Abstract
Genome-wide DNA hypomethylation plays has an important role in genomic instability and colorectal carcinogenesis. However, the relationship between cellular DNA methylation level and patient outcome remains uncertain. Using 643 colon cancers in two independent prospective cohorts, we quantified DNA methylation in repetitive long interspersed nucleotide element-1 (LINE-1) elements using pyrosequencing, which is a good indicator of global DNA methylation level. We used Cox proportional hazard models to calculate hazard ratios (HRs) of colon cancer–specific and overall mortality, adjusting for patient and tumoral features, including CpG island methylator phenotype (CIMP). Statistical tests were two-sided. LINE-1 hypomethylation was linearly associated with a statistically significant increase in colon cancer–specific mortality (for a 30% decrease in LINE-1 methylation: multivariable HR = 2.37, 95% confidence interval [CI] = 1.42 to 3.94; Ptrend < .001) and overall mortality (multivariable HR = 1.85, 95% CI = 1.25 to 2.75; Ptrend = .002). The association was consistent across the two independent cohorts and strata of clinical and molecular characteristics, including sex, age, tumor location, stage, and CIMP, microsatellite instability, KRAS, BRAF, p53, and chromosomal instability status. In conclusion, tumoral LINE-1 hypomethylation is independently associated with shorter survival among colon cancer patients.
CONTEXT AND CAVEATS
Prior knowledge
DNA hypomethylation is involved in genomic instability and colorectal carcinogenesis, but whether it is associated with the survival of colon cancer patients is unknown.
Study design
Prospective study of two colon cancer patient populations to evaluate the association between DNA methylation, which was assayed in long interspersed nucleotide element-1 (LINE-1), and survival. Clinical and molecular characteristics of the tumors were included in the analyses.
Contribution
LINE-1 hypomethylation was associated with higher colon cancer–specific and overall mortality, independent of other tumor characteristics.
Implications
LINE-1 hypomethylation may be a prognostic indicator of patient survival in colon cancer.
Limitations
Treatment data were limited for the patients studied. The patients were all health care professionals, and it is unknown whether the findings are applicable to the general population. The mechanism involved is unknown.
From the Editors
DNA methylation is a major epigenetic mechanism in X-chromosome inactivation, imprinting, and repression of endogenous retroviruses (1,2). Repetitive nucleotide elements (eg, long interspersed nucleotide element-1 [LINE-1]) contain numerous CpG dinucleotides, and previous studies (3–5) have shown that LINE-1 methylation level has been reported to be a good indicator of cellular 5-methylcytosine level (ie, global DNA methylation level). Genome-wide DNA hypomethylation has an important role in genomic instability by reactivating transposable DNA sequences during colorectal carcinogenesis (1,6–15). However, the influence of global DNA hypomethylation on the prognosis of colon cancer patients remains uncertain.
We used two independent prospective cohort studies: the Nurses’ Health Study (121 700 women followed since 1976) (16,17) and the Health Professionals Follow-up Study (51 500 men followed since 1986) (17,18). Every 2 years, participants were sent follow-up questionnaires to update information on potential risk factors and to identify newly diagnosed cancer, including colon cancer (16–18). Informed consent was obtained from all study subjects. This study was approved by the Human Subjects Committees and the Institutional Review Boards of Harvard School of Public Health and Brigham and Women's Hospital. We collected paraffin-embedded tissue blocks from hospitals where patients underwent tumor resections (17). Based on availability of tissue specimens, we included 643 colon cancers that were diagnosed up to June 30, 2002. Patients were observed until death or June 30, 2006, whichever came first (19).
KRAS and BRAF sequencing and analyses for microsatellite instability (MSI) and loss of heterozygosity (chromosomal instability, CIN) were performed on DNA from paraffin-embedded tissue (20–22). Immunohistochemistry for p53 was performed using a mouse monoclonal antibody (clone DO-1, dilution 1:50; Calbiochem, San Diego, CA) as previously described (23). Bisulfite DNA treatment and real-time polymerase chain reaction [MethyLight (24)] were validated and performed (25). We quantified promoter methylation in eight CpG island methylator phenotype (CIMP)–specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) (26–28). CIMP-high was defined as methylation in at least six of the eight promoters (26).
