unconditional logistic regression modeling. The association between the TLR2 variant rs3804100 and MALT lymphoma, the most common type of MZL, was also investigated using unconditional logistic regression.
Results
The distributions of cases and controls were similar with respect to sex, age, race or ethnicity and study site, both overall and within each study (Table II). Approximately 90% of the overall study population were non-Hispanic Caucasian (87% of controls and 90% of cases), 5% were Black and 6% were members of other racial/ethnic groups. The NCI-SEER and Yale studies had similar distributions of NHL subtypes, whereas the NSW study had a higher frequency of FL (37 versus 24%) and a lower frequency of CLL/SLL (3 versus 10%) and not otherwise specified (NOS) tumors (7 versus 15%) compared with the other studies.
Table II.
Study-specific and pooled demographic and pathology characteristics of study participants in the NCI-SEER, Yale and NSW NHL case–control studies
| NCI | Yale | NSW | Pooled | |||||
| Control, N = 828, n (%) | Case, N = 990, n (%) | Control, N = 515, n (%) | Case, N = 436, n (%) | Control, N = 465, n (%) | Case, N = 520, n (%) | Control, N = 1808, n (%) | Case, N = 1946, n (%) | |
| Sex | ||||||||
| Male | 443 (53) | 536 (54) | — | — | 268 (58) | 304 (58) | 711 (39) | 840 (43) |
| Female | 385 (47) | 454 (46) | 515 (100) | 436 (100) | 197 (42) | 216 (42) | 1097 (61) | 1106 (57) |
| Age (years) | ||||||||
| <50 | 203 (25) | 277 (28) | 98 (19) | 86 (20) | 107 (23) | 121 (23) | 408 (23) | 484 (25) |
| 50–59 | 177 (21) | 235 (24) | 97 (19) | 89 (20) | 135 (29) | 171 (33) | 409 (23) | 495 (25) |
| 60–69 | 285 (34) | 311 (31) | 120 (23) | 110 (25) | 151 (32) | 154 (30) | 556 (31) | 575 (30) |
| 70+ | 163 (20) | 167 (17) | 200 (39) | 151 (35) | 72 (16) | 74 (14) | 435 (24) | 392 (20) |
| Race/ethnicity | ||||||||
| White, non-Hispanic | 646 (78) | 829 (84) | 473 (92) | 415 (95) | 459 (99) | 507 (98) | 1578 (87) | 1751 (90) |
| Black | 112 (13) | 64 (6) | 14 (3) | 13 (3) | — | — | 126 (7) | 77 (4) |
| Other/unknown | 70 (8) | 97 (10) | 28 (5) | 8 (2) | 6 (1) | 13 (3) | 104 (6) | 118 (6) |
| Study site | ||||||||
| Detroit | 139 (17) | 197 (20) | — | — | — | — | 139 (8) | 197 (10) |
| Iowa | 246 (30) | 301 (30) | — | — | — | — | 246 (14) | 301 (16) |
| Los Angeles | 199 (24) | 234 (24) | — | — | — | — | 199 (11) | 234 (12) |
| Seattle | 244 (29) | 258 (26) | — | — | — | — | 244 (13) | 258 (13) |
| Connecticut | — | — | 515 (100) | 436 (100) | — | — | 515 (28) | 436 (22) |
| NSW | — | — | — | — | 446 (96) | 496 (95) | 446 (25) | 496 (26) |
| ACT | — | — | — | — | 19 (4) | 24 (5) | 19 (1) | 24 (1) |
| NHL subtype | ||||||||
| DLBCL | — | 294 (30) | — | 137 (31) | — | 169 (33) | — | 600 (31) |
| Follicular | — | 246 (25) | — | 103 (24) | — | 191 (37) | — | 540 (28) |
| CLL/SLL | — | 101 (10) | — | 43 (10) | — | 17 (3) | — | 161 (8) |
| Mantle cell | — | 40 (4) | — | 10 (2) | — | 19 (4) | — | 69 (4) |
| Marginal zone | — | 82 (8) | — | 29 (7) | — | 49 (9) | — | 160 (8) |
| LPL | — | 24 (2) | — | 9 (2) | — | 23 (4) | — | 56 (3) |
| MF/SS | — | 18 (2) | — | 10 (2) | — | 3 (1) | — | 31 (2) |
| Burkitt | — | 11 (1) | — | 0 | — | 3 (1) | — | 14 (1) |
| Peripheral T | — | 41 (4) | — | 14 (3) | — | 7 (1) | — | 62 (3) |
| NOS | — | 133 (13) | — | 81 (19) | — | 39 (7) | — | 253 (13) |
| DNA source | ||||||||
| Blood | 598 (72) | 688 (70) | 515 (100) | 436 (100) | 465 (100) | 520 (100) | 1578 (87) | 1644 (85) |
| Buccal | 230 (28) | 302 (30) | — | — | — | — | 230 (13) | 302 (15) |
ACT, Australian Capital Territory; LPL, lymphoplasmacytic lymphoma; MF/SS, mycosis fungoides/sézary syndrome.
