Abstract
Vascular endothelial growth factor (VEGF) is a major regulator of angiogenesis in the process of tumor growth and metastasis in esophageal adenocarcinoma (EA). Polymorphisms in the VEGF gene have been associated with altered VEGF expression and plasma VEGF levels. We hypothesized that polymorphisms of VEGF may contribute to EA risk. Functional polymorphisms in the VEGF gene (−460C/T, +405C/G and +936C/T) were determined in 308 patients with EA and 546 healthy controls. Logistic regression analysis was employed to assess the associations between genotypes, haplotypes of VEGF and EA risk, adjusting for multiple confounding factors. Compared with the +936CC genotype, the combined +936CT+TT genotypes were significantly associated with increased risk of developing EA, with adjusted odds ratio (OR) = 1.49 [95% confidence interval (CI), 1.05–2.12; P = 0.027]. The −460CT+CC were associated with increased risk of EA in smokers (adjusted OR = 1.57; 95% CI, 1.07–2.30; P = 0.021), whereas the −460CT/CC were associated with decreased risk of EA (adjusted OR = 0.47; 95% CI, 0.25–0.91; P = 0.025) in non-smokers. Compared with non-smokers with the +460TT, smokers with the +460CT+CC had significantly higher risk of EA (adjusted OR = 3.32; 95% CI, 1.56–7.10; P = 0.002). No overall or interacting association with EA risk was found for the +405C/G polymorphism. Haplotype CGT (−460C/+405G/+936T) was significantly associated with higher risk of EA (adjusted OR = 1.70; 95% CI, 1.04–2.73; P = 0.034). These results suggested that cigarette smoking modifies the association between VEGF polymorphisms and EA risk among Caucasians.
Introduction
The incidence of esophageal adenocarcinoma (EA) has increased 300–400% in western countries over the past four decades (1). It is widely accepted that EA develops on a background of Barrett’s esophagus, a metaplastic columnar-lined epithelium that replaces the normal squamous cell epithelium of the distal esophagus (2). Progression of Barrett’s esophagus into invasive EA is reflected histologically by the metaplasia–dysplasia–carcinoma sequence (3,4). Although smoking, Barrett’s esophagus, obesity and gastroesophageal reflux disease (GERD) are identified as risk factors for EA, only a fraction of individuals with these risk factors develop EA, which suggests that both genetic factors and lifestyle risk factors may modulate individual susceptibility to EA risk.
Angiogenesis is a process in which endothelial cells divide and migrate to form new capillaries that support the continuous growth of tumor through blood flow (5). There is increasing evidence that angiogenesis is an essential process in cancer development and metastasis (6). Also, angiogenesis has been associated with the metaplasia–dysplasia–carcinoma sequence of Barrett’s carcinoma (7,8). Vascular endothelial growth factor (VEGF) is a critical angiogenic factor, and inhibition of VEGF has been correlated with suppression of tumor growth and angiogenesis (9–13). Excessive VEGF expression and VEGF-induced angiogenesis have been involved in the precancerous lesions of esophagus and EA (14–18), suggesting that VEGF-driven angiogenesis might be an early event in EA carcinogenesis. Functional genetic polymorphisms at positions −460C/T, +405C/G and +936C/T in the VEGF gene have been correlated to VEGF promoter activity, gene expression, protein production (19–21) and risks of other cancers (22–26). The aims of the present study were to investigate whether these functional VEGF polymorphisms are associated with increased risk of EA and whether the associations between VEGF polymorphisms and EA risk are modulated by smoking or other risk factors.