To accurately quantify relatively high LINE-1 methylation levels, we used pyrosequencing (29). LINE-1 methylation as measured by pyrosequencing has been reported to be a good indicator of cellular 5-methylcytosine level (ie, global DNA methylation level) (3,5,30). LINE-1 methylation levels in the 643 tumors were approximately normally distributed (mean = 61.2%; standard deviation = 9.4%) (Supplementary Figure 1, available online).
Cox proportional hazard models were used to calculate hazard ratios (HRs) of death according to LINE-1 methylation level, adjusted for age, sex, year of diagnosis, tumor location, stage, grade, and CIMP, MSI, CIN, KRAS, BRAF, and p53 status. The proportionality of hazards assumption was satisfied by evaluating time-dependent variables, which were the cross product of LINE-1 methylation level and survival time (P = .31 for colon cancer–specific mortality and P = .73 for overall mortality). The primary statistical analysis was the linear test for trend with LINE-1 methylation as a continuous variable, to avoid the possibility of selecting cut points with maximal P values. We also performed analyses using categories (Supplementary Table 1, available online) that were based on the overall distribution of LINE-1 methylation levels. We used SAS version 9.1 (SAS Institute, Cary, NC) for statistical analyses. All statistical tests were two-sided, and P values less than .05 were considered statistically significant.
LINE-1 hypomethylation was associated with a statistically significant increase in colon cancer–specific mortality (univariate analysis Ptrend < .001 and multivariable analysis Ptrend < .001) (Table 1). For a 30% decrease in LINE-1 methylation, the multivariable HR for colon cancer–specific mortality was 2.37 (95% confidence interval [CI] = 1.42 to 3.94). LINE-1 hypomethylation was also associated with a statistically significant increase in overall mortality (univariate analysis Ptrend = .008 and multivariable analysis Ptrend = .002).
Table 1.
LINE-1 methylation level and survival analysis in colon cancer*
| LINE-1 methylation level | Total N | Colon cancer–specific mortality |
Overall mortality |
||||
| Deaths/person-years | Univariate HR (95% CI) | Multivariable HR (95% CI) | Deaths/person-years | Univariate HR (95% CI) | Multivariable HR (95% CI) | ||
| ≥75% | 40 | 8/352 | 1 (referent) | 1 (referent) | 14/352 | 1 (referent) | 1 (referent) |
| 60%–75% | 334 | 66/3008 | 0.99 (0.47 to 2.05) | 1.79 (0.81 to 3.97) | 130/3008 | 1.10 (0.63 to 1.91) | 2.19 (1.19 to 4.02) |
| 45%–60% | 238 | 71/2129 | 1.52 (0.73 to 3.16) | 2.43 (1.08 to 5.49) | 113/2129 | 1.36 (0.78 to 2.37) | 2.60 (1.39 to 4.84) |
| <45% | 31 | 15/231 | 3.15 (1.34 to 7.44) | 5.00 (1.92 to 13.1) | 19/231 | 2.20 (1.10 to 4.39) | 4.73 (2.20 to 10.2) |
| 30% decrease† | 2.58 (1.62 to 4.12) | 2.37 (1.42 to 3.94) | 1.67 (1.15 to 2.42) | 1.85 (1.25 to 2.75) | |||
| Ptrend | <.001 | <.001 | .008 | .002 | |||
The multivariable Cox model includes age at diagnosis, year of diagnosis, sex, tumor location, stage, tumor grade, and status of microsatellite instability, CpG island methylator phenotype, chromosomal instability, KRAS, BRAF, and p53 (cut points shown in Supplementary Table 1, available online). Ptrend values (two-sided) were calculated using the Wald test. HR = hazard ratio; CI = confidence interval; LINE-1 = long interspersed nucleotide element-1.