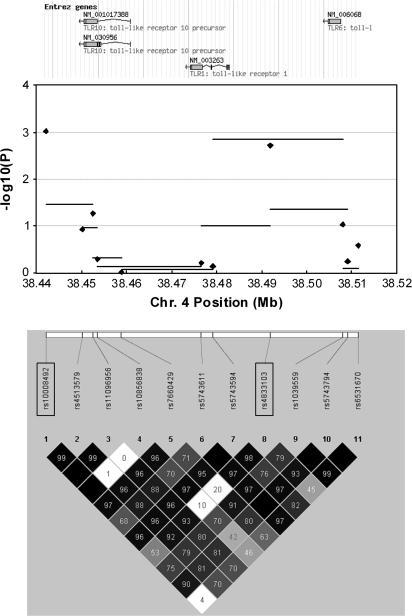
Figure 1 summarizes findings from tests of association involving the 11 investigated SNPs within the TLR10–TLR1–TLR6 gene cluster. Two SNPs were associated with NHL risk: rs10008492 (ORCT 1.12, 95% CI 0.97–1.30; ORTT 1.40, 95% CI 1.15–1.71; Ptrend = 0.001) and rs4833103 (ORAC 0.75, 95% CI 0.64–0.88; ORAA 0.74, 95% CI 0.62–0.90; Ptrend = 0.002; Pdominant = 0.0002). When analyses were restricted to non-Hispanic Caucasians, both SNPs remained associated with NHL (rs10008492, Ptrend = 0.008; rs4833103, Ptrend = 0.01, Pdominant = 0.003). Associations with these SNPs were consistent across the three studies (Figure 2). These SNPs, located 50 kb apart, are in moderate LD (D′ = 0.54; r2 = 0.19 among Caucasian controls; Figure 1).
Fig. 1.
A summary of findings from analyses investigating variation in the TLR10–TLR1–TLR6 gene cluster and risk of NHL. The top of the figure illustrates the chromosomal positions of TLR10, TLR1 and TLR6. The middle portion of the figure is a plot of −log10 P-values from tests of association with risk of all NHL across the region. Horizontal bars represent the −log10 P-values from global tests of 3-SNP haplotype windows moving sequentially across the region (conducted among non-Hispanic Caucasians only). Diamonds represent the −log10 P-values from tests of trend for each individual SNP (conducted among all subjects). The bottom of the figure illustrates the chromosomal position of the 11 tag SNPs and a plot of pairwise D′ within the Caucasian (CEU) population sample of HapMap (Data Release 20/Phase II, NCBI B35 assembly, dbSNP b125). The SNPs rs10008492 and rs4833103 are enclosed in rectangles.
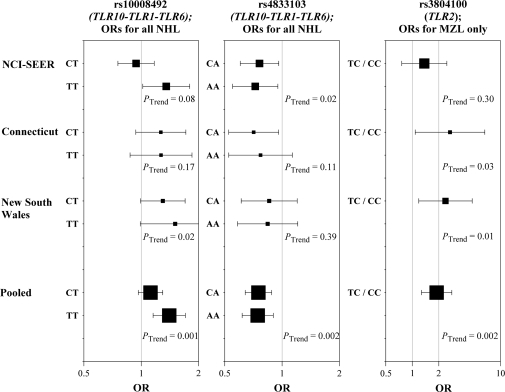
Fig. 2.
Study-specific associations with NHL for rs10008492 and rs4833103 (TLR10–TLR1–TLR6) and with MZL for rs3804100 (TLR2). Square symbols represent ORs; symbol size is proportional to number of cases. Horizontal lines represent 95% CIs.
A complete summary of findings for all investigated SNPs in the TLR10–TLR1–TLR6 region is provided in supplementary Table I (available at Carcinogenesis Online). Marginally significant associations with NHL risk were also observed for two other SNPs in the region, rs11096956 (Ptrend = 0.05) and rs1039559 (Ptrend = 0.09). Global tests of association for the entire region were statistically significant (MinP test, P = 0.008; likelihood ratio test, P = 0.006). Haplotype-based analyses further supported an association between TLR10–TLR1–TLR6 variants and NHL risk; findings from analyses of consecutive 3-SNP haplotype windows across the region suggested associations with rs10008492 and rs4833103 (Figure 1), and a global test of haplotypes across all 11 genotyped variants in the region was statistically significant (P = 0.01).
