Abstract
Summary:: The assessment of data quality is a major concern in microarray analysis. arrayQualityMetrics is a Bioconductor package that provides a report with diagnostic plots for one or two colour microarray data. The quality metrics assess reproducibility, identify apparent outlier arrays and compute measures of signal-to-noise ratio. The tool handles most current microarray technologies and is amenable to use in automated analysis pipelines or for automatic report generation, as well as for use by individuals. The diagnosis of quality remains, in principle, a context-dependent judgement, but our tool provides powerful, automated, objective and comprehensive instruments on which to base a decision.
Availability:: arrayQualityMetrics is a free and open source package, under LGPL license, available from the Bioconductor project at www.bioconductor.org. A users guide and examples are provided with the package. Some examples of HTML reports generated by arrayQualityMetrics can be found at http://www.microarray-quality.org
Contact:: audrey@ebi.ac.uk
Supplementary information:: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
As microarray data quality can be affected at each step of the microarray experiment processing (Schuchhardt et al., 2000), quality assessment is an integral part of the analysis. There are freely available tools allowing quality assessment for a specific microarray type, such as Affymetrix (Parman and Halling, 2005), Illumina (Dunning et al., 2007) and two-colour cDNA arrays (Buness et al., 2005). Other free tools are designed to identify a particular problem among which are spot quality (Li et al., 2005) or hybridization quality (Petri et al., 2004). Some tools perform outlier detection from quality metrics done before (Freue et al.,2007), or propose interactive quality plots (Lee et al., 2006). We developed a Bioconductor (Gentleman et al., 2004) package, arrayQualityMetrics, with the aim to provide a comprehensive tool that works on all expression arrays and platforms and produces a self-contained report which can be web-delivered. The Supplementary table shows a comparison with the functionality and scope of other Bioconductor packages concerned with quality assessment or outlier detection.
2 DESCRIPTION
Input: to perform an analysis using the arrayQualityMetrics package, one needs to provide the matrix of microarray intensities and optionally, information about the samples and the probes in a Bioconductor object of class AffyBatch, ExpressionSet, NChannelSet or BeadLevelList. These classes are widely used and well documented. The manner of calling the arrayQualityMetrics function to create a report is the same for all of these classes, and it can be applied to raw array intensities as well as to normalized data. Applied to raw intensities, the quality metrics can help with monitoring experimental procedures and with the choice of normalization procedure; application to the normalized data is more relevant for assessing the utility of the data in downstream analyses.
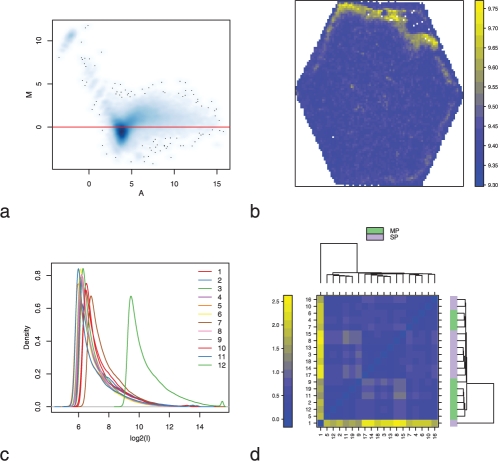
Individual array quality: the MA-plot allows the evaluation of the dependence between the intensity levels and the distribution of the ratios (Fig. 1a) (Dudoit et al., 2002). For two-colour arrays, a probe's M-value is the log-ratio of the two intensities and the A-value is the mean of their logarithms. In the case of one colour arrays, the M-value is computed by dividing the intensity by the median intensity of the same probe across all arrays. A false colour representation of each array's spatial distribution of feature intensities (Fig. 1b) helps in identifying spatial effects that may be caused by, for example, gradients in the hybridization chamber, air bubbles or printing problems.
Homogeneity between arrays: to assess the homogeneity between the arrays, boxplots of the log2 intensities and density estimate plots (Fig. 1c) are presented.
Between array comparison: Figure 1d shows a heatmap of between array distances, computed as the mean absolute difference of the M-value for each pair of arrays
| (1) |
where Mxi is the M-value of the i-th probe on the x-th array.
Consider the decomposition of Mxi.
| (2) |
where zi is the probe effect for probe i (the same across all arrays), εxi are i.i.d random variables with mean zero and βxi is a sparse matrix representing differential expression effects. Under these assumptions, all values dxy are approximately the same and deviations from this can be used to identify outlier arrays. The dendrogram can serve to check if the experiments cluster in accordance with the sample classes.