LINE-1 methylation level was used as a continuous variable.
To facilitate display of the results, we also grouped patients according to four categories of LINE-1 methylation (≥75%, 60%–75%, 45%–60%, and <45%). Compared with patients who had 75% or greater LINE-1 methylated tumors, those with less than 45% LINE-1 methylation had higher colon cancer–specific mortality (multivariable HR = 5.00; 95% CI = 1.92 to 13.1) and a similar increase in overall mortality (multivariable HR = 4.73; 95% CI = 2.20 to 10.2) (Table 1).
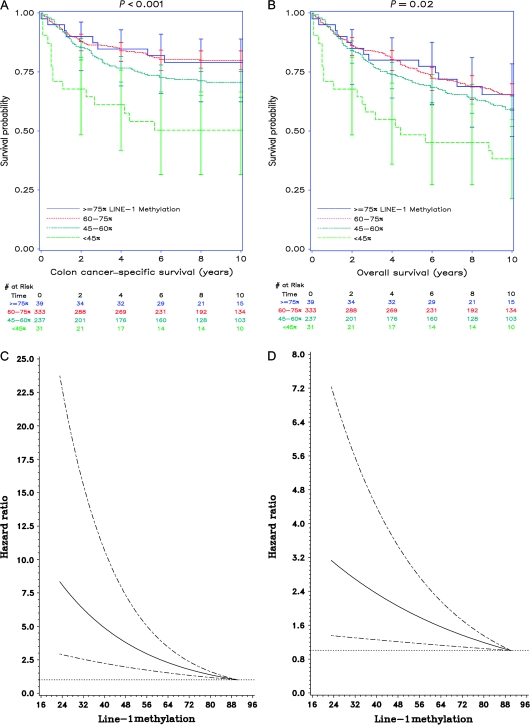
The colon cancer–specific survival probability at 5 years was progressively lower as LINE-1 methylation level declined (≥75% LINE-1 methylation: survival probability = 0.85, 95% CI = 0.69 to 0.93; 60%–75% methylation: survival probability = 0.83, 95% CI = 0.79 to 0.87; 45%–60% methylation: survival probability = 0.74, 95% CI = 0.68 to 0.80; and <45% methylation: survival probability = 0.54, 95% CI = 0.35 to 0.70; P < .001) (Figure 1, A). Similarly, overall survival at 5 years was statistically significantly lower for patients with relatively hypomethylated LINE-1 (≥75% LINE-1 methylation: survival probability = 0.80, 95% CI = 0.64 to 0.89; 60%–75% methylation: survival probability = 0.77, 95% CI = 0.72 to 0.81; 45%–60% methylation: survival probability = 0.71, 95% CI = 0.64 to 0.76; and <45% methylation: survival probability = 0.48, 95% CI = 0.30 to 0.64; P = .02) (Figure 1, B).
Figure 1.
Long interspersed nucleotide element-1 (LINE-1) methylation level and patient survival in colon cancer. Colon cancer samples from 640 participants in the Health Professionals Follow-up Study and the Nurses’ Health Study were assayed for LINE-1 methylation by pyrosequencing. A) Kaplan–Meier analysis for colon cancer–specific survival (≥75% LINE-1 methylation: survival probability at 5 years = 0.85, 95% CI = 0.69 to 0.93; 60%–75% methylation: survival probability at 5 years = 0.83, 95% CI = 0.79 to 0.87; 45%–60% methylation: survival probability at 5 years = 0.74, 95% CI = 0.68 to 0.80; and <45% methylation: survival probability at 5 years = 0.54, 95% CI = 0.35 to 0.70). B) Kaplan–Meier analysis for overall survival (≥75% LINE-1 methylation: survival probability at 5 years = 0.80, 95% CI = 0.64 to 0.89; 60%–75% methylation: survival probability at 5 years = 0.77, 95% CI = 0.72 to 0.81; 45%–60% methylation: survival probability at 5 years = 0.71, 95% CI = 0.64 to 0.76; and <45% methylation: survival probability at 5 years = 0.48, 95% CI = 0.30 to 0.64). Vertical bars, 95% confidence intervals at 2-year intervals. P values (two-sided) were calculated using the log-rank test. C) Smoothing spline plot of unadjusted hazard ratios (HRs) for colon cancer–specific mortality according to LINE-1 methylation level (%). D) Smoothing spline plot of unadjusted HRs for overall mortality according to LINE-1 methylation level (%). Hatched lines, 95% confidence intervals.