Associations with rs10008492 and rs4833103 for specific NHL subtypes are shown in Table III (complete subtype-specific findings for all SNPs are in supplementary Tables II–VI, available at Carcinogenesis Online). Only some subtype-specific associations were statistically significant (FL for rs10008492; DLBCL and MZL for rs4833103). However, for each SNP, the associations were in the same direction for all subtypes. Subtype-specific sliding-window haplotype analyses (supplementary Table VII is available at Carcinogenesis Online) and global tests of association (supplementary Table VIII is available at Carcinogenesis Online) of the TLR10–TLR1–TLR6 region supported associations with DLBCL and, less consistently, FL and CLL/SLL.
Table III.
Subtype-specific associations with rs10008492, rs4833103 (TLR10–TLR1–TLR6) and rs3804100 (TLR2)
| Number of minor alleles | ||||||||||||
| 0 (Reference) | 1 | 2 | 1 or 2 | |||||||||
| n | n | ORa (95% CI) | P | n | OR (95% CI) | P | n | OR (95% CI) | P | Ptrend | Pheterogeneity | |
| TLR10–TLR1–TLR6 region | ||||||||||||
| rs10008492 | CC | CT | TT | CT/TT | ||||||||
| Controls | 654 | 799 | 341 | 1140 | ||||||||
| Cases | 652 | 865 | 1.12 (0.97–1.30) | 0.14 | 409 | 1.40 (1.15–1.71) | 0.0007 | 1274 | 1.19 (1.03–1.36) | 0.02 | 0.001 | |
| DLBCL | 201 | 285 | 1.21 (0.98–1.50) | 0.07 | 106 | 1.23 (0.92–1.64) | 0.16 | 391 | 1.22 (1.00–1.49) | 0.05 | 0.09 | |
| FL | 183 | 242 | 1.17 (0.94–1.46) | 0.17 | 110 | 1.58 (1.18–2.12) | 0.002 | 352 | 1.26 (1.02–1.56) | 0.03 | 0.003 | |
| CLL/SLL | 53 | 66 | 0.99 (0.68–1.45) | 0.96 | 41 | 1.53 (0.96–2.43) | 0.07 | 107 | 1.12 (0.79–1.60) | 0.53 | 0.11 | |
| MZL | 59 | 62 | 0.90 (0.62–1.31) | 0.59 | 37 | 1.37 (0.86–2.19) | 0.19 | 99 | 1.01 (0.71–1.44) | 0.94 | 0.32 | 0.68 |
| rs4833103 | CC | CA | AA | CA/AA | ||||||||
| Controls | 478 | 880 | 441 | 1321 | ||||||||
| Cases | 584 | 893 | 0.75 (0.64–0.88) | 0.0005 | 459 | 0.74 (0.62–0.90) | 0.002 | 1352 | 0.75 (0.64–0.87) | 0.0002 | 0.002 | |
| DLBCL | 176 | 284 | 0.78 (0.62–0.97) | 0.03 | 136 | 0.70 (0.54–0.92) | 0.01 | 420 | 0.75 (0.61–0.93) | 0.01 | 0.01 | |
| FL | 140 | 264 | 0.86 (0.67–1.10) | 0.23 | 134 | 0.82 (0.62–1.09) | 0.18 | 398 | 0.85 (0.67–1.07) | 0.16 | 0.18 | |
| CLL/SLL | 51 | 67 | 0.68 (0.46–1.01) | 0.06 | 42 | 0.86 (0.55–1.36) | 0.53 | 109 | 0.74 (0.51–1.07) | 0.11 | 0.51 | |
| MZL | 62 | 58 | 0.48 (0.33–0.71) | 0.0003 | 40 | 0.64 (0.41–0.99) | 0.04 | 98 | 0.53 (0.37–0.76) | 0.0005 | 0.03 | 0.54 |
| TLR2 region | ||||||||||||
| rs3804100 | TT | TC | CC | TC/CC | ||||||||
| Controls | 1556 | 233 | 9 | 242 | ||||||||
| Cases | 1658 | 272 | 1.07 (0.89–1.30) | 0.46 | 12 | 1.17 (0.49–2.80) | 0.73 | 284 | 1.08 (0.89–1.30) | 0.43 | 0.42 | |
| DLBCL | 513 | 82 | 1.04 (0.79–1.36) | 0.79 | 4 | 1.26 (0.38–4.13) | 0.71 | 86 | 1.05 (0.80–1.37) | 0.74 | 0.70 | |
| FL | 469 | 66 | 0.91 (0.68–1.23) | 0.55 | 5 | 2.04 (0.67–6.22) | 0.21 | 71 | 0.95 (0.71–1.27) | 0.73 | 0.95 | |
| CLL/SLL | 146 | 15 | 0.66 (0.38–1.15) | 0.15 | 0 | 15 | 0.63 (0.36––1.10) | 0.11 | 0.09 | |||
| MZL | 123 | 35 | 1.91 (1.28–2.87) | 0.002 | 1 | 36 | 1.89 (1.27–2.81) | 0.002 | 0.003 | 0.007 | ||
Statistically significant (P < 0.05) results in boldface type. n, number of subjects.
