Abstract
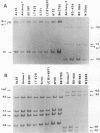
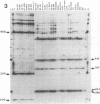
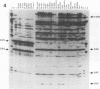
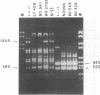
Leptospira serovar hardjo isolates of the hardjoprajitno and hardjobovis genotypes were characterized by ribotyping, arbitrarily primed PCR (AP-PCR) fingerprinting, and the study of mapped restriction site polymorphisms (MRSPs) in rrs and rrl genes. After restriction of chromosomal DNA with BglII, EcoRI, or HindIII, each genotype was individualized with a distinct ribotype. The fingerprints produced by AP-PCR with seven primers clearly separated the two groups; primers KF and RSP produced species-specific products which assigned hardjoprajitno and hardjobovis isolates to the species L. interrogans sensu stricto and L. borgpetersenii, respectively. Furthermore, AP-PCR fingerprints gave evidence of a considerable genomic heterogeneity at the strain level among the hardjobovis group. Conversely, the hardjoprajitno group was homogeneous. MRSP profiles in ribosomal genes indicated that hardjoprajitno and hardjobovis isolates belonged to L. interrogans MRSP group B and L. borgpetersenii group C, respectively. AP-PCR and determination of MRSPs in ribosomal genes proved to be quick and reliable methods for typing Leptospira strains and for studying intraspecific population structures.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brenner D. J., McWhorter A. C., Knutson J. K., Steigerwalt A. G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982 Jun;15(6):1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney B. G., Colley J., Djordjevic S. P., Whittington R., Graham G. C. Rapid identification of some Leptospira isolates from cattle by random amplified polymorphic DNA fingerprinting. J Clin Microbiol. 1993 Nov;31(11):2927–2932. doi: 10.1128/jcm.31.11.2927-2932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. J. Leptospira interrogans serovar Hardjo: an abortifacient in New Zealand? A review of the literature. N Z Vet J. 1983 Jul;31(7):107–109. doi: 10.1080/00480169.1983.34986. [DOI] [PubMed] [Google Scholar]
- ELLINGHAUSEN H. C., Jr, MCCULLOUGH W. G. NUTRITION OF LEPTOSPIRA POMONA AND GROWTH OF 13 OTHER SEROTYPES: FRACTIONATION OF OLEIC ALBUMIN COMPLEX AND A MEDIUM OF BOVINE ALBUMIN AND POLYSORBATE 80. Am J Vet Res. 1965 Jan;26:45–51. [PubMed] [Google Scholar]
- Ellis W. A., Montgomery J. M., Thiermann A. B. Restriction endonuclease analysis as a taxonomic tool in the study of pig isolates belonging to the Australis serogroup of Leptospira interrogans. J Clin Microbiol. 1991 May;29(5):957–961. doi: 10.1128/jcm.29.5.957-961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis W. A., O'Brien J. J., Bryson D. G., Mackie D. P. Bovine leptospirosis: some clinical features of serovar hardjo infection. Vet Rec. 1985 Aug 3;117(5):101–104. doi: 10.1136/vr.117.5.101. [DOI] [PubMed] [Google Scholar]
- Ellis W. A., Songer J. G., Montgomery J., Cassells J. A. Prevalence of Leptospira interrogans serovar hardjo in the genital and urinary tracts of non-pregnant cattle. Vet Rec. 1986 Jan 4;118(1):11–13. doi: 10.1136/vr.118.1.11. [DOI] [PubMed] [Google Scholar]
- Ellis W. A., Thiermann A. B., Montgomery J., Handsaker A., Winter P. J., Marshall R. B. Restriction endonuclease analysis of Leptospira interrogans serovar hardjo isolates from cattle. Res Vet Sci. 1988 May;44(3):375–379. [PubMed] [Google Scholar]
