Abstract
Motivation: DNA sequence reads from Sanger and pyrosequencing platforms differ in cost, accuracy, typical coverage, average read length and the variety of available paired-end protocols. Both read types can complement one another in a ‘hybrid’ approach to whole-genome shotgun sequencing projects, but assembly software must be modified to accommodate their different characteristics. This is true even of pyrosequencing mated and unmated read combinations. Without special modifications, assemblers tuned for homogeneous sequence data may perform poorly on hybrid data.
Results: Celera Assembler was modified for combinations of ABI 3730 and 454 FLX reads. The revised pipeline called CABOG (Celera Assembler with the Best Overlap Graph) is robust to homopolymer run length uncertainty, high read coverage and heterogeneous read lengths. In tests on four genomes, it generated the longest contigs among all assemblers tested. It exploited the mate constraints provided by paired-end reads from either platform to build larger contigs and scaffolds, which were validated by comparison to a finished reference sequence. A low rate of contig mis-assembly was detected in some CABOG assemblies, but this was reduced in the presence of sufficient mate pair data.
Availability: The software is freely available as open-source from http://wgs-assembler.sf.net under the GNU Public License.
Contact: jmiller@jcvi.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
New DNA sequencing technologies demand new assembly software to stitch together short strings of nucleotide bases—as determined by a sequencer—called reads. Many mature assemblers were developed when virtually all DNA sequence data were generated using Sanger chemistry to produce high-fidelity long reads. De novo assemblers, for which sequence data are the only input, include: Phrap (www.phrap.org), TIGR Assembler (Sutton et al., 1995), Celera Assembler (Myers et al., 2000), Euler (Pevzner et al., 2001), PCAP (Huang and Yang, 2005) and Arachne (Jaffe et al., 2003). The pyrosequencing platform produced by 454 Life Sciences is sold with Newbler, an assembler specifically for 454's medium-length reads (Margulies et al., 2005). New assemblers including Velvet (Zerbino and Birney, 2008) offer functionality specifically for short-read sequencing technologies, such as Solexa (Bentley, 2006).
Hybrid sequencing: hybrid sequencing strategies leverage the strengths of two or more sequencing platforms and may require assembly software tuned for specific-read type combinations (Hall, 2007). At least three groups have introduced software for hybrids of pyrosequencing and other read types. We introduce a package that best exploits paired-end mate information.
The first protocols for assembly of hybrid data applied a multiple-assembler pipeline. Newbler combined ‘pyro’ reads into initial contigs that were shredded to produce overlapping pseudo-Sanger reads. These were processed with Sanger reads by a Sanger-specific assembler: using Celera Assembler (Goldberg et al., 2006) or Arachne for whole genomes, or using Phrap (Wicker et al., 2006) for cloned targets. Recent protocols use a single assembler tuned for hybrid data. Newbler was updated to accept non-pyro data (Roche, 2007), and Euler was modified to accept pyro data in a version called Euler-SR (Chaisson and Pevzner, 2008). Now, Celera Assembler has been modified to accept pyrosequencing data natively, alone or in combination with Sanger data.
Modified Celera Assembler: the Celera Assembler software has modules for successive phases of assembly: pairwise overlap detection; initial ungapped multiple sequence alignments called unitigs; unitig consensus calculation; combination of unitigs with mate constraints to form contigs and scaffolds, which are ungapped and gapped multiple sequence alignments, respectively; and finally, scaffold consensus determination (Myers et al., 2000). Our approach to hybrid data assembly reuses the Celera Assembler scaffold and consensus modules. Independent of the hybrid problem, the scaffold module was revised to recover trimmed base calls confirmed by co-locating reads, and the consensus module was revised to determine alternate consensus sequences in regions of apparent polymorphism (Denisov et al., 2008). Our analysis narrowed the source of hybrid assembly problems to the overlap and unitig stages.
For speed, Celera Assembler relies on short exact matches between reads as seeds for overlap detection. Its exact-match algorithms were sensitive to the different proclivities for stutter observed between platforms. Stutter, that is, incorrect determination of the number of bases in homopolymer (single-letter) runs, is more prevalent in pyro reads than Sanger reads. We therefore modified the software to search for matches in compressed sequence, in which all single-letter repeats are reduced to a single base. The uncompressed sequence is consulted later before the seeds become overlaps.