ORs adjusted for age, sex, race and study center.
Investigations of the 11 tag SNPs summarizing the TLR2 region did not provide strong evidence of association with NHL risk (supplementary Table I is available at Carcinogenesis Online), although two tag SNPs, located within the neighboring gene RNF175, were associated with risk at a moderate level of statistical significance (rs11935252, Ptrend = 0.01; rs7695605, Ptrend = 0.03). Of the global tests of TLR2 variation performed, the likelihood ratio test was statistically significant (P = 0.005), but the MinP test and region-wide haplotype global test were not (P = 0.12 and 0.84, respectively).
While subtype-specific analyses of TLR2 SNPs were generally null, one notable exception was observed for rs3804100 (Table III), which was strongly associated with MZL risk (CT/CC versus TT: OR 1.89, 95% CI 1.27–2.81; P = 0.002). Statistically significant associations with this SNP were observed in two of the three studies (Figure 2). Analyses of rs3804100 among non-Hispanic Caucasians only yielded identical findings (P = 0.001), and sliding-window haplotype analysis also supported an association with this SNP (supplementary Table VII is available at Carcinogenesis Online). Additional analyses restricted to MALT lymphoma (n = 118), the most common type of MZL, yielded an even stronger association (ORCT/CC 2.21, 95% CI 1.42–3.43; P = 0.0005). This variant was not associated with other subtypes or with NHL risk overall; a test of heterogeneity in SNP effects across subtypes was statistically significant (P = 0.007). The MinP test of TLR2 variants approached statistical significance for MZL (P = 0.06), but the likelihood ratio test and region-wide haplotype global test did not (P = 0.15 and 0.14, respectively).
The 15 investigated SNPs in TLR4 were not associated with NHL (supplementary Table I is available at Carcinogenesis Online). Gene region tests, haplotype-based analyses and investigations by subtype were similarly null.
Discussion
In this pooled investigation of TLR gene polymorphisms in three case–control studies of NHL, we found consistent evidence that variation in the TLR10–TLR1–TLR6 region is associated with disease risk. Global tests of association across all genotyped variants in this region were statistically significant. In particular, two SNPs within the region, rs10008492 and rs4833103, were significantly associated with NHL; these SNP associations were observed in all the three studies and did not appreciably differ by histologic subtype. We did not observe clear evidence of association with variation in TLR2, although the variant rs3804100 was significantly associated with MZL. No associations with TLR4 variants were observed.
TLR10, TLR1 and TLR6 are located in a gene cluster spanning 57 kb on chromosome 4p14. TLR-1 and -6 are important elements of TLR-2 signaling; these polypeptides, which bind tri-acyl and di-acyl lipoproteins, respectively, interact with TLR-2 to create heterodimer receptors capable of recognizing a broad spectrum of pathogen ligands (2). The ligand and function of TLR-10 are not known, although this receptor has been shown to be expressed in normal and malignant B lymphocytes (8). There is evidence that genetic variation in the TLR10–TLR1–TLR6 region may influence risk of inflammatory diseases; associations with asthma (10,11) and aspergillosis following allogeneic stem cell transplantation (28) have been reported. Associations with TLR10–TLR1–TLR6 variants were also observed in a large case–control study of prostate cancer (12), although a subsequent study of this region reported null findings (29).
