Abstract
Summary: BioCaster is an ontology-based text mining system for detecting and tracking the distribution of infectious disease outbreaks from linguistic signals on the Web. The system continuously analyzes documents reported from over 1700 RSS feeds, classifies them for topical relevance and plots them onto a Google map using geocoded information. The background knowledge for bridging the gap between Layman's terms and formal-coding systems is contained in the freely available BioCaster ontology which includes information in eight languages focused on the epidemiological role of pathogens as well as geographical locations with their latitudes/longitudes. The system consists of four main stages: topic classification, named entity recognition (NER), disease/location detection and event recognition. Higher order event analysis is used to detect more precisely specified warning signals that can then be notified to registered users via email alerts. Evaluation of the system for topic recognition and entity identification is conducted on a gold standard corpus of annotated news articles.
Availability: The BioCaster map and ontology are freely available via a web portal at http://www.biocaster.org.
Contact: collier@nii.ac.jp
1 INTRODUCTION
Informal data on the distribution of disease outbreaks is published in many forms and languages on the World Wide Web, but manual surveillance methods are costly and time consuming. Identifying positive linguistic signals and correctly interpreting the geo-temporal dynamics of pathogen spread remain key challenges for text mining. The difficulty of the task is characterized by (i) the massive volume of data, (ii) the need to interpret information as early as possible in the outbreak cycle when reliable facts tend to be scarce, (iii) the need to understand texts in many languages and (iv) the ambiguity inherent in natural language text.
To meet these challenges, several publicly supported Web-surveillance projects have been established that involve a greater or lesser degree of automated monitoring of public health threats including MedISys (http://medusa.jrc.it) (EU), GPHIN (Canada) and Argus (http://biodefense.georgetown.edu/projects/argus.aspx) (USA). Additionally, ProMed-mail (http://www.promedmail.org) is a widely acknowledged manually curated system that provides reports by public health experts. Other systems which are close to the one we present are EpiSpider (http://www.epispider.org) and HealthMap (http://www.healthmap.org) both of which collect news from the Internet about human and animal health and plot the data on a Google Maps mashup.
BioCaster is a non-governmental public health surveillance system characterized by its open ontology-centered approach and a priority for Asia-Pacific languages and health hazards. Specific advantages of BioCaster are that it brings together within a single system (i) text mining techniques, such as entity recognition which aim to generalize to previously unseen terms and expressions, (ii) text-level recognition of severity indicators, such as international travel or the contamination of blood products, (iii) ontology-based inferencing to fill in the gaps, e.g. between a mentioned pathogen and the unmentioned disease that caused it or between symptoms and diseases and (iv) direct knowledge of term equivalence within and across languages.
2 METHODS
Figure 1 shows a high-level view of the information flow through the system which is built on a Linux-based NPACI Rocks cluster for high-throughput semantic analysis.
Fig. 1.
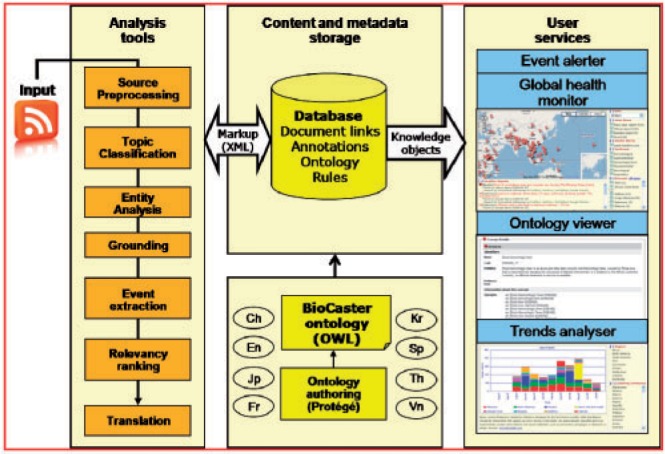
Overview of the BioCaster system showing the stages of text-to-knowledge conversion followed by user service provision.
Currently BioCaster ingests documents through RSS feeds. Each hour a purpose built news aggregator script written in Perl identifies novel links from over 1700 feeds. Sourced documents are then cleansed and put into the cluster queue. Automatic classification of the reports for topical relevance using a naïve Bayes algorithm then acts as the gate-keeper for further levels of processing. For relevant documents named entity recognition is then performed for 18 term types based on the BioCaster ontology (BCO) (Collier et al., 2007).