Materials and methods
Study population
This study was approved by the Human Subjects Committees of Massachusetts General Hospital, Dana Farber Cancer Institute and the Harvard School of Public Health (Boston, MA). All participants provided informed, written consent. Incident patients with histologically confirmed EA were recruited prospectively at Massachusetts General Hospital between 1999 and 2005 and at Dana Farber Cancer Institute between 2004 and 2005. All cases were >18 years of age and were diagnosed within 6 months prior to study entry. All patients had tumors that were deemed endoscopically or at the time of resection to have a tumor center located at or above the gastroesophageal junction (midpoint of length located at or above the gastroesophageal junction) and to have at least two-thirds of the bulk tumor located in the esophagus. The controls were comprised of healthy non-blood-related family members (usually spouses) and friends of other thoracic cancer/surgical patients who were visiting patients, also over the age of 18 years. Controls who reported a previous diagnosis of any cancer (except non-melanoma skin cancer) were excluded from participation. For both cases and controls, interviewer-administered questionnaires collected information on age, gender, race, body mass index, smoking status (non-, ex- and current smokers) and pack-years of smoking. Smoking habits were defined at 1 year prior to diagnosis for cases or 1 year prior to interview for controls. Non-smokers smoked <100 cigarettes in their lifetime. Questions on GERD symptoms in controls were added midway through recruitment. Chronic GERD was defined as having reflux, heartburn or regurgitation symptoms at least once a month for at least a 6 month continuous period in one’s lifetime.
Genotyping assays
Genome DNA was extracted from peripheral blood samples using the Puregene DNA Isolation Kit (Gentra Systems/Qiagen, Valencia, CA) following the manufacturer’s protocol. The allelic discrimination of the VEGF gene was assessed using Taqman and minor groove binder assays. The primers and probes for the −460C/T (rs833061) and −405C/G (rs2010963) were ordered from Applied Biosystems (Foster city, CA) and the +936C/T (rs3025039) from Nanogen (Bothell, WA). The wild-type genotype probes were FAM labeled and the variant genotype probes were VIC labeled. Genotyping was performed by laboratory personnel blinded to subject status, and a random 10% of the samples were repeated to validate genotyping procedures. Two authors reviewed independently all genotyping results.
Statistical analysis
We restricted the analysis to Caucasians, because Caucasians composed >90% of our study population. Demographic characteristics between cases and controls were compared using the chi-square, Fisher exact and Student’s t-test, where appropriate. Hardy–Weinberg disequilibrium of the VEGF polymorphisms was checked in controls. Detection of linkage disequilibrium among the three polymorphisms was based on Lewontin’s D′ in controls. Haplotypes of the three VEGF polymorphisms were generated using the SAS macro HAPPY program (27). Missing variables were analyzed using the Multiple Imputation process.
Genotype and haplotype associations with EA risk were analyzed using unconditional logistic regression models. Logistic regression models were fit to examine the relationship between the log odds of EA and each polymorphism, after adjusting for possible confounding factors such as age, gender, smoking status, pack-years of smoking, years since smoking cessation (if ex-smoker), body mass index at age 18 and GERD. Firstly, we investigated the associations between individual VEGF polymorphisms and the risk of EA in separate logistic regression models. Secondly, we estimated gene–environment interactions using genotype–smoking interaction models. Lastly, we investigated the associations between VEGF haplotypes and the risk of EA, treating expected haplotype scores as observed covariates in a standard unconditional logistic analysis, instead of assigning each subject with the most likely haplotype pair. All statistical analyses were undertaken using the SAS statistical packages (SAS version 9.1 Institute, Cary, NC).
Results
Population characteristics
Basic demographic characteristics for cases and controls are summarized in Table I. All cases and controls were Caucasians. Participation rates for both cases and controls exceeded 85%. Mean ages were not significantly different for cases and controls. There were more males in cases than in controls. Body mass index at 18 years old in cases was significantly higher than that in controls (P = 0.003). Not surprisingly, more smokers, higher smoking pack-years and higher GERD prevalence were seen in the cases, as these are known risk factors for the development of EA (P < 0.0001).
Table I.