The functional relevance of the tag SNPs rs10008492, located 8.5 kb telomeric of TLR10, and rs4833103, located 9.1 kb centromeric of TLR1 and 12.9 kb telomeric of TLR6, has not been investigated. If our observed associations are real, it is probably that these two variants do not directly influence NHL susceptibility but rather are markers for one or more underlying causal variants. Given the strong LD throughout the TLR10–TLR1–TLR6 region (Figure 1), it is difficult to infer a specific effect of any one of these genes from these SNP findings. It is worth noting though that, within the HapMap CEU sample, both variants exhibit LD with the non-synonymous TLR1 SNP I602S, which has been shown to exhibit reduced activity toward tri-acyl lipoproteins in two studies (30,31). It is possible that I602S and/or other functionally relevant TLR1 variants such as P315L (32) directly influence NHL risk. However, we cannot rule out TLR10 and TLR6 as susceptibility loci; indeed, a rare non-synonymous TLR6 SNP was significantly associated with NHL risk in a recent case–control analysis (9). Additional investigations involving a fine-mapping approach to TLR10–TLR1–TLR6 will be important for identifying the underlying causal variants in this region.
We did not observe clear evidence of association between TLR2 variants and risk of all NHL; the likelihood ratio test was statistically significant, but findings from other analyses were null or equivocal. The TLR2 region tag SNPs rs11935252 and rs7695605 were associated with NHL at a moderate level of statistical significance; however, as both variants actually reside within the neighboring gene RNF175, involved in metal ion binding, it is unclear what gene effect would be responsible for those associations (if real). We did observe a strong association with MZL risk for the variant rs3804100, both overall and within two of the three studies. No TLR2 variants were associated with other NHL subtypes. Nieters et al. (6) reported a statistically significant association with FL for the TLR2 −16933T>A (rs4696480) variant. This SNP was not genotyped in our study, although we note that it resides within a LD block shared by three SNPs included in our study (rs6835636, rs4696187 and rs13150331) in the HapMap CEU sample.
TLR2 haplotypes including rs3804100 have been previously associated with risk of type 1 diabetes and severity of genital herpes simplex virus type 2 infection (33,34). If our observed association with rs3804100 is real, it is most probably that this SNP, which is synonymous (S450S), is a marker for another causal variant. Several non-synonymous SNPs within the TLR2 exon have been identified, including the putatively functional variant Arg753Gln (rs5743708) (35–37), although most are rare. An association between TLR2 and MZL is biologically plausible given the strong evidence linking specific infectious organisms to the pathogenesis of MZL, MALT lymphoma in particular (38), and the importance of TLR-2 in recognizing these organisms. There is convincing evidence that H.pylori infection is a causal factor for gastric MALT lymphoma, the most common type of extranodal MZL (39,40). Other infectious organisms have also been linked to MALT lymphomas at other anatomic sites, including Borrelia burgdorferi (41,42), Chlamydia psittaci (43), Campylobacter jejuni (44) and the hepatitis C virus (45,46). Interestingly, TLR-2 has been shown to play an important role in mediating immune responses to H.pylori (47,48), B.burgdorferi (49) and hepatitis C virus (50).
Genetic variation in TLR4 was not associated with NHL risk in our study. There is conflicting evidence from smaller studies regarding an association of the TLR4 polymorphism Asp299Gly (rs4986790) with NHL; this variant was associated with increased risk of MALT lymphoma in one study (6) and with decreased risk of MALT lymphoma (7) and DLBCL (5) in two others. Our findings for this SNP were consistently null, both overall and for specific subtypes.
Strengths of this pooled analysis include its large size and the population-based design of the three participating case–control studies. It is unlikely that our findings are the result of bias due to population stratification, as race was adjusted for in regression modeling, and analyses restricted to non-Hispanic Caucasians yielded virtually identical findings. Given that we investigated several variants across the three gene regions for both all NHL and different subtypes, we must consider the possibility that our observed associations are false-positive findings. The consistency of our key findings across the three participating studies suggests that they are not due to chance; however, these results require replication in other studies before meaningful inferences regarding causation are drawn.
In conclusion, this pooled investigation of TLR gene variants across the three case–control studies of NHL provides strong evidence that variation in the TLR10–TLR1–TLR6 region is associated with risk and suggests that TLR2 variants may influence susceptibility to MZL. Additional studies are needed to replicate these findings and, more generally, to explore further the relevance of TLR pathways to the pathogenesis of NHL. Pooled investigations within the InterLymph Consortium (51) will be especially informative in this regard.
Supplementary material
Supplementary Tables I–VIII can be found at http://carcin.oxfordjournals.org/
Funding
Intramural Research Program of the National Institutes of Health (National Cancer Institute) to all genotyping and statistical analysis for this work and NCI-SEER study; Public Health Service (N01-PC-65064, N01-PC-67008, N01-PC-67009, N01-PC-67010, N02-PC-71105) to NCI-SEER study; National Cancer Institute National Institutes of Health grant (CA62006) to Connecticut study; National Health and Medical Research Council of Australia (990920 to B.K.A), Cancer Council New South Wales (NSW), University of Sydney Medical Foundation to NSW study; NSW Health Department to NSW Central Cancer Registry.