Affymetrix specific plots: four Affymetrix-specific metrics are evaluated if the input object is an AffyBatch. The RNA degradation plot from the affy package (Gautier et al., 2004),, the relative log expression (RLE) boxplots and the normalized unscaled standard error (NUSE) boxplots from the affyPLM package (Brettschneider et al., 2007) and the QC stat plot from the simpleaffy package (Wilson and Miller, 2005) are represented.
Fig. 1.
(a) MA-plot for an Agilent microarray. The M-values are not centered on zero meaning that there is a dependency between the intensities and the log-ratio. (b) Spatial distribution of the background of the green channel for an Illumina chip. There is an abnormal distribution of high intensities at the top border of the array. (c) Density plot of the log-intensities of an Affymetrix set of arrays (E-GEOD-349 ArrayExpress set). The density of one of the arrays is shifted on the x-axis. (d) Heatmap of the ArrayExpress Affymetrix data set E-GEOD-1571. Array 18 is an outlier.
Scores: to guide the interpretation of the report, we have included the computation of numeric scores associated with the plots. Outliers are detected on the MA-plot, spatial distributions of the features’ intensities, boxplot, heatmap, RLE and NUSE. The mean of the absolute value of M is computed for each array and those that lie beyond the extremes of the boxplot's whiskers are considered as possible outliers arrays. The same approach, i.e. using the whiskers of the boxplot, is applied to the following: the mean and interquartile range (IQR) from the boxplots and NUSE, the sums of the rows of the distance matrix, and the relative amplitude of low versus high frequence components of the Fourier transformation. In the case of the RLE plot, any array with a median RLE higher than 0.1 is considered an outlier.
Report: the metrics are rendered as figures with legends in a detailed report and the scores are used to provide a summary table. Examples of reports are provided at http://www.microarray-quality.org/quality_metrics.html.
3 CONCLUSION
arrayQualityMetrics supports the quality assessment of many types of microarrays in R. After preparation of the data, a single command line is used to create the report. The main benefits of arrayQualityMetrics are its simplicity of use, the ability to have the same report for different types of platforms, and the opportunity for users or developers to extend it for their needs. This tool can be used for individual data analyses or in routine data production pipelines, to provide fast uniform reporting.
Supplementary Material
Acknowledgments
We would like to thank the developers of the R and Bioconductor packages that we are using, especially Ben Bolstad, Mark Dunning, Crispin Miller, Gregoire Pau and Deepayan Sarkar.
Funding: EU FP6 (EMERALD, Project no. LSHG-CT-2006-037686 to A.K.). National Institutes of Health (P41HG004059 R.G.)
Conflict of Interest: none declared.
References
- Brettschneider J, et al. arXiv:0710.0178v2. 2007. Quality assessment for short oligonucleotide arrays. [Google Scholar]
- Buness A. array{M}agic: two-colour c{DNA} microarray quality control and preprocessing. Bioinformatics. 2005;21:554–556. doi: 10.1093/bioinformatics/bti052. [DOI] [PubMed] [Google Scholar]
- Dudoit S. Statistical methods for identifying differentially expressed genes in replicated cdna microarray experiments. Stat. Sinica. 2002;12:111–139. [Google Scholar]
- Dunning MJ. beadarray: R classes and methods for {I}llumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- Freue GVC, et al. MDQC: a new quality assessment method for microarrays based on quality control reports. Bioinformatics. 2007;23:3162–3169. doi: 10.1093/bioinformatics/btm487. [DOI] [PubMed] [Google Scholar]
- Gautier L. affy – analysis of affymetrix genechip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gentleman RC. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E-K, et al. array{QC}plot: software for checking the quality of microarray data. Bioinformatics. 2006;22:2305–2307. doi: 10.1093/bioinformatics/btl367. [DOI] [PubMed] [Google Scholar]
- Li Q. Donuts, scratches and blanks: robust model-based segmentation of microarray images. Bioinformatics. 2005;21:2875–2882. doi: 10.1093/bioinformatics/bti447. [DOI] [PubMed] [Google Scholar]
- Parman C, Halling C. affyQCReport: QC Report Generation for affyBatch objects. 2005. R package version 1.17.0. [Google Scholar]
- Petri A. Array-a-lizer: a serial DNA microarray quality analyzer. BMC Bioinformatics. 2004;5:12. doi: 10.1186/1471-2105-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchhardt J. Normalization strategies for c{DNA} microarrays. Nucleic Acids Res. 2000;28:E47. doi: 10.1093/nar/28.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Miller CJ. Simpleaffy: a bioconductor package for {A}ffymetrix quality control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.