Demographic characteristics of cases and controls
| Characteristics | Cases (n = 308) | Controls (n = 546) | P-value |
| Age (years) | 63.3 ± 11.6 | 62.7 ± 12.0 | 0.558 |
| Gender | 0.008 | ||
| Female | 34 (11.0%) | 98 (17.9%) | |
| Male | 274 (89.0%) | 448 (82.1%) | |
| BMI (kg/m2) at 18 years old | 23.4 ± 3.7 | 22.5 ± 3.4 | 0.003a |
| GERDb, N (%) | 157 (51.1%) | 98 (26.7%) | <0.0001 |
| Smoking status | 0.001 | ||
| Non-smokers | 60 (19.5%) | 166 (30.4%) | |
| Ever-smokers | 172 (55.8%) | 278 (50.9%) | |
| Current smokers | 76 (24.7%) | 102 (18.7%) | |
| Pack-years (median, range) | 24 (0.1–212) | 15 (0.1–218) | <0.0001 |
BMI, body mass index.
Median test.
GERD data was missing in 179 controls and 1 cases.
Associations between VEGF genotypes and EA risk
Each of the VEGF polymorphisms in the controls was consistent with Hardy–Weinberg equilibrium (P > 0.05, chi-squared goodness of fit). Genotype frequencies of the −460C/T, +405C/G and +936C/T polymorphisms in controls were in close agreement with those previously published for healthy Caucasian individuals (28,29). Table II shows VEGF genotype distributions between EA cases and controls. There were no statistically significant overall differences between cases and controls for the distributions of −460C/T, +405C/G and +936C/T polymorphisms. When the combined variant genotypes were compared with the corresponding wild-type genotypes, the +936CT+TT and the +405CG+CC genotypes in cases were significantly differed between cases and controls (Table II).
Table II.
Genotype and haplotype distributions between EA cases and controls
| Polymorphism | Cases (n = 308) | Controls (n = 546) | Pa-value |
| VEGF−460C/T | |||
| TT | 72 (23.4%) | 149 (27.3%) | 0.296 |
| CT | 155 (50.3%) | 275 (50.4%) | |
| CC | 81 (26.3%) | 122 (22.3%) | |
| CT/CC | 236 (76.6%) | 397 (72.7%) | 0.223 |
| VEGF +405C/G | |||
| GG | 155 (50.3%) | 233 (42.7%) | 0.097 |
| CG | 124 (40.3%) | 251 (46.0%) | |
| CC | 29 (9.4%) | 62 (11.4%) | |
| CG/CC | 153 (49.7%) | 313 (57.3%) | 0.032 |
| VEGF +936C/T | |||
| CC | 228 (74.0%) | 441 (80.8%) | 0.068 |
| CT | 73 (23.7%) | 97 (17.8%) | |
| TT | 7 (2.3%) | 8 (1.4%) | |
| CT/TT | 80 (25.9%) | 105 (19.2%) | 0.025 |
| Haplotype | |||
| CGC | 42.3% | 41.5% | 0.022 |
| TCC | 25.3% | 29.3% | |
| TGC | 18.2% | 18.0% | |
| CGT | 9.1% | 5.2% | |
| TCT | 4.2% | 4.2% | |
| Othersb | 0.9% | 1.8% |
Fisher's exact test.
Haplotypes with frequencies < 4%.
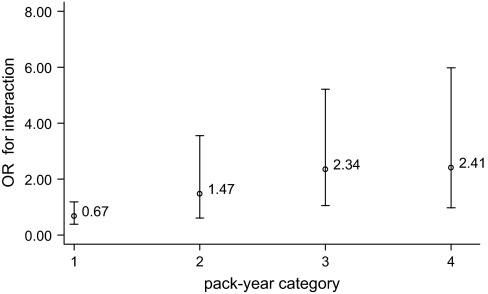
There were no overall associations between individual −460C/T genotypes and the risk of EA. However, in non-smokers, the −460CT [adjusted odds ratio (OR) = 0.36; 95% confidence interval (CI), 0.18–0.73; P = 0.005] and the −460CT+CC genotypes were significantly associated with lower risk of developing EA (adjusted OR = 0.49; 95% CI, 0.25–0.91; P = 0.025). In contrast, the −460CT (adjusted OR = 1.59; 95% CI, 1.06–2.38; P = 0.025) and the −460CT+CC (adjusted OR = 1.57; 95% CI, 1.07–2.30; P = 0.021) were associated with higher risk of EA in smokers (Table III). We then evaluated a genotype–smoking interaction model using non-smokers with the wild-type genotypes as reference groups. We observed a gene–smoking interaction between the −460C/T polymorphism and smoking status (non-smoker versus ever-smoker). The adjusted OR for gene–smoking interactions was 3.32 (95% CI, 1.56–7.10; interaction P = 0.002), (Table IV). To further quantify gene–smoking interaction, we calculated ORs for interaction based on different smoking pack-year categories. We observed that the ORs of −460C/T–pack-year interaction were consistently correlated to the pack-year levels in a dose-response manner (Figure 1).