Celera Assembler was sensitive to the different average read lengths between platforms. The shorter reads are more likely to be entirely contained within genomic repeats. Over-collapsed alignments of short repeat reads induce true and false overlaps to the interior of longer reads. Where the longer reads extend beyond the genomic repeats, they do not all overlap each other. The result is short reads with containment overlaps to multiple long reads that do not overlap each other. These overlap tangles were triggering Celera heuristics designed to detect mis-assembly, leading to unnecessarily short contigs.
Celera Assembler was also sensitive to the higher coverage typical of lower cost pyrosequencing. Higher coverage leads to increased collisions of reads with exactly the same prefix sequence. The assembler's arbitrary tie-breaking heuristics, sufficient for infrequent ties, had the potential to lead the assembler away from the global optimum in hybrid data. To address these problems we developed an aggressive approach to unitig construction that builds unitigs in greedy fashion, always following a read's best overlap (by an appropriate criterion), and ignoring contained reads at first. The aggressive unitigs initially incorporate mistakes that, ideally, are caught and corrected later by pattern analysis applied to best overlaps and mate constraints.
High coverage could also increase the number of spurs, that is, reads with invalid sequence at one end. These seemed to contribute to fractured unitigs on hybrid data. We realized the software could turn higher coverage to its advantage by carefully trimming reads of unconfirmed sequence.
The new pipeline for hybrid data assembly is named CABOG (Celera Assembler with the Best Overlap Graph). It was challenged to assemble small genomes from 454 GS FLX reads in combination with paired-end mates from either FLX, or Sanger sequencing, or both. It was compared with other hybrid assembly protocols for continuity, accuracy and performance.
2 ALGORITHM
CABOG parses the native SFF files produced by the 454 FLX pyrosequencing machines. It discards 454 reads that include at least one unresolved base (the letter N). It recognizes mated reads as those whose sequence contains 454 linker sequences. From these mated reads it generates one or two shorter, linker-free pseudo-reads, plus a distance constraint set to the estimated mean separation (default 3 kb).
Overlap-based trimming: to exploit the increased expected read coverage, CABOG employs a read-trimming step. This functionality has been explored previously in Lucy (Chou and Holmes, 2001), PCAP, Arachne, UMD Overlapper (Roberts et al., 2004), and Figaro (White et al., 2008). The new read trimmer first computes for all read pairs local alignments, or partial overlaps that may not span the end of either read. On reads with sequence beyond the initially specified clear range, it extends the clear range to the extent confirmed by overlaps. It flags regions with discontinuous overlap coverage and trims the clear range to the longest covered span, possibly length zero, using heuristics to identify the precise boundaries. It identifies probable spurs and chimers (reads that join discontinuous genomic loci) and trims each to one trusted clear range.
Anchors and overlaps: CABOG uses exact-match seeds to detect possibly overlapping reads quickly. It finds these seed matches in compressed sequence where consecutive instances of the same base are reduced to one base. Compression compensates for the stutter that is observed more frequently in pyro reads. Each seed is a k-mer (a substring of k bases, with k=22 by default). CABOG counts the number of instances of each distinct k-mer observed in the compressed input sequence. To avoid highly repetitive k-mers, it dynamically tabulates a threshold M so that k-mers with more than M occurrences constitute at most 1% of all k-mer occurrences. Only k-mers with between two and M occurrences are used for overlap seeds. CABOG identifies read pairs as likely to overlap if they share sufficiently many k-mers, with a sliding threshold that favors rare k-mers but accepts more common k-mers if they cover longer spans. For the selected read pairs, it chooses a single 0-length anchor position from the rarest k-mer shared by that pair of reads.
CABOG then determines which anchors extend to overlaps. Iteratively and in parallel, it considers each read as a reference to which it aligns all other reads anchored to it. It calculates pairwise alignments by first extending from the anchor in one direction to find the last aligning position X. If X is at the end of either read, the full-alignment extension is computed from X back across the anchor. Calculating all alignments in a consistent direction with respect to the reference read produces a more accurate multialignment, particularly near homopolymer runs. A modified version of the Landau–Vishkin algorithm (Gusfield, 1997) is used to efficiently find the position X. The multialignment of overlapping reads to each reference read is used to detect likely sequencing errors and modify alignments accordingly. It is also used to average the homopolymer run lengths, applying different weighting criteria for Sanger and pyro reads. Finally, it outputs directed overlaps from the corrected reference sequences, with each overlap region required to span at least 40 bases covering two-read ends, and to have 94% or better sequence identity.
