Abstract
Agrobacterium tumefaciens can transfer part of its Ti plasmid, the T-DNA, to plant cells where it integrates into the nuclear genome via illegitimate recombination. Integration of the T-DNA results in small deletions of the plant target DNA, and may lead to truncation of the T-DNA borders and the production of filler DNA. We showed previously that T-DNA can also be transferred from A. tumefaciens to Saccharomyces cerevisiae and integrates into the yeast genome via homologous recombination. We show here that when the T-DNA lacks homology with the S. cerevisiae genome, it integrates at random positions via illegitimate recombination. From 11 lines the integrated T-DNA was cloned back to Escherichia coli along with yeast flanking sequences. The T-DNA borders and yeast DNA flanking the T-DNA were sequenced and characterized. It was found that T-DNA integration had resulted in target DNA deletions and sometimes T-DNA truncations or filler DNA formation. Therefore, the molecular mechanism of illegitimate recombination by which T-DNA integrates in higher and lower eukaryotes seems conserved.
The soil bacterium Agrobacterium tumefaciens can transfer part of its Ti plasmid, the T-DNA, to plants (1). Virulence (vir) genes located on the Ti plasmid effect this transfer to the plant cell. The transfer system encoded by the vir genes resembles that used by bacteria for conjugation (2). However, some Vir proteins enter plant cells during T-DNA transfer to protect the T-DNA and mediate its transport to the nucleus (3–5). The T-DNA is integrated into the plant genome by illegitimate recombination (IR), a mechanism that joins two DNA molecules that do not share extensive homology, in this case the plant DNA and T-DNA. In higher eukaryotic organisms such as plants, IR is the predominant mechanism of DNA integration (6, 7). IR of T-DNA in the plant genome has been described (8–10), but little is known about the plant factors involved.
We have shown that A. tumefaciens can also transfer T-DNA to the yeast Saccharomyces cerevisiae (11). This result was recently confirmed by Piers et al. (12). S. cerevisiae has served as a good model system for studying DNA recombination leading to the identification of many genes and proteins involved. Segments of DNA carrying homology with the S. cerevisiae genome integrate very efficiently via homologous recombination. IR events occur at a low frequency in S. cerevisiae, but such events can be selected for after transformation with a stretch of DNA lacking homology with the yeast genome (13). Although T-DNA integrates only rarely by homologous recombination in plants (6, 7), T-DNA integration into the S. cerevisiae genome occurs via homologous recombination (11). These results indicate that the preferred mechanism of T-DNA integration is determined by the host organism and not by the complex of T-DNA and Vir proteins.
In this paper we studied the fate of non-homologous T-DNA in S. cerevisiae and show that such T-DNA integrates into the yeast genome by a process of IR that is reminiscent of T-DNA integration in plants. This finding allows the analysis of the process of T-DNA integration using all the assets of yeast genetics.
MATERIALS AND METHODS
Bacterial Strains and Yeast Strains.
Cocultivations were carried out between binary vector-containing derivatives of A. tumefaciens strain LBA1126 and S. cerevisiae strain RSY12 (MATa leu2-3, 112 his3-11, 15 ura3Δ::HIS3) as described previously (11), except that after washing the yeast cells in induction medium the yeast cells were pelleted and resuspended in their own volume to ensure as high an input number of yeast cells as possible. A 50-μl aliquot of the yeast cell suspension was mixed with 50 μl of Agrobacterium culture. A. tumefaciens strain LBA1126 was constructed by integration of the mutated vir genes encoding the VirG I77V (14) and VirA-TAR proteins (15) at the wild-type virG and virA loci of pAL1100 in A. tumefaciens strain LBA1100. Plasmid rescues were done using the Escherichia coli strain NM554 [F-araD139 Δ(ara-leu)7696 Δ(lac)X74 galU galK hsdR2 (rk− mk−) mcrB1 rpsL(Strr) recA13]. All cloning steps were done in E. coli strain DH5α.
Plasmid Constructions.
pRAL7102 was constructed by insertion of a 1.1-kb URA3 HindIII fragment (16) into the HindIII site of pBIN19 (17). For the plasmid rescues, the binary vector pRAL7103 was constructed by ligation of the 2.6-kb pUC9 plasmid linearized with BamHI into the BamHI site of pRAL7102.
T-DNA Rescue from the RSY12 Genome.