Table III.
Associations of VEGF polymorphisms with EA risks
| Polymorphism | Major effects | Non-smokers | Ever-smokers |
| ORa (95% CI) | ORa (95% CI) | ORa (95% CI) | |
| VEGF−460C/T | |||
| TT | 1.0 | 1.0 | 1.0 |
| CT | 1.22 (0.78–1.93) | 0.36 (0.18–0.73) | 1.59 (1.06–2.38) |
| CC | 1.22 (0.78–1.71) | 0.79 (0.36–1.75) | 1.54 (0.96–2.46) |
| CT/CC | 1.18 (0.81–1.71) | 0.47 (0.25–0.91) | 1.57 (1.07–2.30) |
| VEGF+405C/G | |||
| GG | 1.0 | 1.0 | 1.0 |
| CG | 0.75 (0.55–1.02) | 0.54 (0.29–1.04) | 0.83 (0.59–1.17) |
| CC | 0.78 (0.48–1.28) | 1.73 (0.64–4.69) | 0.60 (0.34–1.06) |
| CG/GG | 0.76 (0.57–1.01) | 0.67 (0.37–1.23) | 0.78 (0.56–1.08) |
| VEGF+936C/T | |||
| CC | 1.0 | 1.0 | 1.0 |
| CT | 1.50 (1.07–2.11) | 2.12 (1.03–4.40) | 1.32 (0.89–1.96) |
| TT | 1.62 (0.57–4.66) | 2.06 (0.32–13.36) | 1.53 (0.43–5.44) |
| CT/TT | 1.49 (1.05–2.12) | 2.12 (1.05–4.25) | 1.33 (0.91–1.96) |
| Haplotype | Global test P < 0.001 | ||
| CGC | 1.0 | 1.0 | 1.0 |
| TCC | 0.95 (0.72–1.26) | 1.19 (0.65–2.17) | 0.85 (0.62–1.17) |
| TGC | 1.00 (0.74–1.38) | 1.28 (0.64–2.56) | 0.90 (0.63–1.29) |
| CGT | 1.70 (1.05–2.69) | 2.46 (0.91–6.63) | 1.47 (0.83–2.60) |
| TCT | 0.94 (0.51–1.74) | 1.00 (0.26–3.83) | 0.92 (0.46–1.83) |
Adjusted for age, gender, body mass index at 18 years, GERD and pack-years.
Table IV.
Interaction of VEGF genotypes and smoking status in EA risk
| Genotype–smoking interaction | Adjusted ORa | Pb-value |
| VEGF−460C/Tb smoking | 3.32 (1.56–7.10) | 0.002 |
| VEGF+405C/Gb smoking | 1.45 (0.97–2.17) | 0.074 |
| VEGF+936C/Tb smoking | 0.63 (0.28–1.40) | 0.254 |
OR for interaction.
Adjusted for age, gender, body mass index at 18 years and GERD.
Fig. 1.
The ORs of smoking and VEGF–460C/T interaction for the development of EA at different categories of pack-years. Category 1 = pack-years ≤1; Category 2 = pack-years 1–20; Category 3 = pack-years 20–50 and Category 4 = pack-years >50.
For the +936C/T polymorphism, the adjusted OR of the variant +936CT+TT genotype versus the wild-type +936CC was 1.49; 95% CI, 1.05–2.12; P = 0.027 (Table III). No significant interaction was detected between the +936C/T polymorphism and smoking in the risk of EA. No significant association was found between the +405C/G polymorphism and EA risk.