Supplementary Material
Acknowledgments
We thank Mary McAdams, Peter Hui, Michael Stagner and Zeynep Kalaylioglu of Information Management Services for their programming support. For the NCI-SEER study, we also gratefully acknowledge the contributions of the staff and scientists at the SEER centers of Iowa, Los Angeles, Detroit and Seattle for the conduct of the study’s field effort. The NSW study was made possible by access to new notifications to the NSW Central Cancer Registry. Ann-Maree Hughes oversaw conduct of the study and Melisa Litchfield, Maria Agaliotis, Chris Goumas, Jackie Turner and staff of the Hunter Valley Research Foundation contributed to the data collection. Jenny Turner, study pathologist, reviewed all pathology reports and original slides as necessary.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence interval
- CLL
chronic lymphocytic leukemia
- DLBCL
diffuse large B-cell lymphoma
- FL
follicular lymphoma
- LD
linkage disequilibrium
- MALT
mucosa-associated lymphoid tissue
- MZL
marginal zone lymphoma
- NCI
National Cancer Institute
- NHL
non-Hodgkin lymphoma
- NSW
New South Wales
- OR
odds ratio
- SEER
Surveillance Epidemiology and End Results
- SLL
small lymphocytic lymphoma
- SNP
single-nucleotide polymorphism
- TLR
Toll-like receptor
References
- 1.Hartge P, et al. Non-Hodgkin lymphoma. In: Schottenfeld D, Fraumeni JF Jr, editors. Cancer Epidemiology and Prevention. New York, NY: Oxford University Press; 2006. pp. 898–918. [Google Scholar]
- 2.Chen K, et al. Toll-like receptors in inflammation, infection and cancer. Int. Immunopharmacol. 2007;7:1271–1285. doi: 10.1016/j.intimp.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Finberg RW, et al. Toll like receptors and viruses. Rev. Med. Virol. 2007;17:35–43. doi: 10.1002/rmv.525. [DOI] [PubMed] [Google Scholar]
- 4.Rifkin IR, et al. Toll-like receptors, endogenous ligands and systemic autoimmune disease. Immunol. Rev. 2005;204:27–42. doi: 10.1111/j.0105-2896.2005.00239.x. [DOI] [PubMed] [Google Scholar]
- 5.Forrest MS, et al. Polymorphisms in innate immunity genes and risk of non-Hodgkin lymphoma. Br. J. Haematol. 2006;134:180–183. doi: 10.1111/j.1365-2141.2006.06141.x. [DOI] [PubMed] [Google Scholar]
- 6.Nieters A, et al. Gene polymorphisms in Toll-like receptors, interleukin-10 and interleukin-10 receptor alpha and lymphoma risk. Genes Immun. 2006;7:615–624. doi: 10.1038/sj.gene.6364337. [DOI] [PubMed] [Google Scholar]
- 7.Hellmig S, et al. Association study of a functional Toll-like receptor 4 polymorphism with susceptibility to gastric mucosa-associated lymphoid tissue lymphoma. Leuk. Lymphoma. 2005;46:869–872. doi: 10.1080/1042819050086451. [DOI] [PubMed] [Google Scholar]
- 8.Bourke E, et al. The toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood. 2003;102:956–963. doi: 10.1182/blood-2002-11-3355. [DOI] [PubMed] [Google Scholar]
- 9.Cerhan JR, et al. Genetic variation in 1253 immune and inflammation genes and risk of non-Hodgkin lymphoma. Blood. 2007;110:4455–4463. doi: 10.1182/blood-2007-05-088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tantisira K, et al. Toll-like receptor 6 gene (TLR6): single-nucleotide polymorphism frequencies and preliminary association with the diagnosis of asthma. Genes Immun. 2004;5:343–346. doi: 10.1038/sj.gene.6364096. [DOI] [PubMed] [Google Scholar]
- 11.Lazarus R, et al. TOLL-like receptor 10 genetic variation is associated with asthma in two independent samples. Am. J. Respir. Crit Care Med. 2004;170:594–600. doi: 10.1164/rccm.200404-491OC. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, et al. Sequence variants in Toll-like receptor gene cluster (TLR6-TLR1-TLR10) and prostate cancer risk. J. Natl Cancer Inst. 2005;97:525–532. doi: 10.1093/jnci/dji070. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee N, et al. Risk of non-Hodgkin's lymphoma and family history of lymphatic, hematologic and other cancers. Cancer Epidemiol. Biomarkers Prev. 2004;13:1415–1421. [PubMed] [Google Scholar]