Best overlap graph: conceptually, reads and overlaps are represented in a multigraph, G, with both directed and undirected edges. Each read is represented by a pair of nodes, corresponding to the two ends of the read, connected by an undirected edge. Directed edges represent dovetail overlaps, that is, those that span exactly one end of each read. Dovetail paths in G are acyclic paths that include an undirected edge immediately before and after every directed edge. Path length is the number of implicit reads traversed.
When CABOG creates G, it disregards overlaps from reads with containment overlaps. It also disregards overlaps that do not satisfy a quality threshold (by default, at most 1.5% alignment error and at least 40 bp spanned). It loads at most one directed edge per node, which represents the corresponding read end's best overlap. By default ‘best’ is measured as most bases spanned by the overlap alignment, although other criteria can be used. Ties are broken by alignment percentage error, or failing that, arbitrarily by read ID. G is a best overlap graph, or BOG. It is implemented as multiple linked lists in an array of reads, where each array element includes one left and one right (possibly null) pointer to a particular end and strand of another read. The BOG represents a drastic and lossy data reduction of the overlap set. It is a greedy heuristic to avoid the overlap tangles expected in high-coverage hybrid data that present a wide mix of read lengths.
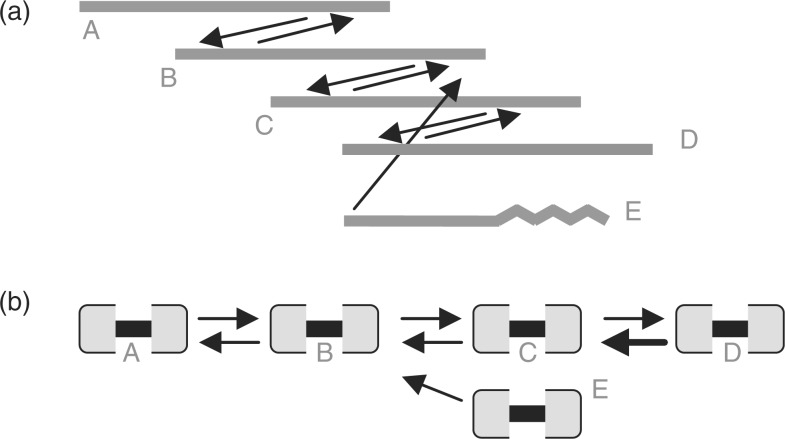
Unitig construction: any cycles in G are eliminated by deletion of one edge, chosen arbitrarily. The resulting BOG paths cannot diverge because each node has out-degree of at most one in the overlap edges. The BOG paths can converge due to overlaps that are not mutually best for both reads involved. See Figure 1.
Fig. 1.
Two representations of a best overlap graph. In (a), the layout resembles a multiple sequence alignment. In (b) each read is represented by two nodes joined by an undirected edge. Arrows represent best overlaps, where best means covering the most sequence. There are mutual best overlaps between successive pairs of reads A through D. Due to erroneous bases at one end (wavy line), read E has a non-mutual best overlap to B. Paths span undirected and directed edges alternately. Path EBA converges on path ABCD. CABOG scores read E lower than the others since only three reads are on paths from it. Starting with any one of the high-scoring reads, CABOG would build initial unitig ABCD, then E. Using saved information about each path intersection, CABOG would discount the intersection at B because the path from E spanned only one read before B. It would break ABCD only if there were also a change in read arrival rate at B, which is not the case here. Although linear-time directed-path following finds the longest possible unitig in this constructed case, it is not guaranteed to do so when paths span multiple intersections.
CABOG scores each read by the number of other reads reachable from it along BOG paths. Each read's score is the sum of the lengths of the paths from both of the read's nodes. CABOG finds path lengths efficiently by reusing saved path lengths to short-circuit path following.