Total DNA was isolated from Ura+ RSY12 strains (18) and 4 μg was digested with BglII. Rescue of the T-DNA plus flanking DNA was carried out as described (19). Restriction digestion was done on the rescued plasmids to confirm that they contained the 1.1-kb HindIII URA3 and 2.6-kb pUC9 fragments present in pRAL7103 (data not shown).
T7 Polymerase Sequencing and CHEF Gels.
Sequencing was carried out using the T7 Polymerase Sequencing kit (Pharmacia) following the manufacturer’s instructions. The following primers were used: p1, 5′-CGTTGCGGTTCTGTCAGTTCC-3′; p2, 5′-CACTCAACCCTATCTCGGGC-3′. For the CHEF gels, complete intact chromosomes were isolated in agarose blocks as described (20, 21). The chromosomes were separated using a CHEF apparatus (Bio-Rad). The agarose blocks were placed in the wells of a 0.25× TBE 1% agarose gel. Electrophoresis was done in 0.25× TBE at 14°C with an initial switch time of 40 sec and a final switch time of 90 sec at 200 V for 20 hr. The DNA was transferred to a Hybond N+ membrane (Amersham) using a LKB2016 VacuGene vacuum blotting apparatus with 1× blot buffer (0.6 M NaCl/0.4 M NaOH). The membrane was probed with a 1.1-kb HindIII fragment of the URA3 gene. Autoradiography was done for 7 days using Kodak XAR film.
RESULTS
Stretches of DNA carrying homology with the S. cerevisiae genome integrate preferentially via homologous recombination into the genome of this yeast. This is also true for T-DNA introduced into this yeast by A. tumefaciens (11). In plants the T-DNA integrates preferentially by a process of IR. Therefore, we wanted to study what would happen to a T-DNA that lacked homology with the S. cerevisiae genome. To select for such a T-DNA in yeast we constructed the binary vector pRAL7102, which carries the URA3 gene. The T-DNA of this binary vector carries no homology with the genome of yeast strain RSY12 (URA3Δ) and it lacks a yeast origin of replication.
Integration of Nonhomologous T-DNA into the Genome of S. cerevisiae Is Random at the Chromosome Level.
Cocultivations between the Agrobacterium strain LBA1126(pRAL7102) and the yeast strain RSY12 were carried out as described previously (11). The results are shown in Table 1. Ura+ yeast colonies were obtained from the cocultivations at a low frequency. As a positive control, the A. tumefaciens T-DNA donor LBA1126(pRAL7100) was also used. The T-DNA of pRAL7100 carries the S. cerevisiae PDA1 locus disrupted with the URA3 gene. After T-DNA transfer homologous recombination can occur at the PDA1 locus on chromosome V. By comparing the frequencies of Ura+ colonies obtained from cocultivations using either LBA1126(pRAL7100) or LBA1126(pRAL7102), the ratio of homologous recombination versus IR of T-DNA in S. cerevisiae can be calculated. In our experiments, T-DNA from pRAL7100 was ≈200 times more likely to integrate than T-DNA from pRAL7102 (Table 1). Because RSY12 is a haploid yeast strain, a percentage of the random T-DNA integrations in the yeast genome may have been lethal or have generated a mutant yeast strain unable to grow on the selective medium used. To test for this a diploid derivative of RSY12 was used in a cocultivation with LBA1126(pRAL7102), but no increase in the frequency of Ura+ colonies was observed (data not shown).
Table 1.
T-DNA transfer to S. cerevisiae strain RSY12 (URA3Δ)
| Plasmid present in Agrobacterium donor strain LBA1126 | Medium | RSY12 colonies on medium lacking uracil | RSY12 colonies (108/ml) on medium with uracil | Frequency of Ura+ colonies per output recipient |
|---|---|---|---|---|
| pRAL7102 | +AS | 12 | 1.7 | 7 × 10−8 |
| −AS | 0 | 1.5 | <6.6 × 10−9 | |
| pRAL7103 | +AS | 16 | 1.2 | 1.3 × 10−7 |
| −AS | 0 | 1.5 | <6.6 × 10−9 | |
| pRAL7100 | +AS | 2790 | 0.8 | 3.4 × 10−5 |
| −AS | 0 | 1.1 | <9 × 10−9 |
To show that the Ura+ yeast colonies generated from the LBA1126(pRAL7102)/RSY12 cocultivations contained integrated T-DNA, the chromosomes of these strains were separated on a CHEF gel, blotted onto a membrane, and probed with the 1.1-kb URA3 HindIII fragment carried on the T-DNA. The results are shown in Fig. 1. T-DNA was detected on a single different chromosome in each Ura+ yeast strain. We can therefore conclude that nonhomologous T-DNA does integrate in the yeast genome and that this integration appears random at the chromosome level.