Association between VEGF haplotypes and EA risk
There were six common haplotypes (>2%) among both cases and controls. The distribution of different haplotypes was similar between cases and controls (Table II) and were comparable with previous reports in Caucasians (28,29). The most common haplotype was the −460C/+405G/+936C (CGC), with the frequencies of 41% in cases and 42% in controls. In overall analysis, haplotype frequencies in cases were significantly different from that in controls (P = 0.022). In haplotype association analysis where the most common haplotype CGC was treated as reference group, the CGT haplotype was significantly associated with higher risk of EA development (adjusted OR = 1.70; 95% CI, 1.05–2.69; P = 0.032; global test P < 0.0001). No interactions between any haplotypes and smoking status were found in stratified analyses.
Discussion
VEGF-induced angiogenesis is an early event of EA development. Polymorphisms that can alter VEGF expression and protein production may contribute to the risk of EA. We tested this hypothesis by evaluating the relationship between three functional polymorphisms of VEGF gene and the risk of EA. We found that the +936C/T polymorphism and specific VEGF haplotypes were significantly associated with higher risk of EA. In addition, we found a significant interaction between smoking and −460C/T polymorphism and a greater risk of developing EA.
The +936C/T polymorphism has been studied in relation to various cancers with disparate results. It has been reported to be associated with decreased risk of breast cancer and small cell lung cancer (22,24), but another lung cancer study found no relationship (29). In separate studies, this polymorphism has been studied in several cancers with mixed results (28,30–32). We know of no previous studies investigating the role of +936C/T polymorphism in EA risk. In the present study, we found that the variant T allele of the +936C/T polymorphism was significantly associated with increased risk of EA. Consistent with this result, haplotype CGT that contains the +936T allele showed a similar association with the risk of EA. The observed +936CT+TT–EA associations in our study were consistent with reports of the function of +936CT+TT in other cancers that are characterized histologically by adenocarcinomas, such as colorectal, breast and gastric cancers (30,31,33).
The strong interaction effects of the −460C/T polymorphism and smoking suggest that this polymorphism is not an independent risk factor, but probably an effect modifier that acts synergistically with smoking on EA risk. Previous studies that explored the relationship between VEGF −460C/T genotypes and cancer risk found inconsistent results. The −460C/T polymorphism was found to be associated with prostate cancer risk (34), but no associations were observed in malignant melanoma, lung and renal cell cancers (29,35,36). The reasons for these discrepancies are not completely understood, but if the effect of this polymorphism is only through an interaction with smoking, then main effects may not be observed depending on the distribution of smoking variables across cases and controls. Further, the relevant interactions with smoking may be different as the magnitudes of relationship between cigarette smoking and various cancer risks are different.
Mechanisms for the interaction between smoking and the −460C/T polymorphism could be at the level of gene expression or protein activity through angiogenesis pathways (37,38). The −460C/T polymorphisms have been associated with variation in VEGF expression and protein production (20,21). Smoking can stimulate both angiogenesis and VEGF expression, which exacerbates the procarcinogenic effect of angiogenesis (37–40). Additionally, it has been shown that smoke-induced VEGF expression and release were mediated by other pathways, such as the epidermal growth factor receptor–extracellular signal-regulated kinase, 5-lipoxygenase, oxidative stress, inflammatory responses and beta-adrenoceptors pathways (38,39,41,42). It is possible that cigarette smoke and VEGF activate multiple effects on EA carcinogenesis.
We found no overall association of +405C/G or its interaction with smoking on EA risk. There are conflicting reports in the literature on the exact function of the +405G/C polymorphism. The variant C allele of the +405G/C polymorphism has been associated with lower VEGF production, and the presence of a C allele may disrupt the myeloid zinc finger 1 transcription factor-binding site (20). However, some groups reported that higher VEGF level or no association with the +405C/C genotype (43–45). Any association between +405C/G and EA may be through its linkage disequilibrium with −460C/T and +936C/T.