- Ezeh A. O., Adesiyun A. A., Addo P. B., Ellis W. A., Makinde A. A., Bello C. S. Serological and cultural examination for human leptospirosis in Plateau State, Nigeria. Cent Afr J Med. 1991 Jan;37(1):11–15. [PubMed] [Google Scholar]
- Fukunaga M., Horie I., Mifuchi I. Nucleotide sequence of a 23S ribosomal RNA gene from Leptospira interrogans serovar canicola strain Moulton. Nucleic Acids Res. 1989 Mar 11;17(5):2123–2123. doi: 10.1093/nar/17.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga M., Horie I., Okuzako N., Mifuchi I. Nucleotide sequence of a 16S rRNA gene for Leptospira interrogans serovar canicola strain Moulton. Nucleic Acids Res. 1990 Jan 25;18(2):366–366. doi: 10.1093/nar/18.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont F., Chevrier D., Grimont P. A., Lefevre M., Guesdon J. L. Acetylaminofluorene-labelled ribosomal RNA for use in molecular epidemiology and taxonomy. Res Microbiol. 1989 Sep;140(7):447–454. doi: 10.1016/0923-2508(89)90065-x. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Herrmann J. L., Baril C., Bellenger E., Perolat P., Baranton G., Saint Girons I. Genome conservation in isolates of Leptospira interrogans. J Bacteriol. 1991 Dec;173(23):7582–7588. doi: 10.1128/jb.173.23.7582-7588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J. L., Bellenger E., Perolat P., Baranton G., Saint Girons I. Pulsed-field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovar identification. J Clin Microbiol. 1992 Jul;30(7):1696–1702. doi: 10.1128/jcm.30.7.1696-1702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu D., Pan M. J., Zee Y. C., LeFebvre R. B. Unique ribosome structure of Leptospira interrogans is composed of four rRNA components. J Bacteriol. 1990 Jun;172(6):3478–3480. doi: 10.1128/jb.172.6.3478-3480.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFebvre R. B. DNA probe for detection of the Leptospira interrogans serovar hardjo genotype hardjo-bovis. J Clin Microbiol. 1987 Nov;25(11):2236–2238. doi: 10.1128/jcm.25.11.2236-2238.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFebvre R. B., Thiermann A. B., Foley J. Genetic and antigenic differences of serologically indistinguishable leptospires of serovar hardjo. J Clin Microbiol. 1987 Nov;25(11):2094–2097. doi: 10.1128/jcm.25.11.2094-2097.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R. B., Wilton B. E., Robinson A. J. Identification of Leptospira serovars by restriction-endonuclease analysis. J Med Microbiol. 1981 Feb;14(1):163–166. doi: 10.1099/00222615-14-1-163. [DOI] [PubMed] [Google Scholar]
- Marshall R. B., Winter P. J., Thiermann A. B., Ellis W. A. Genotypes of Leptospira interrogans serovar hardjo in cattle in the UK. Vet Rec. 1985 Dec 21;117(25-26):669–670. doi: 10.1136/vr.117.25-26.669. [DOI] [PubMed] [Google Scholar]
- Perolat P., Lecuyer I., Postic D., Baranton G. Diversity of ribosomal DNA fingerprints of Leptospira serovars provides a database for subtyping and species assignation. Res Microbiol. 1993 Jan;144(1):5–15. doi: 10.1016/0923-2508(93)90210-s. [DOI] [PubMed] [Google Scholar]
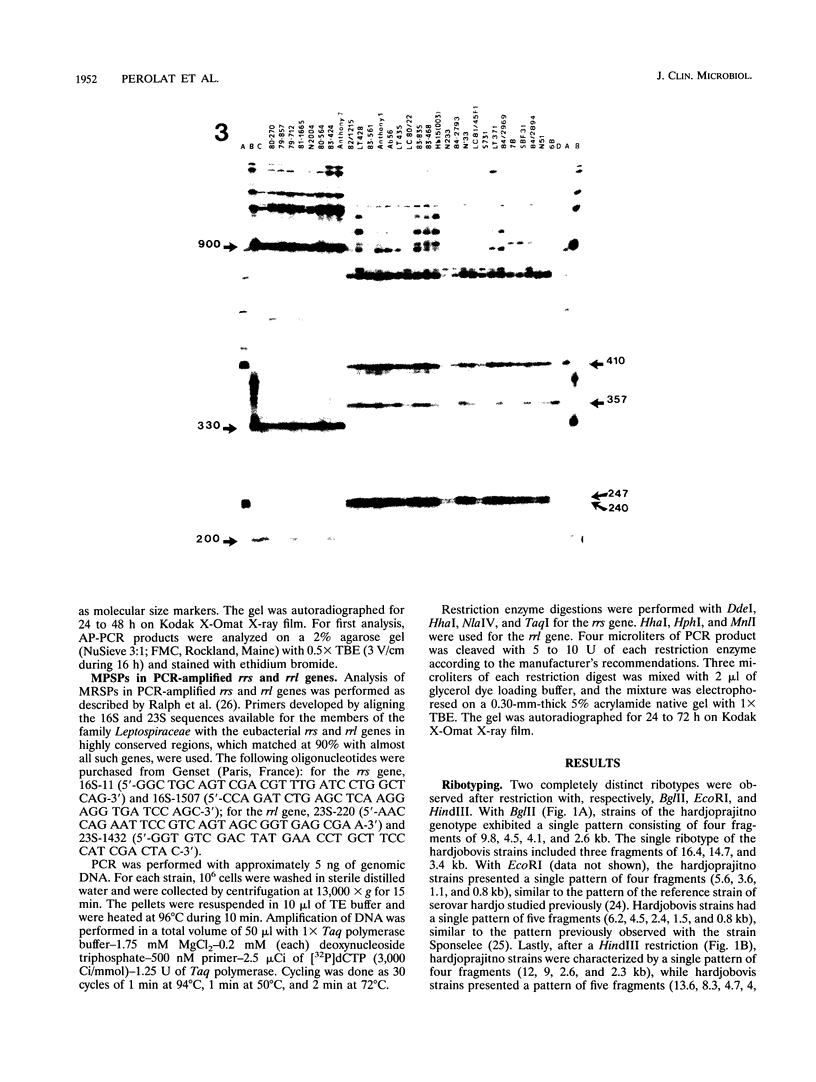
- Pérolat P., Grimont F., Regnault B., Grimont P. A., Fournié E., Thevenet H., Baranton G. rRNA gene restriction patterns of Leptospira: a molecular typing system. Res Microbiol. 1990 Feb;141(2):159–171. doi: 10.1016/0923-2508(90)90025-l. [DOI] [PubMed] [Google Scholar]