- 14.Wang SS, et al. Common genetic variants in proinflammatory and other immunoregulatory genes and risk for non-Hodgkin lymphoma. Cancer Res. 2006;66:9771–9780. doi: 10.1158/0008-5472.CAN-06-0324. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Hair-coloring product use and risk of non-Hodgkin's lymphoma: a population-based case-control study in Connecticut. Am. J. Epidemiol. 2004;159:148–154. doi: 10.1093/aje/kwh033. [DOI] [PubMed] [Google Scholar]
- 16.Lan Q, et al. Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood. 2006;107:4101–4108. doi: 10.1182/blood-2005-10-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes AM, et al. Pigmentary characteristics, sun sensitivity and non-Hodgkin lymphoma. Int. J. Cancer. 2004;110:429–434. doi: 10.1002/ijc.20128. [DOI] [PubMed] [Google Scholar]
- 18.Purdue MP, et al. Polymorphisms in immune function genes and risk of non-Hodgkin lymphoma: findings from the New South Wales non-Hodgkin lymphoma study. Carcinogenesis. 2007;28:704–712. doi: 10.1093/carcin/bgl200. [DOI] [PubMed] [Google Scholar]
- 19.Turner JJ, et al. Use of the WHO lymphoma classification in a population-based epidemiological study. Ann. Oncol. 2004;15:631–637. doi: 10.1093/annonc/mdh140. [DOI] [PubMed] [Google Scholar]
- 20.Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Vol. 352. Lyon: IARC Press; 2001. [Google Scholar]
- 21.Morton LM, et al. Proposed classification of lymphoid neoplasms for epidemiologic research from the Pathology Working Group of the International Lymphoma Epidemiology Consortium (InterLymph) Blood. 2007;110:695–708. doi: 10.1182/blood-2006-11-051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Closas M, et al. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol. Biomarkers Prev. 2001;10:687–696. [PubMed] [Google Scholar]
- 23.Bhatti P, et al. Genetic variation and willingness to participate in epidemiologic research: data from three studies. Cancer Epidemiol. Biomarkers Prev. 2005;14:2449–2453. doi: 10.1158/1055-9965.EPI-05-0463. [DOI] [PubMed] [Google Scholar]
- 24.Carlson CS, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westfall PH, et al. Multiple tests for genetic effects in association studies. Methods Mol. Biol. 2002;184:143–168. doi: 10.1385/1-59259-242-2:143. [DOI] [PubMed] [Google Scholar]
- 26.Chen BE, et al. Resampling-based multiple hypothesis testing procedures for genetic case-control association studies. Genet. Epidemiol. 2006;30:495–507. doi: 10.1002/gepi.20162. [DOI] [PubMed] [Google Scholar]
- 27.Chapman JM, et al. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum. Hered. 2003;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- 28.Kesh S, et al. TLR1 and TLR6 polymorphisms are associated with susceptibility to invasive aspergillosis after allogeneic stem cell transplantation. Ann. N. Y. Acad. Sci. 2005;1062:95–103. doi: 10.1196/annals.1358.012. [DOI] [PubMed] [Google Scholar]
- 29.Chen YC, et al. Association between Toll-like receptor gene cluster (TLR6, TLR1 and TLR10) and prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16:1982–1989. doi: 10.1158/1055-9965.EPI-07-0325. [DOI] [PubMed] [Google Scholar]
- 30.Hawn TR, et al. A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. Eur. J. Immunol. 2007;37:2280–2289. doi: 10.1002/eji.200737034. [DOI] [PubMed] [Google Scholar]
- 31.Johnson CM, et al. Cutting edge: a common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J. Immunol. 2007;178:7520–7524. doi: 10.4049/jimmunol.178.12.7520. [DOI] [PubMed] [Google Scholar]
- 32.Omueti KO, et al. The polymorphism P315L of human toll-like receptor 1 impairs innate immune sensing of microbial cell wall components. J. Immunol. 2007;178:6387–6394. doi: 10.4049/jimmunol.178.10.6387. [DOI] [PubMed] [Google Scholar]
- 33.Park Y, et al. Association of the polymorphism for Toll-like receptor 2 with type 1 diabetes susceptibility. Ann. N. Y. Acad. Sci. 2004;1037:170–174. doi: 10.1196/annals.1337.028. [DOI] [PubMed] [Google Scholar]