We examined the possibility of a nonlinear relationship between LINE-1 hypomethylation and mortality by using a nonparametric method with restricted cubic splines (31), which was independent of predetermined LINE-1 categorizations. The HRs for colon cancer–specific and overall mortality increased continuously with decreasing LINE-1 methylation (Figure 1, C and D). Notably, the lower limits of the 95% confidence intervals remained above 1.0 for all values of LINE-1 methylation, suggesting a robust association between LINE-1 hypomethylation and mortality.
We also generated a receiver operator characteristics curve for the prediction of 5-year colon cancer–specific survival using all possible cutoffs (positive vs negative) for LINE-1 hypomethylation (area under the curve = 0.56) (Supplementary Figure 2, available online). This result implies that there were limitations to using LINE-1 methylation as a binary diagnostic test to predict patient outcome at a fixed time point.
Finally, we examined the influence of LINE-1 hypomethylation on colon cancer–specific mortality across strata of other clinical and molecular characteristics (Supplementary Figure 3, available online). The association between LINE-1 hypomethylation and colon cancer–specific mortality was not modified by any of the clinical or molecular variables nor was it different between the two cohorts (the Health Professionals Follow-up Study and the Nurses’ Health Study; Pinteraction = .74).
Although prognostic factors in colon cancer have been extensively studied (32–38), little is known regarding the prognostic value of global DNA hypomethylation. The relationship between genome-wide DNA hypomethylation and clinical outcome has been examined in prostate, liver, and ovarian cancers and leukemia (39–42). Among those studies, only the study of chronic myeloid leukemia (42) showed a statistically significant association between global DNA hypomethylation and poor survival. In colon cancer, Frigola et al. (43) analyzed 93 tumors using the amplification of intermethylated sites (AIMS) method and observed a trend toward poor survival in DNA hypomethylated tumors; however, the results did not reach statistical significance.
The mechanism by which global DNA hypomethylation may confer a poor prognosis remains speculative. Genome-wide DNA hypomethylation has been associated with genomic instability (7–15), which may confer poor prognosis. Transcriptional dysregulation might be another possible mechanism, and activation of proto-oncogenes, endogenous retroviruses, or transposable elements might affect tumor aggressiveness. A third possible mechanism involves inflammatory mediators and oxidative stress; the latter has been associated with genomic DNA hypomethylation (44,45). Activation of the inflammatory pathway has been associated with poor prognosis in colon cancer (46).
The study has potential limitations. In the two cohorts, data on cancer treatment were limited. It is unlikely that chemotherapy use differed according to tumoral LINE-1 methylation level, since such data were not available to patients or treating physicians. Other potential study limitations include a nonperfect correlation (correlation coefficient < 1) between LINE-1 methylation and global DNA methylation level and that these health professionals–based cohort studies may not be completely representative of the general population.
In summary, the results from this large prospective cohort study suggest that genome-wide DNA hypomethylation as measured in LINE-1 is independently associated with poor survival among patients with colon cancer. The adverse effect of LINE-1 hypomethylation on prognosis persisted across the two independent prospective cohort studies as well as strata of other clinical or tumoral characteristics. These findings may have considerable clinical implications. Future studies are needed to confirm this association as well as examine potential mechanisms by which genome-wide DNA hypomethylation affects tumor behavior.
Funding
The US National Institute of Health (P01 CA87969, P01 CA55075, P50 CA127003, and K07 CA122826 to S.O.); the Bennett Family Fund; the Entertainment Industry Foundation. K. Nosho was supported by a fellowship grant from the Japan Society for Promotion of Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of National Cancer Institute or National Institutes of Health. Funding agencies did not have any role in the design of the study; the collection, analysis, or interpretation of the data; the decision to submit the manuscript for publication; or the writing of the manuscript.