CABOG sorts the reads by score. Starting at higher scoring reads, it follows paths and builds a unitig from each path. More precisely, starting at the next-highest scoring read R, it skips R if R is already in a unitig. Otherwise, it begins a new unitig with R and follows the two dovetail paths from R's two nodes, adding reads to the unitig until it encounters a path end or a read already belonging to some unitig.
This completes the greedy, aggressive stage of unitig construction. At this point, the unitigs partition the reads. The initial unitigs are called ‘promiscuous’ because their paths could span non-mutually best overlaps. The read visitation order ensures that the longest path through each intersection becomes a unitig first. Shorter convergent paths become shorter unitigs that terminate at the intersections. On paths with a single intersection, CABOG always selects the longer path first. On paths with multiple intersections, it can miss opportunities for even larger unitigs. Such unitigs would be revealed by making all overlap edges bi-directional in G. However, in such a graph, path following would be a non-linear operation.
Unitig splitting: CABOG breaks promiscuous unitigs at sites corresponding to selected path intersections. Each BOG path intersection can signal a genomic repeat boundary or represent noise. CABOG uses heuristics designed to select most repeat-induced intersections while avoiding noise-induced intersections. Spurs are a common form of noise, and not all spurs would be corrected during the overlap-based trimming step. Spur-induced path intersections produce an ‘intruder’ path of length 1. It would be wrong to ignore all length=1 intruders; a valid read that spans a genomic repeat boundary can have no overlaps at its non-repetitive end due to random low coverage. Therefore, CABOG ignores most, but not all, length=1 intruders, according to the heuristics below.
A BOG path is called ‘long’, if its unitig contains more than one read and has a total sequence length >500 bases. CABOG visits every BOG path P and applies the following operations in order. If it splits P, then it also splits the corresponding unitig. (i) At each point where P is intersected by path L, it breaks P if L is long. (ii) Between every consecutive pair of intersections with P, if neither incident path is long, then it examines the bracketed interval of P. If the interval's read-arrival rate is approximately double or more compared to the surround, it breaks P at both intersection points. A path's arrival rate is measured by the average spacing between read starts in the corresponding unitig. (iii) If P has one or more incident paths that are not long, and if the intersection points correspond to changes in arrival rate in P, then CABOG chooses one such point and breaks P there. It chooses the intersection point across which P's read arrival rate changes most dramatically.
After intersection-based splitting, CABOG breaks unitigs further using mate constraints. First, CABOG incorporates into unitigs the contained reads, according to their best overlaps. It tabulates mate pair satisfaction and violation from mate pairs that co-locate to a unitig. Satisfaction means placement within predicted mean length±5 SDs at the proper orientation relative to each other; anything else is a violation. CABOG ejects from unitigs any contained reads whose placement violates a mate constraint. It breaks unitigs where total mate coverage is sufficient but the number of violations is above a given threshold.
Contigs, scaffolds, consensus: the rest of the Celera Assembler pipeline runs without any special modification for hybrid assembly. Note the scaffold module may re-incorporate reads ejected previously for mate constraint violations; it can use mate constraints to guide individual reads to their appropriate contigs.
3 METHODS
Assemblers: the Celera Assembler was used for the CABOG, Goldberg and traditional Celera pipelines; version 5.0 from 5/2008 was used everywhere except the human trial, which used version 5.2 from 10/2008. The latest production version of Newbler (1.1.03.24) was used on FLX data, with the large option for the human trial. The software for Arachne, PCAP and Euler-SR were current through 5/2008. Velvet version 0.7, from 10/2008, ran with expected coverage set to 24. Assemblies ran under SuSE Linux on 64-bit Intel or AMD processors with 24 GB or 32 GB RAM, although the human assembly also exploited 48 GB of a high-RAM node. CABOG and Newbler were fed 454 reads in SFF format. Arachne was fed files from NCBI, slightly modified to satisfy the input parser. Euler-SR, PCAP and Velvet were fed files generated by CABOG's parser following instructions in each program's documentation.