- 34.Bochud PY, et al. Toll-like receptor 2 (TLR2) polymorphisms are associated with reversal reaction in leprosy. J. Infect. Dis. 2008;197:253–261. doi: 10.1086/524688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroder NW, et al. Heterozygous Arg753Gln polymorphism of human TLR-2 impairs immune activation by Borrelia burgdorferi and protects from late stage Lyme disease. J. Immunol. 2005;175:2534–2540. doi: 10.4049/jimmunol.175.4.2534. [DOI] [PubMed] [Google Scholar]
- 36.Merx S, et al. Characterization and investigation of single nucleotide polymorphisms and a novel TLR2 mutation in the human TLR2 gene. Hum. Mol. Genet. 2007;16:1225–1232. doi: 10.1093/hmg/ddm070. [DOI] [PubMed] [Google Scholar]
- 37.Mrabet-Dahbi S, et al. The Toll-like receptor 2 R753Q mutation modifies cytokine production and Toll-like receptor expression in atopic dermatitis. J. Allergy Clin. Immunol. 2008;121:1013–1019. doi: 10.1016/j.jaci.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 38.Ferreri AJ, et al. Marginal-zone lymphoma. Crit. Rev. Oncol. Hematol. 2007;63:245–256. doi: 10.1016/j.critrevonc.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Wotherspoon AC. Gastric MALT lymphoma and Helicobacter pylori. Yale J. Biol. Med. 1996;69:61–68. [PMC free article] [PubMed] [Google Scholar]
- 40.Hussell T, et al. The response of cells from low-grade B-cell gastric lymphomas of mucosa-associated lymphoid tissue to Helicobacter pylori. Lancet. 1993;342:571–574. doi: 10.1016/0140-6736(93)91408-e. [DOI] [PubMed] [Google Scholar]
- 41.Garbe C, et al. Borrelia burgdorferi-associated cutaneous B cell lymphoma: clinical and immunohistologic characterization of four cases. J. Am. Acad. Dermatol. 1991;24:584–590. doi: 10.1016/0190-9622(91)70088-j. [DOI] [PubMed] [Google Scholar]
- 42.Roggero E, et al. Eradication of Borrelia burgdorferi infection in primary marginal zone B-cell lymphoma of the skin. Hum. Pathol. 2000;31:263–268. doi: 10.1016/s0046-8177(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 43.Ferreri AJ, et al. Regression of ocular adnexal lymphoma after Chlamydia psittaci-eradicating antibiotic therapy. J. Clin. Oncol. 2005;23:5067–5073. doi: 10.1200/JCO.2005.07.083. [DOI] [PubMed] [Google Scholar]
- 44.Lecuit M, et al. Immunoproliferative small intestinal disease associated with Campylobacter jejuni. N. Engl. J. Med. 2004;350:239–248. doi: 10.1056/NEJMoa031887. [DOI] [PubMed] [Google Scholar]
- 45.Seve P, et al. Hepatitis C virus infection and B-cell non-Hodgkin's lymphoma: a cross-sectional study in Lyon, France. Eur. J. Gastroenterol. Hepatol. 2004;16:1361–1365. doi: 10.1097/00042737-200412000-00021. [DOI] [PubMed] [Google Scholar]
- 46.Ramos-Casals M, et al. Characterization of B cell lymphoma in patients with Sjogren's syndrome and hepatitis C virus infection. Arthritis Rheum. 2007;57:161–170. doi: 10.1002/art.22476. [DOI] [PubMed] [Google Scholar]
- 47.Smith MF, Jr, et al. Toll-like receptor (TLR) 2 and TLR5, but not TLR4, are required for Helicobacter pylori-induced NF-kappa B activation and chemokine expression by epithelial cells. J. Biol. Chem. 2003;278:32552–32560. doi: 10.1074/jbc.M305536200. [DOI] [PubMed] [Google Scholar]
- 48.Torok AM, et al. Helicobacter pylori induces interleukin-8 secretion by Toll-like receptor 2- and Toll-like receptor 5-dependent and -independent pathways. Infect. Immun. 2005;73:1523–1531. doi: 10.1128/IAI.73.3.1523-1531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexopoulou L, et al. Hyporesponsiveness to vaccination with Borrelia burgdorferi OspA in humans and in. Nat. Med. 2002;8:878–884. doi: 10.1038/nm732. [DOI] [PubMed] [Google Scholar]
- 50.Dolganiuc A, et al. Hepatitis C core and nonstructural 3 proteins trigger toll-like receptor 2-mediated pathways and inflammatory activation. Gastroenterology. 2004;127:1513–1524. doi: 10.1053/j.gastro.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 51.Rothman N, et al. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncol. 2006;7:27–38. doi: 10.1016/S1470-2045(05)70434-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.