Supplementary Material
Footnotes
We deeply thank the Nurses' Health Study and Health Professionals Follow-up Study cohort participants who have generously agreed to provide us with biologic specimens and information through responses to questionnaires and hospitals and pathology departments throughout the United States for providing us with tumor tissue materials.
References
- 1.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33(21):6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang AS, Doshi KD, Choi SW, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66(10):5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 6.Bestor TH. Transposons reanimated in mice. Cell. 2005;122(3):322–325. doi: 10.1016/j.cell.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Ji W, Hernandez R, Zhang XY, et al. DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat Res. 1997;379(1):33–41. doi: 10.1016/s0027-5107(97)00088-2. [DOI] [PubMed] [Google Scholar]
- 8.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300(5618):489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 9.Yamada Y, Jackson-Grusby L, Linhart H, et al. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci USA. 2005;102(38):13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez J, Frigola J, Vendrell E, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66(17):8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki K, Suzuki I, Leodolter A, et al. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9(3):199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 12.Karpf AR, Matsui S. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 2005;65(19):8635–8639. doi: 10.1158/0008-5472.CAN-05-1961. [DOI] [PubMed] [Google Scholar]
- 13.Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM, III, Jaenisch R. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell. 2005;8(4):275–285. doi: 10.1016/j.ccr.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Chalitchagorn K, Shuangshoti S, Hourpai N, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23(54):8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 15.Matsuzaki K, Deng G, Tanaka H, Kakar S, Miura S, Kim YS. The relationship between global methylation level, loss of heterozygosity, and microsatellite instability in sporadic colorectal cancer. Clin Cancer Res. 2005;11(24):8564–8569. doi: 10.1158/1078-0432.CCR-05-0859. [DOI] [PubMed] [Google Scholar]
- 16.Colditz GA, Hankinson SE. The Nurses' Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5(5):388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 17.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356(21):2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 18.Wu K, Feskanich D, Fuchs CS, Willett WC, Hollis BW, Giovannucci EL. A nested case control study of plasma 25-hydroxyvitamin D concentrations and risk of colorectal cancer. J Natl Cancer Inst. 2007;99(14):1120–1129. doi: 10.1093/jnci/djm038. [DOI] [PubMed] [Google Scholar]
- 19.Bass AJ, Meyerhardt JA, Chan JA, Giovannucci EL, Fuchs CS. Family history and survival after colorectal cancer diagnosis. Cancer. 2008;112(6):1222–1229. doi: 10.1002/cncr.23294. [DOI] [PubMed] [Google Scholar]
- 20.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7(3):413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8(3):582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino S, Kawasaki T, Kirkner GJ, Ohnishi M, Fuchs CS. 18q loss of heterozygosity in microsatellite stable colorectal cancer is correlated with CpG island methylator phenotype-negative (CIMP-0) and inversely with CIMP-low and CIMP-high. BMC Cancer. 2007;7(1):72. doi: 10.1186/1471-2407-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogino S, kawasaki T, Kirkner GJ, Yamaji T, Loda M, Fuchs CS. Loss of nuclear p27 (CDKN1B/KIP1) in colorectal cancer is correlated with microsatellite instability and CIMP. Mod Pathol. 2007;20(1):15–22. doi: 10.1038/modpathol.3800709. [DOI] [PubMed] [Google Scholar]