Figure 1.
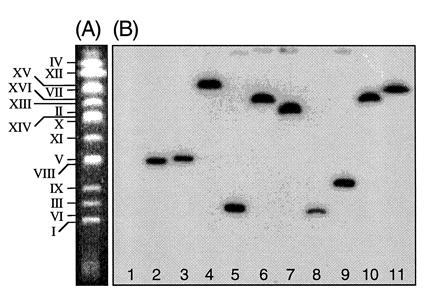
Integration of nonhomologous T-DNA into the genome of S. cerevisiae is random at the chromosome level. (A) A CHEF gel showing the separated chromosomes of an untransformed colony of S. cerevisiae strain RSY12. Each chromosome is indicated. (B) Chromosomes from Ura+ strains obtained after cocultivation of LBA1126(pRAL7102) with RSY12 were separated on a CHEF gel and blotted to a nylon membrane. The blot was probed with a labeled 1.1-kb HindIII URA3 fragment. Lane 1, RSY12 (not cocultivated); lane 2, Ura+ S. cerevisiae M5-1a strain obtained after cocultivation of M5-1a with LBA1100(pRAL7100). The T-DNA has integrated via a double crossover on chromosome V (11). Lane 3, as in lane 2, but the whole binary vector pRAL7100 has integrated via a single crossover on chromosome V, causing a shift in chromosome V mobility. Lanes 4–11, RSY12 Ura+ strains obtained after cocultivation of RSY12 with LBA1126(pRAL7102).
Integration of T-DNA by IR into the Yeast Genome Resembles T-DNA Integration in Plants.
To determine the yeast target sites for T-DNA integration we constructed the binary vector pRAL7103. The T-DNA of this vector carries not only the URA3 fragment for selection in yeast, but also a 2.6-kb DNA fragment containing a gene for carbenicillin resistance and a pUC origin of replication. After integration of T-DNA from pRAL7103 into the yeast genome, the yeast sequences flanking the T-DNA could be rescued as follows. Total yeast DNA was isolated and digested with BglII, which does not cut within the T-DNA. The resulting BglII fragments were self-ligated and electroporated to E. coli. Plasmids from the resulting carbenicillin-resistant colonies were purified. Primers p1 and p2 homologous to the T-DNA were then used with the T7 Sequenase Kit (Pharmacia) to obtain the sequence of the genomic DNA flanking the T-DNA.
The results of cocultivations between LBA1126(pRAL7103) and RSY12 are shown in Table 1 and the integration sites of these Ura+ colonies are shown in Fig. 2. Complete T-DNAs could be found integrated in the S. cerevisiae genome (strains 1 and 3). In most cases, however, truncation of one or both of the borders was observed. This occurred more often at the LB than at the RB, as has been reported previously after sequencing of T-DNAs that had integrated into the plant genome (10, 22, 23).
Figure 2.

Nucleotide sequences of yeast target sites and the insertion points of the T-DNA 3′ end [left border (LB)] and 5′ end [right border (RB)]. The diagram at the top of the figure presents the rationale used to present the sequence data. All the sequences are shown in the 3′ to 5′ orientation. The line marked “T-DNA” shows the termini of the processed bottom strand of the T-region that constitutes the T-DNA. This DNA is transferred to the yeast cell during cocultivations. The extent of T-DNA border truncation found in each strain after T-DNA integration can therefore be calculated by comparison of the integrated borders with the intact T-DNA. Yeast target DNA is shown in lowercase letters, and deleted yeast DNA is shown in boldface type. Filler DNA is shown in lowercase, underlined, italic type. The rescued and sequenced T-DNA borders are shown on the line above (the LB) and below (the RB) the yeast target sequence. The T-DNA borders are positioned to correspond with the left and right ends of the deleted yeast DNA. Numbers on the same line as the T-DNA border sequences indicate the number of bases lost from that T-DNA border. The T-DNA in strain 10 was fused to mitochondrial sequences, and we were therefore unable to determine the extent of the target DNA deletion.