Analysis: continuity statistics were gathered from each assembler using Perl analysis of the FASTA output files. Assembly alignments were generated with MUMmer (Kurtz et al., 2004), ATAC (http://kmer.sf.net) and Stretcher (http://emboss.sf.net). Repeat annotation was generated with REPuter (Kurtz et al., 2001) with a post-process to aggregate repeat classes by overlapping sequence. EST alignments were generated with the ESTmapper (http://kmer.sf.net) extension to Sim4 (Florea et al., 1998).
Reference: the Psychromas sp. CNPT3 reference (RefSeq NZ_AAPG00000000), with 2 945 265 bases in one linear contig, had been produced at JCVI using Celera Assembler plus finishing. The Porphyromonas gingivalis W83 reference (GenBank NC_AE015924), with 2 343 476 bases in one circular contig, had been sequenced by Sanger chemistry, assembled with TIGR Assembler, and finished at TIGR/JCVI (Nelson et al., 2003). The Sanger reads assembled here were a distinct set. The Escherichia coli K12 MG1655 reference (GenBank NC_000913), with 4 639 221 bases in a circular contig, had been produced independently by a method other than whole-genome shotgun sequencing (Blattner et al., 1997). There was no reference for Cryptosporidium muris RN66, a eukaryotic genome estimated at 9 Mb. The ESTs were obtained from NCBI via CryptoDB.
Reads: many reads were obtained directly from JCVI. All reads are available at the NCBI Trace Archive or Short Read Archive (see Supplementary Material for detail). The homogeneous component sets of reads were combined to make hybrid datasets with realistic levels of genome coverage.
4 RESULTS
Reads were combined from several genome sequencing projects as shown in Table 1. Pyrosequencing reads were used in the half-plate units provided by 454 FLX sequencers. Half-plates of unpaired reads (∼250 bases/read) were combined with Sanger mate pairs (∼800 bases/read) to make hybrid sets. Half-plates from paired-end libraries were considered hybrid sets in themselves because they consist of mostly unpaired reads mixed with some (∼30%) mate pairs (∼100 bases/read).
Table 1.
Homogeneous components for hybrid datasets
| Sp | Cmp | Library | #Unmated | Len | #Mated | Len | Cov |
|---|---|---|---|---|---|---|---|
| P.gingivalis | |||||||
| F1 | FLX unmated | 2 55 329 | 259 | 0 | - | 28.2 | |
| F2 | FLX unmated | 2 54 703 | 259 | 0 | - | 28.2 | |
| M1 | FLX 3-6Kbp | 1 84 680 | 243 | 80 304 | 116 | 23.1 | |
| M2 | FLX 3-6Kbp | 1 87 012 | 243 | 81 926 | 116 | 23.4 | |
| S1 | Sanger 40Kbp | 90 | 601 | 2786 | 728 | 1.0 | |
| E. coli | |||||||
| F1 | FLX unmated | 2 30 517 | 253 | 0 | - | 12.6 | |
| F2 | FLX unmated | 2 16 458 | 253 | 0 | - | 11.8 | |
| M1 | FLX 3-6Kbp | 2 34 299 | 232 | 65 118 | 115 | 13.3 | |
| P. CNPT3 | |||||||
| F1 | FLX unmated | 2 98 610 | 266 | 0 | - | 26.0 | |
| F2 | FLX unmated | 2 78 142 | 267 | 0 | - | 24.3 | |
| S1 | Sanger 40Kbp | 38 | 537 | 1522 | 830 | 0.4 | |
| C. muris | |||||||
| F1 | FLX unmated | 4 34 956 | 243 | 0 | - | 11.7 | |
| S1 | Sanger 40Kbp | 3272 | 434 | 21 092 | 713 | 1.7 | |
| Sanger 6-8Kbp | 4108 | 727 | 17 382 | 892 | 1.7 | ||
| Sanger 2-3Kbp | 2652 | 508 | 27 296 | 826 | 2.7 |
Sequence contribution from each component dataset. Sp, species name; Cmp, component name; Unmated/Mated, number of non-paired or paired-end reads; Len, for unmated and mated, the average clear range per read in bases; Cov, fold coverage of the genome by reads; FLX reads originate from the 454 GS FLX sequencer. Sanger reads originate from the ABI 3730 sequencer.