- 24.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28(8):E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogino S, kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8(2):209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9(3):305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55(7):1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38(7):787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 29.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG methylator phenotype in colorectal cancer. Int J Cancer. 2008;122(12):2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estecio MR, Gharibyan V, Shen L, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS ONE. 2007;2(5):e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durrleman S, Simon R. Flexible regression models with cubic splines. Stat Med. 1989;8(5):551–561. doi: 10.1002/sim.4780080504. [DOI] [PubMed] [Google Scholar]
- 32.Mudduluru G, Medved F, Grobholz R, et al. Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer. 2007;110(8):1697–1707. doi: 10.1002/cncr.22983. [DOI] [PubMed] [Google Scholar]
- 33.Chang GJ, Rodriguez-Bigas MA, Skibber JM, Moyer VA. Lymph node evaluation and survival after curative resection of colon cancer: systematic review. J Natl Cancer Inst. 2007;99(6):433–441. doi: 10.1093/jnci/djk092. [DOI] [PubMed] [Google Scholar]
- 34.Freedman DM, Looker AC, Chang SC, Graubard BI. Prospective study of serum vitamin D and cancer mortality in the United States. J Natl Cancer Inst. 2007;99(21):1594–1602. doi: 10.1093/jnci/djm204. [DOI] [PubMed] [Google Scholar]
- 35.Derks S, Postma C, Carvalho B, et al. Integrated analysis of chromosomal, microsatellite and epigenetic instability in colorectal cancer identifies specific associations between promoter methylation of pivotal tumour suppressor and DNA repair genes and specific chromosomal alterations. Carcinogenesis. 2008;29(2):434–439. doi: 10.1093/carcin/bgm270. [DOI] [PubMed] [Google Scholar]
- 36.Govindarajan A, Coburn NG, Kiss A, Rabeneck L, Smith AJ, Law CH. Population-based assessment of the surgical management of locally advanced colorectal cancer. J Natl Cancer Inst. 2006;98(20):1474–1481. doi: 10.1093/jnci/djj396. [DOI] [PubMed] [Google Scholar]
- 37.Dignam JJ, Polite BN, Yothers G, et al. Body mass index and outcomes in patients who receive adjuvant chemotherapy for colon cancer. J Natl Cancer Inst. 2006;98(22):1647–1654. doi: 10.1093/jnci/djj442. [DOI] [PubMed] [Google Scholar]
- 38.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10(1):13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brothman AR, Swanson G, Maxwell TM, et al. Global hypomethylation is common in prostate cancer cells: a quantitative predictor for clinical outcome? Cancer Genet Cytogenet. 2005;156(1):31–36. doi: 10.1016/j.cancergencyto.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Calvisi DF, Simile MM, Ladu S, et al. Altered methionine metabolism and global DNA methylation in liver cancer: relationship with genomic instability and prognosis. Int J Cancer. 2007;121(11):2410–2420. doi: 10.1002/ijc.22940. [DOI] [PubMed] [Google Scholar]
- 41.Pattamadilok J, Huapai N, Rattanatanyong P, et al. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int J Gynecol Cancer. 2008;18(4):711–717. doi: 10.1111/j.1525-1438.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- 42.Roman-Gomez J, Jimenez-Velasco A, Agirre X, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24(48):7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 43.Frigola J, Sole X, Paz MF, et al. Differential DNA hypermethylation and hypomethylation signatures in colorectal cancer. Hum Mol Genet. 2005;14(2):319–326. doi: 10.1093/hmg/ddi028. [DOI] [PubMed] [Google Scholar]
- 44.Shahrzad S, Bertrand K, Minhas K, Coomber BL. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics. 2007;2(2):119–125. doi: 10.4161/epi.2.2.4613. [DOI] [PubMed] [Google Scholar]
- 45.Bagnyukova TV, Luzhna LI, Pogribny IP, Lushchak VI. Oxidative stress and antioxidant defenses in goldfish liver in response to short-term exposure to arsenite. Environ Mol Mutagen. 2007;48(8):658–665. doi: 10.1002/em.20328. [DOI] [PubMed] [Google Scholar]
- 46.Soumaoro LT, Uetake H, Higuchi T, Takagi Y, Enomoto M, Sugihara K. Cyclooxygenase-2 expression: a significant prognostic indicator for patients with colorectal cancer. Clin Cancer Res. 2004;10(24):8465–8471. doi: 10.1158/1078-0432.CCR-04-0653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.