The identity of the yeast sequences fused to the T-DNA borders were obtained by homology searches of the GenBank data base. In this way we could accurately establish the T-DNA insertion point in the yeast genome. Comparisons of the nucleotide sequences of the yeast target sites did not reveal any obvious homology between them. The lack of large homology between target sequences, and between the target sequences and the incoming DNA, is indicative of an IR mechanism.
Many of the strains show microhomology between either one or both of the T-DNA ends and the deleted genomic DNA. Such microhomology has been implicated in IR in mammalian (24) and plant (8–10) species. Microhomology has not been documented for every analyzed T-DNA insertion in plants, and in line with this, microhomology could not be seen in all of the T-DNA insertions in yeast.
Possible deletions of the yeast target DNA in each Ura+ yeast strain were calculated after determining to which yeast nucleotides the borders of the T-DNA had been ligated. The yeast DNA between these two T-DNA insertion points was therefore considered to have been deleted during the process of T-DNA integration. Target deletions were found in all strains, but the length of deletions was variable (3–65 bp), and on average shorter than the deletions reported in plants (8–10).
In strains 4 and 8 we also observed filler DNA linking one of the borders to the yeast DNA. Strain 8 has filler DNA containing two direct repeats (GGTGtgGGTG). In strain 4 the filler DNA shares homology with the 5′ end of the deleted yeast DNA. So far, filler DNA has not been reported after IR in S. cerevisiae (13), suggesting that it may be formed specifically during T-DNA integration.
In strain 10, the yeast DNA flanking the T-DNA borders had no homology with the yeast nuclear genome but imperfect homology with the yeast mitochondrial genome. According to the mitochondrial genome map of Zamaroczy and Bernardi (25), the RB is fused to the nucleotide at position 56240 and the LB at position 12014. As gel electrophoresis shows the rescued plasmid to be only 12 kb, each border must be fused to different small mitochondrial DNA fragments rather than a single fragment of 44 kb. The URA3 marker is not functional in the mitochondria (26) and therefore this integrant was probably produced by ligation of the T-DNA to nuclear-located mitochondrial DNA fragments. Migration of mitochondrial DNA to the yeast nucleus has been demonstrated (26), but the mechanism by which this occurs is unknown. Mitochondrial DNA contains many autonomously replicating sequences, thus allowing the T-DNA to be present in the yeast cell as an autonomously replicating plasmid. Similar events were previously reported to occur at a high frequency (5–10% total transformants) after transformation of yeast with a nonhomologous segment of double-stranded DNA (dsDNA) (13). The stability of the Ura+ marker in strain 10 was checked by growth of the strain in nonselective medium. Surprisingly, the Ura+ marker was stable after growth of the strain in nonselective medium for 72 hr. One of the 12 Ura+ strains, generated from the LBA1126(pRAL7102)/RSY12 cocultivation and tested on the CHEF gel, showed a band on the blot that was smaller than any chromosome (data not shown). This strain may also have contained T-DNA fused to linear mitochondrial fragments, thus forming a plasmid in yeast.
DISCUSSION
The mechanisms underlying the transformation of plant species by A. tumefaciens are complex. Lately our understanding of the processes involved in the early events of T-DNA transfer has increased considerably, but the fate of the T-DNA in the plant cell and the host factors involved in T-DNA integration remain a mystery. In this report we show that T-DNA can integrate via IR in S. cerevisiae, and that the T-DNA integration occurs in a similar way as observed previously in dicotyledonous plant species. T-DNA integration in the plant genome has been found (9, 10) to (i) not be site specific, (ii) lead to small deletions of the plant DNA at the insertion sites, (iii) the 3′ end of the T-DNA is less conserved compared with the 5′ end, (iv) some IR events may be mediated by microhomology, and (v) in some cases generate filler DNA during integration. As is shown in Fig. 2 all these features also occur after IR of T-DNA into the S. cerevisiae genome. Therefore, higher and lower eukaryotes seem to have retained the same mechanisms of IR throughout their evolution.