We recognize several limitations to our study. Firstly, we only evaluated three functional polymorphisms of the VEGF gene. Whether other genetic variants in the VEGF gene would also be associated with EA risk requires further investigation. Secondly, although we adjusted for various confounding variables in our analysis, diet, environmental and occupational exposure data were not included in our logistic regression models because of incomplete or missing information. However, given the consistent results of individual genotypes, gene–environment interaction effects and haplotype analysis, these potential confounders would probably not substantially change the results. Thirdly, the significant associations between VEGF polymorphisms and EA risk were observed in only one study population. These findings should be validated in independent studies, as well as expanded to cover more completely the entire angiogenic pathway. Finally, multiple comparisons may have led to spuriously significant results. Even if we corrected the level of significance for the number of main and gene–smoking interaction analyses (12 in total) using the Bonferroni method, the interaction between −460C/T and smoking status would still be significant.
In summary, this is the first report to our knowledge of the associations between VEGF genotypes, haplotypes and gene–smoking interaction with the risk of EA. Our findings emphasize the importance of considering joint effects of genetic polymorphisms and environmental factors when investigating the risks of EA carcinogenesis. Confirmatory studies and in vitro functional assays are needed to confirm the results and identify the mechanisms underlining the associations.
Funding
National Institute of Health (CA92824, CA74386, CA90578, ES/CA 06409 and CA119650); Flight Attendant Medical Research Institute (062459_YCSA); Kevin Jackson Memorial Fund and Alan Brown Chair of Molecular Genomics.
Acknowledgments
We thank Andrea Shafer, Richard Rivera-Massa, Feng Chen, Monica Ter-Minassian, Chau-Chyun Sheu and Maureen Convery for their contributions to this study.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence interval
- EA
esophageal adenocarcinoma
- GERD
gastroesophageal reflux disease
- OR
odds ratio
- VEGF
vascular endothelial growth factor
References
- 1.Pera M, et al. Epidemiology of esophageal adenocarcinoma. J. Surg. Oncol. 2005;92:151–159. doi: 10.1002/jso.20357. [DOI] [PubMed] [Google Scholar]
- 2.Spechler SJ. Clinical practice. Barrett's esophagus. N. Engl. J. Med. 2002;346:836–842. doi: 10.1056/NEJMcp012118. [DOI] [PubMed] [Google Scholar]
- 3.Jankowski JA, et al. Gastro-oesophageal cancer: death at the junction. Br. Med. J. 2000;321:463–464. doi: 10.1136/bmj.321.7259.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jankowski JA, et al. Barrett's metaplasia. Lancet. 2000;356:2079–2085. doi: 10.1016/S0140-6736(00)03411-5. [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 6.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin. Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 7.Mobius C, et al. The ‘angiogenic switch’ in the progression from Barrett's metaplasia to esophageal adenocarcinoma. Eur. J. Surg. Oncol. 2003;29:890–894. doi: 10.1016/j.ejso.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Stein HJ, et al. Angiogenesis and neoplastic transformation of Barrett's epithelium. Br. J. Surg. 2004;91:941–942. doi: 10.1002/bjs.4633. [DOI] [PubMed] [Google Scholar]
- 9.Saad RS, et al. Endoglin (CD105) and vascular endothelial growth factor as prognostic markers in esophageal adenocarcinoma. Hum. Pathol. 2005;36:955–961. doi: 10.1016/j.humpath.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Kleespies A, et al. Vascular endothelial growth factor in esophageal cancer. J. Surg. Oncol. 2004;87:95–104. doi: 10.1002/jso.20070. [DOI] [PubMed] [Google Scholar]