- Ralph D., McClelland M., Welsh J., Baranton G., Perolat P. Leptospira species categorized by arbitrarily primed polymerase chain reaction (PCR) and by mapped restriction polymorphisms in PCR-amplified rRNA genes. J Bacteriol. 1993 Feb;175(4):973–981. doi: 10.1128/jb.175.4.973-981.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadass P., Jarvis B. D., Corner R. J., Penny D., Marshall R. B. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int J Syst Bacteriol. 1992 Apr;42(2):215–219. doi: 10.1099/00207713-42-2-215. [DOI] [PubMed] [Google Scholar]
- Ramadass P., Marshall R. B., Jarvis B. D. Species differentiation of Leptospira interrogans serovar hardjo strain Hardjobovis from strain Hardjoprajitno by DNA slot blot hybridisation. Res Vet Sci. 1990 Sep;49(2):194–197. [PubMed] [Google Scholar]
- Robinson A. J., Ramadass P., Lee A., Marshall R. B. Differentiation of subtypes within Leptospira interrogans serovars Hardjo, Balcanica and Tarassovi, by bacterial restriction-endonuclease DNA analysis (BRENDA). J Med Microbiol. 1982 Aug;15(3):331–338. doi: 10.1099/00222615-15-3-331. [DOI] [PubMed] [Google Scholar]
- Segers R. P., van Gestel J. A., van Eys G. J., van der Zeijst B. A., Gaastra W. Presence of putative sphingomyelinase genes among members of the family Leptospiraceae. Infect Immun. 1992 Apr;60(4):1707–1710. doi: 10.1128/iai.60.4.1707-1710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiermann A. B., Handsaker A. L., Foley J. W., White F. H., Kingscote B. F. Reclassification of North American leptospiral isolates belonging to serogroups Mini and Sejroe by restriction endonuclease analysis. Am J Vet Res. 1986 Jan;47(1):61–66. [PubMed] [Google Scholar]
- Van Eys G. J., Gerritsen M. J., Korver H., Schoone G. J., Kroon C. C., Terpstra W. J. Characterization of serovars of the genus Leptospira by DNA hybridization with hardjobovis and icterohaemorrhagiae recombinant probes with special attention to serogroup sejroe. J Clin Microbiol. 1991 May;29(5):1042–1048. doi: 10.1128/jcm.29.5.1042-1048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh T. U., Shi M. H., Adler B., Faine S. Characterization and taxonomic significance of lipopolysaccharides of Leptospira interrogans serovar hardjo. J Gen Microbiol. 1989 Oct;135(10):2663–2673. doi: 10.1099/00221287-135-10-2663. [DOI] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., Pretzman C., Postic D., Saint Girons I., Baranton G., McClelland M. Genomic fingerprinting by arbitrarily primed polymerase chain reaction resolves Borrelia burgdorferi into three distinct phyletic groups. Int J Syst Bacteriol. 1992 Jul;42(3):370–377. doi: 10.1099/00207713-42-3-370. [DOI] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuerner R. L., Bolin C. A. Nucleic acid probe characterizes Leptospira interrogans serovars by restriction fragment length polymorphisms. Vet Microbiol. 1990 Sep;24(3-4):355–366. doi: 10.1016/0378-1135(90)90183-v. [DOI] [PubMed] [Google Scholar]
- Zuerner R. L., Bolin C. A. Repetitive sequence element cloned from Leptospira interrogans serovar hardjo type hardjo-bovis provides a sensitive diagnostic probe for bovine leptospirosis. J Clin Microbiol. 1988 Dec;26(12):2495–2500. doi: 10.1128/jcm.26.12.2495-2500.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuerner R. L., Ellis W. A., Bolin C. A., Montgomery J. M. Restriction fragment length polymorphisms distinguish Leptospira borgpetersenii serovar hardjo type hardjo-bovis isolates from different geographical locations. J Clin Microbiol. 1993 Mar;31(3):578–583. doi: 10.1128/jcm.31.3.578-583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]