At this stage disease-location pairs are plotted onto a public portal called the Global Health Monitor. Visualization, using Google Maps, allows users to gain a geographically contextualized view of an outbreak anywhere in the world which can be filtered by pathogen, syndrome or text type. Users can drill down to source evidence by clicking on map points which display associated headlines for the event along with links to scientific databases, such as PubMed, HighWire and Google Scholar.
In event extraction, additional levels of semantic analysis allow the system to gain a deeper level of understanding about the public health significance of the event through rules for disambiguating the geo-temporal and linguistic context in which a term is used. This is done by using a simple rule language (SRL) motivated by DIAL (Feldman et al., 2001) with a capability to match entity classes, skipwords, string literals, regular expressions, entity types as well as guard lists. Examples of lists include verbs of infection, common victim expressions, occupation names and so on. The advantage we perceive in using regular expression patterns in SRL is that they can be easily adapted by users with little linguistics training and also that they are amenable to languages with few extant resources, such as parsers or chunkers.
Analysis then focuses on detecting domain-specific signals, such as cases of drug resistance, malformed blood products, international travel, zoonosis or newly emerging strains. Access to this information is limited to registered users who can create rules for receiving email alerts on topics of interest.
At the core of BioCaster is the BCO, developed by a multi-disciplinary team of experts. The BCO is organized around an application taxonomy with root terms representing key domain concepts. The BCO encoded as an openly available OWL file gives access to term definitions, synonyms and translations in eight languages as well as mappings to external ontologies, such as ICD-10, MedDRA, MeSH and SNOMED-CT. Version 2 of the BCO released in April 2008 includes information on 102 infectious diseases as well as geo-locations for two administrative levels.
3 PERFORMANCE EVALUATION
The development of the named entity recognition schema and module was reported in Kawazoe et al. (2006) based on formal concept analysis. We used an annotated corpus of 200 news articles as training data and the NER system using a support vector machine (SVM) achieved an F-score of 76.97% for all NE classes.
Evaluation of topic classification was presented in Conway et al. (2008). The experiments used the BioCaster gold standard corpus which includes 1000 annotated news stories as training data. The classification model we found to achieve highest accuracy is based on naïve Bayes using raw text, n-grams, semantic tag-based features and χ2 feature selection. The system achieved an accuracy score of 94.8% (F-score 0.93, Recall 0.97, Precision 0.89), outperforming an SVM with accuracy of 92.1% (F-score 0.89, Recall 0.90, Precision 0.88) due to its superior recall—a key requirement for a surveillance system.
Evaluation of the ontology was done informally using corpus coverage of terms. During a 32-week period we observed the percentage of English terms mentioned in 29 443 positive reports with coverage estimated at 78% of diseases, 69% of pathogens and 76% of symptom terms found at least once.
Future quantitative evaluation will focus on the effectiveness of domain-specific signals against human standards, such as ProMed and WHO reports.
4 CONCLUSIONS
BioCaster, in operation since 2006, is an ontology-enabled text mining system developed to enhance early detection of infectious disease outbreaks by experts. Additionally, it offers an intuitive mapping interface for the general reader as well as an openly available ontology for community reuse. Future work will focus on extending coverage to new languages and public health threats.
ACKNOWLEDGEMENTS
We would like to thank John McCrae at NII for his work on implementing and extending SRL in Java.
Funding: Japan Science and Technology Agency (JST); the Japan Society for the Promotion of Science (18049071); Research Organization of Information Systems (ROIS).
Conflict of Interest: none declared.
REFERENCES
- Collier N, et al. A multilingual ontology for infectious disease outbreak surveillance: rationale, design and challenges. J. Lang. Resour. Eval. 2007 doi: 10.1007/s10579-007-9019-7. DOI: 10.1007/s10579-007-9019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway M, et al. Proceedings of the 3rd International Symposium on Semantic Mining in Biomedicine (SMBM 2008). in press; 2008. Classifying disease outbreak reports using n-grams and semantic features. [DOI] [PubMed] [Google Scholar]
- Feldman R, et al. Proceedings of the International Conference on Information and Knowledge Management (CIKM-01). 2001. A domain independent environment for creating information extraction modules; pp. 586–588. [Google Scholar]
- Kawazoe A. Proceedings of the International Workshop on Biomedical Ontology in Action (KR-MED 2006). 2006. The development of a schema for the annotation of terms in the BioCaster disease detection/tracking system; pp. 77–85. [Google Scholar]