- 11.Kim KJ, et al. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 12.Asano M, et al. An anti-human VEGF monoclonal antibody, MV833, that exhibits potent anti-tumor activity in vivo. Hybridoma. 1998;17:185–190. doi: 10.1089/hyb.1998.17.185. [DOI] [PubMed] [Google Scholar]
- 13.Asano M, et al. Inhibition of tumor growth and metastasis by an immunoneutralizing monoclonal antibody to human vascular endothelial growth factor/vascular permeability factor121. Cancer Res. 1995;55:5296–5301. [PubMed] [Google Scholar]
- 14.Torres C, et al. Prognostic significance and effect of chemoradiotherapy on microvessel density (angiogenesis) in esophageal Barrett's esophagus-associated adenocarcinoma and squamous cell carcinoma. Hum. Pathol. 1999;30:753–758. doi: 10.1016/s0046-8177(99)90135-1. [DOI] [PubMed] [Google Scholar]
- 15.Couvelard A, et al. Angiogenesis in the neoplastic sequence of Barrett's oesophagus. Correlation with VEGF expression. J. Pathol. 2000;192:14–18. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH709>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 16.Auvinen MI, et al. Incipient angiogenesis in Barrett's epithelium and lymphangiogenesis in Barrett's adenocarcinoma. J. Clin. Oncol. 2002;20:2971–2979. doi: 10.1200/JCO.2002.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Mobius C, et al. Vascular endothelial growth factor expression and neovascularization in Barrett's carcinoma. World J. Surg. 2004;28:675–679. doi: 10.1007/s00268-004-7286-7. [DOI] [PubMed] [Google Scholar]
- 18.Lord RV, et al. Vascular endothelial growth factor and basic fibroblast growth factor expression in esophageal adenocarcinoma and Barrett esophagus. J. Thorac. Cardiovasc. Surg. 2003;125:246–253. doi: 10.1067/mtc.2003.203. [DOI] [PubMed] [Google Scholar]
- 19.Renner W, et al. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J. Vasc. Res. 2000;37:443–448. doi: 10.1159/000054076. [DOI] [PubMed] [Google Scholar]
- 20.Watson CJ, et al. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000;12:1232–1235. doi: 10.1006/cyto.2000.0692. [DOI] [PubMed] [Google Scholar]
- 21.Stevens A, et al. Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter. Cancer Res. 2003;63:812–816. [PubMed] [Google Scholar]
- 22.Krippl P, et al. A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int. J. Cancer. 2003;106:468–471. doi: 10.1002/ijc.11238. [DOI] [PubMed] [Google Scholar]
- 23.Ku KT, et al. Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for oral cancer. Oral. Oncol. 2005;41:497–502. doi: 10.1016/j.oraloncology.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Lee SJ, et al. Vascular endothelial growth factor gene polymorphisms and risk of primary lung cancer. Cancer Epidemiol. Biomarkers Prev. 2005;14:571–575. doi: 10.1158/1055-9965.EPI-04-0472. [DOI] [PubMed] [Google Scholar]
- 25.Lin CC, et al. Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for prostate cancer. Urology. 2003;62:374–377. doi: 10.1016/s0090-4295(03)00268-1. [DOI] [PubMed] [Google Scholar]
- 26.Ray D, et al. Association of the VEGF gene with proliferative diabetic retinopathy but not proteinuria in diabetes. Diabetes. 2004;53:861–864. doi: 10.2337/diabetes.53.3.861. [DOI] [PubMed] [Google Scholar]
- 27.Kraft P, et al. Accounting for haplotype uncertainty in matched association studies: a comparison of simple and flexible techniques. Genet. Epidemiol. 2005;28:261–272. doi: 10.1002/gepi.20061. [DOI] [PubMed] [Google Scholar]
- 28.Lu H, et al. Association of genetic polymorphisms in the VEGF gene with breast cancer survival. Cancer Res. 2005;65:5015–5019. doi: 10.1158/0008-5472.CAN-04-2786. [DOI] [PubMed] [Google Scholar]
- 29.Zhai R, et al. Vascular endothelial growth factor genotypes, haplotypes, gender, and the risk of non-small cell lung cancer. Clin. Cancer Res. 2008;14:612–617. doi: 10.1158/1078-0432.CCR-07-1655. [DOI] [PubMed] [Google Scholar]