CABOG and other assemblers were run on each combination dataset. Contig and scaffold statistics were tabulated for every assembly by an automated process. CABOG assemblies were compared with reference genomes and also to the outputs from other assemblers. The comparisons included the recent version of Newbler designed to handle FLX mate and Sanger mate hybrid sets, and Euler-SR which had been demonstrated on a hybrid set of 454 GS 20 reads plus simulated Sanger mates (Chaisson and Pevzner, 2008). Velvet was tested on one dataset; it was designed for short reads but recent versions also accept long reads. PCAP, Arachne and the traditional Celera Assembler were included though they were designed for Sanger reads only. The last two were abandoned part way through testing after they produced fractured or no assemblies of several datasets. The Goldberg pipeline (Goldberg et al., 2006), which applies Newbler to pyrosequencing reads and Celera Assembler to Sanger mates, was run on those sets that included Sanger data.
4.1 Contig analysis
Contig size is one measure of assembly utility. Table 2 presents four contig size statistics for assemblies of selected hybrid datasets, with CABOG assemblies compared with Newbler assemblies.
Table 2.
CABOG and Newbler assemblies of hybrid data sets
| Assembler | #Contigs | Contig N50 | Contig Max | Contig Sum |
|---|---|---|---|---|
| P.gingivalis / FLX reads+FLX mates (F1+M2) | ||||
| CABOG | 48 | 67 993 | 205 585 | 2 332 097 |
| Newbler | 119 | 27 561 | 134 859 | 2 183 278 |
| P.gingivalis / FLX reads+Sanger mates (F1+S1) | ||||
| CABOG | 65 | 51 745 | 169 923 | 2 266 305 |
| Newbler | 104 | 32 377 | 154 008 | 2 184 009 |
| P.gingivalis / FLX mates+Sanger mates (M2+S1) | ||||
| CABOG | 34 | 101 101 | 307 732 | 2 314 836 |
| Newbler | 115 | 29 216 | 110 686 | 2 179 717 |
| E.coli / FLX reads+FLX mates (F2+M1) | ||||
| CABOG | 22 | 440 632 | 861 331 | 4 642 198 |
| Newbler | 87 | 87 223 | 240 232 | 4 516 116 |
| P.sp CNPT3 / FLX reads+Sanger mates (F1+S1) | ||||
| CABOG | 39 | 126 165 | 336 216 | 2 992 650 |
| Newbler | 70 | 79 879 | 203 365 | 2 963 428 |
| P.sp CNPT3 / FLX reads+Sanger mates (F2+S1) | ||||
| CABOG | 42 | 138 508 | 365 104 | 2 983 118 |
| Newbler | 99 | 45 693 | 171 391 | 2 951 683 |
| C.muris / FLX reads+Sanger mates (F1+S1) | ||||
| CABOG | 69 | 323 162 | 819 035 | 9 186 849 |
| Newbler | 73 | 247 897 | 731 211 | 9 097 078 |
The analysis included all contigs 2 kb or longer found in each assembler's FASTA output. N50, the length of the shortest contig required to span 50% of the genome length; Max, the length of the longest contig, Sum, the total contig span. Contig size statistics are shown in bases. The codes in parentheses refer to component datasets described in Table 1. Assemblies are compared by contig size statistics. Selected combinations are shown; others are provided in the Supplementary Material.
The differences in Table 2 are clearly significant. On average, CABOG's largest contig was twice the Newbler's. Its N50 contig was more than twice as large. CABOG consistently assembled more total bases into fewer (larger) contigs. Thus, CABOG demonstrated greater continuity than Newbler on these data.
In Table 2, the rows for P.gingivalis F1+S1 and M2+S1 offer a comparison between sets containing the same Sanger reads (S1) but distinct FLX reads. The FLX paired-end reads in M2 give that set slightly shorter reads on average, but also additional mate constraints. In the combination with M2, the CABOG contig N50 doubled but the Newbler value actually dropped. CABOG may exploit mate constraints more fully during contig construction. It has been observed that Newbler uses mate constraints mostly to join contigs in scaffolds (Jarvie and Harkins, 2008).