Sequencing of the rescued T-DNA insertions from yeast showed that, as in plants, the sequence of the RB was more often intact compared with the LB sequences. This is probably due to the presence of the VirD2 protein attached to the 5′ end of the T-DNA, which may protect the RB against host exonuclease activity (27). Previously, we have shown that T-DNAs carrying the URA3 gene and the yeast 2μ origin of replication can circularize in the yeast cell by ligation of the borders, forming a T-circle (11). One protein that may mediate this ligation reaction is the VirD2 protein as it is already covalently attached to the 5′ end of the T-DNA and has in vitro border nickase/ligase activity (28). The ligated borders were sequenced and found not to be truncated, but to encompass precise RB/LB fusion products. Therefore, truncations of the borders seen after integration of the T-DNA in the yeast genome may be due to the mechanism of IR used. Alternatively, border truncations may also be formed before integration, but such molecules might not be able to circularize and therefore were not found among the rescued T-circles. The T-DNA copies in strains 1 and 3 have integrated intact borders, showing that border truncation does not always occur and that intact borders are not always ligated.
The possible role of microhomology during IR in higher eukaryotes has been reported (8–10, 24). In S. cerevisiae, microhomology is present between single-strand overhangs at the ends of restriction enzyme-digested dsDNA and the yeast target sequences before integration (13). During this restriction enzyme-mediated integration (REMI) mechanism, nucleotides can be lost from such single-strand overhangs of the dsDNA transforming fragment. This can be considered equivalent to the truncation of the T-DNA borders that we have observed. Deletion of yeast target sequences has also been observed after transformation of yeast with blunt-ended nonhomologous dsDNA and may be a general property of integration mechanisms using IR. Deletions have also been observed after double-strand break formation at the S. cerevisiae MATα locus (29).
In S. cerevisiae two genes have been implicated in IR, RAD50 and TOP1. In RAD50 mutants the frequency of IR is reduced (30) and mating type switching is delayed (31). The Rad50 protein seems to be multifunctional, and it will be interesting to see how RAD50 mutations affect T-DNA integration and if a functional homologue of Rad50 can be found in higher eukaryotes.
IR of dsDNA shows preferential integration at S. cerevisiae genomic sites with the consensus nick site for topoisomerase I (Top1) [(G/C)(A/T)T]. In fact, overexpression of Top1 leads to a greater percentage of integration events in sites with this consensus sequence (32). Class I topoisomerases are involved in changing the superhelical state of DNA, allowing transcription and replication of DNA. Whether Top1 is involved in IR in higher eukaryotes is not known, but T-DNA seems to integrate preferentially into transcriptionally active regions of the plant genome (33). A number of the T-DNA insertions in yeast have a Top1 consensus nick site adjacent to either the left or the right T-DNA border. This could be coincidental, and therefore we shall try to establish if TOP1 plays a role in IR of T-DNA in yeast by varying the levels of Top1 in the cell as was done before (32).
Much work has been done in plants to target T-DNA to a wild-type or introduced locus located in the plant genome. To maximize the frequency of homologous recombination at the genomic locus, positive/negative selection and large regions of homology have been used (6, 7, 34). Unfortunately, the frequency of gene targeting in plants remained low, perhaps because of the very efficient system of IR. Identification of yeast and plant factors involved in IR of T-DNA should help to answer why higher eukaryotes have such an efficient IR system. Solving the mysteries of dynamic DNA may lead to the development of both monocot and dicot plant lines with higher frequencies of gene targeting.
Acknowledgments
The S. cerevisiae strain RSY12 was a kind gift from Dr. R. H. Schiestl. We would like to thank Amke den Dulk-Ras and Emma Scheeren-Groot for construction of the A. tumefaciens strain LBA1126, Yde Steensma and Aloys Teunissen for technical help with the CHEF gels, and Kaska Mroczek for construction of the diploid derivative of RSY12. This work was supported by a grant to P.B. from the Biological and Biotechnological Sciences Research Council (Swindon, U.K.).
Footnotes
Abbreviations: IR, illegitimate recombination; LB, left border; RB, right border; dsDNA, double-strand DNA.