- 30.Tzanakis N, et al. Vascular endothelial growth factor polymorphisms in gastric cancer development, prognosis, and survival. J. Surg. Oncol. 2006;94:624–630. doi: 10.1002/jso.20619. [DOI] [PubMed] [Google Scholar]
- 31.Kim JG, et al. Vascular endothelial growth factor gene polymorphisms associated with prognosis for patients with gastric cancer. Ann. Oncol. 2007;18:1030–1036. doi: 10.1093/annonc/mdm085. [DOI] [PubMed] [Google Scholar]
- 32.Kim DH, et al. Vascular endothelial growth factor (VEGF) gene (VEGFA) polymorphism can predict the prognosis in acute myeloid leukaemia patients. Br. J. Haematol. 2008;140:71–79. doi: 10.1111/j.1365-2141.2007.06887.x. [DOI] [PubMed] [Google Scholar]
- 33.Kataoka N, et al. Population-based case-control study of VEGF gene polymorphisms and breast cancer risk among Chinese women. Cancer Epidemiol. Biomarkers Prev. 2006;15:1148–1152. doi: 10.1158/1055-9965.EPI-05-0871. [DOI] [PubMed] [Google Scholar]
- 34.Fukuda H, et al. Clinical implication of vascular endothelial growth factor T-460C polymorphism in the risk and progression of prostate cancer. Oncol. Rep. 2007;18:1155–1163. [PubMed] [Google Scholar]
- 35.Howell WM, et al. Influence of vascular endothelial growth factor single nucleotide polymorphisms on tumour development in cutaneous malignant melanoma. Genes Immun. 2002;3:229–232. doi: 10.1038/sj.gene.6363851. [DOI] [PubMed] [Google Scholar]
- 36.Abe A, et al. Single nucleotide polymorphisms in the 3’ untranslated region of vascular endothelial growth factor gene in Japanese population with or without renal cell carcinoma. Tohoku J. Exp. Med. 2002;198:181–190. doi: 10.1620/tjem.198.181. [DOI] [PubMed] [Google Scholar]
- 37.Conklin BS, et al. Nicotine and cotinine up-regulate vascular endothelial growth factor expression in endothelial cells. Am. J. Pathol. 2002;160:413–418. doi: 10.1016/S0002-9440(10)64859-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanda Y, et al. Nicotine-induced vascular endothelial growth factor release via the EGFR-ERK pathway in rat vascular smooth muscle cells. Life Sci. 2007;80:1409–1414. doi: 10.1016/j.lfs.2006.12.033. [DOI] [PubMed] [Google Scholar]
- 39.Wong HP, et al. Nicotine promotes colon tumor growth and angiogenesis through beta-adrenergic activation. Toxicol. Sci. 2007;97:279–287. doi: 10.1093/toxsci/kfm060. [DOI] [PubMed] [Google Scholar]
- 40.Ye YN, et al. A mechanistic study of colon cancer growth promoted by cigarette smoke extract. Eur. J. Pharmacol. 2005;519:52–57. doi: 10.1016/j.ejphar.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 41.Shin VY, et al. Nicotine promotes gastric tumor growth and neovascularization by activating extracellular signal-regulated kinase and cyclooxygenase-2. Carcinogenesis. 2004;25:2487–2495. doi: 10.1093/carcin/bgh266. [DOI] [PubMed] [Google Scholar]
- 42.Edirisinghe I, et al. VEGFR-2 inhibition augments cigarette smoke-induced oxidative stress and inflammatory responses leading to endothelial dysfunction. FASEB J. 2008;22:2297–2310. doi: 10.1096/fj.07-099481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Awata T, et al. Functional VEGF C-634G polymorphism is associated with development of diabetic macular edema and correlated with macular retinal thickness in type 2 diabetes. Biochem. Biophys. Res. Commun. 2005;333:679–685. doi: 10.1016/j.bbrc.2005.05.167. [DOI] [PubMed] [Google Scholar]
- 44.Awata T, et al. A common polymorphism in the 5′-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes. 2002;51:1635–1639. doi: 10.2337/diabetes.51.5.1635. [DOI] [PubMed] [Google Scholar]
- 45.Balasubramanian SP, et al. Influence of VEGF-A gene variation and protein levels in breast cancer susceptibility and severity. Int. J. Cancer. 2007;121:1009–1016. doi: 10.1002/ijc.22772. [DOI] [PubMed] [Google Scholar]