References
- 1.Hooykaas P J J, Beijersbergen A. Annu Rev Phytopathol. 1994;32:157–179. [Google Scholar]
- 2.Lessl M, Lanka E. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 3.Citovsky V, Zupan J, Warnick D, Zambryski P. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 4.Rossi L, Hohn B, Tinland B. Proc Natl Acad Sci USA. 1996;93:126–130. doi: 10.1073/pnas.93.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Regensburg-Tuink A J G, Hooykaas P. Nature (London) 1993;363:69–70. doi: 10.1038/363069a0. [DOI] [PubMed] [Google Scholar]
- 6.Offringa R, de Groot M J A, Haagsman H J, Does M P, van den Elzen P J M, Hooykaas P J J. EMBO J. 1990;9:3077–3084. doi: 10.1002/j.1460-2075.1990.tb07504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paszkowski J, Baur M, Bogucki A, Potrykus I. EMBO J. 1988;7:4021–4026. doi: 10.1002/j.1460-2075.1988.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumoto S, Ito Y, Hosoi T, Takahashi Y, Machida Y. Mol Gen Genet. 1990;224:309–316. doi: 10.1007/BF00262423. [DOI] [PubMed] [Google Scholar]
- 9.Gheysen G, Villarroel R, Van Montagu M. Genes Dev. 1991;5:287–297. doi: 10.1101/gad.5.2.287. [DOI] [PubMed] [Google Scholar]
- 10.Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei G P, Schell J, Hohn B, Koncz C. EMBO J. 1991;10:697–704. doi: 10.1002/j.1460-2075.1991.tb07999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas P J J. EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piers K L, Heath J D, Liang X, Stephens K M, Nester E W. Proc Natl Acad Sci USA. 1996;93:1613–1618. doi: 10.1073/pnas.93.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiestl R, Dominska M, Petes T D. Mol Cell Biol. 1993;13:2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheeren-Groot E P, Rodenburg K W, den Dulk-Ras A, Turk S C H J, Hooykaas P J J. J Bacteriol. 1994;176:6418–6426. doi: 10.1128/jb.176.21.6418-6426.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turk S C H J, van Lange R P, Sonneveld E, Hooykaas P J J. J Bacteriol. 1993;175:5706–5709. doi: 10.1128/jb.175.17.5706-5709.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steensma H Y, Holterman L, Dekker I, Van Sluis C A, Wenzel T J. Eur J Biochem. 1990;191:769–774. doi: 10.1111/j.1432-1033.1990.tb19186.x. [DOI] [PubMed] [Google Scholar]
- 17.Bevan M. Nucleic Acids Res. 1984;22:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm C, Meeks-Wagner D W, Fangman W L, Botstein P. Gene. 1986;42:169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- 19.Gibson S I, Sommerville C. In: Methods in Arabidopsis Research. Koncz C, Chua N-H, Schell J, editors. Singapore: World Scientific; 1992. pp. 119–143. [Google Scholar]
- 20.de Jonge P, de Jongh F C M, Meijers R, Steensma H Y, Scheffers W A. Yeast. 1986;2:193–204. doi: 10.1002/yea.320020307. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz D C, Cantor C R. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 22.Tinland B, Schoumacher F, Gloeckler V, Bravo-Angel A M, Hohn B. EMBO J. 1995;14:3585–3595. doi: 10.1002/j.1460-2075.1995.tb07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Graaf E, den Dulk-Ras A, Hooykaas P J J. Plant Mol Biol. 1996;31:677–681. doi: 10.1007/BF00042239. [DOI] [PubMed] [Google Scholar]
- 24.Roth D B, Wilson J H. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zamaroczy M, Bernardi G. Gene. 1986;47:155–177. doi: 10.1016/0378-1119(86)90060-0. [DOI] [PubMed] [Google Scholar]
- 26.Thorsness P E, Fox T D. Nature (London) 1990;346:376–379. doi: 10.1038/346376a0. [DOI] [PubMed] [Google Scholar]
- 27.Dürrenberger F, Crameri A, Hohn B, Koukolíková-Nicola Z. Proc Natl Acad Sci USA. 1989;86:9154–9158. doi: 10.1073/pnas.86.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pansegrau W, Schröder W, Lanka E. Proc Natl Acad Sci USA. 1993;90:2925–2929. doi: 10.1073/pnas.90.7.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore J K, Haber J E. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiestl R, Zhu J, Petes T D. Mol Cell Biol. 1994;14:4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov E L, Sugawara N, White C I, Fabre F, Haber J E. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J, Schiestl R H. Mol Cell Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kertbundit S, De Greve H, Deboeck F, Van Montagu M, Hernalsteens J P. Proc Natl Acad Sci USA. 1991;88:5212–5216. doi: 10.1073/pnas.88.12.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Risseeuw E, Offringa R, Franke-van Dijk M E I, Hooykaas P J J. Plant J. 1995;7:109–119. doi: 10.1046/j.1365-313x.1995.07010109.x. [DOI] [PubMed] [Google Scholar]