In Table 2, the rows for P.gingivalis F1+M2 and E.coli F2+M1 are devoid of Sanger data. On both of these sets, CABOG assembled over 120 000 more total bases into 1/2 to 1/4 as many contigs. Thus, CABOG provided more continuity than Newbler on 454-only sets that included mate data. To address the question of whether the mates were critical, CABOG and Newbler were further tested on homogeneous sets of just 454 unpaired reads. Here, the statistics were similar between assemblers and the assembler ranking varied. This provides additional support for the observation that CABOG better exploits mate constraints during contig construction.
The genomes in Table 2 include three prokaryotes and a small eukaryotic genome from C.muris. Thus, CABOG provided more continuity across both domains. CABOG's gain over Newbler was smallest for the eukaryote, possibly because coverage was lowest on that dataset.
CABOG's extra sequence may represent genomic repeats. To investigate this hypothesis, the P.gingivalis reference was annotated for repeats. CABOG and Newbler contigs for six datasets were mapped to the annotated reference. The repeat and contig spans were compared for overlap. The result indicated that CABOG contigs spanned more repeats and longer repeats in all the assemblies. In one example, using the F1+M2 combination, CABOG spanned 34 repeats of average length 1099 bases, but Newbler spanned 14 repeats of average length 703 bases. The difference was more pronounced in the M2+S1 combination, which included long-range Sanger mates. CABOG contigs spanned 26 repeats of average length 1981. Newbler contigs spanned nine repeats of average length 815. Thus, some of CABOG's continuity gain is attributable to increased resolution of repetitive sequence inside assembled contigs, which increases with mate availability.
Table 3 shows contig size statistics on one dataset for all assemblers tested. PCAP produced surprisingly large contigs considering it was not designed for hybrid data. The table is representative of results on other datasets, provided as Supple-mentary Material. The statistics consistently ranked CABOG first, followed by Goldberg (when run), Newbler, PCAP and Euler-SR.
Table 3.
Assemblies of one hybrid data set by all assemblers
| Assembler | #Contigs | Contig N50 | Contig max | Contig sum |
|---|---|---|---|---|
| E.coli / FLX reads+FLX mates (F1+M1) | ||||
| CABOG | 27 | 285 910 | 833 636 | 4 629,501 |
| Newbler | 89 | 82 668 | 209 279 | 4 519,532 |
| PCAP | 152 | 50 897 | 175 160 | 4 554 652 |
| Euler-SR | 328 | 22 159 | 71 505 | 4 343 338 |
| Velvet | 490 | 11 510 | 53 664 | 4 230 559 |
The analysis is described in Table 2. Only CABOG and Newbler were designed for FLX hybrid datasets. Euler-SR had been introduced for 454 GS 20 reads+Sanger mates. PCAP was designed for Sanger mates only. Velvet was designed for short reads. The Goldberg method was not run since it requires Sanger mates to improve Newbler contigs. Arachne and the traditional Celera Assembler did not assemble this dataset. The assemblies are summarized and compared using contig length statistics.
4.2 Scaffold analysis
Scaffold size is another measure of assembly utility. Table 4 presents scaffold size statistics for CABOG and Newbler assemblies of selected combinations of P.gingivalis data. The table indicates that CABOG scaffolds were significantly larger. All but two measurements favored CABOG. In one of the exceptions, Newbler's largest scaffold was longer than CABOG's on the M2 set. It may be significant that this was a low-coverage dataset.
Table 4.
Scaffold analysis of CABOG and Newbler assemblies
| Assembler | #Scaf. | Scaf. N50 | Scaf. max | Scaf. sum | Cov. (%) |
|---|---|---|---|---|---|
| P.gingivalis / FLX mates (M2) | |||||
| CABOG | 7 | 392 892 | 661 267 | 2 324,483 | 98.7 |
| Newbler | 9 | 268 678 | 718 704 | 2 187 430 | 94.1 |
| P.gingivalis / FLX mates+FLX mates (M1+M2) | |||||
| CABOG | 7 | 417 898 | 758 093 | 2 339 970 | 98.9 |
| Newbler | 11 | 266 698 | 718 559 | 2 183 668 | 93.9 |
| P.gingivalis / FLX reads+FLX mates (F1+M2) | |||||
| CABOG | 9 | 450 308 | 758 275 | 2 335 950 | 98.8 |
| Newbler | 382 223 | 720 519 | 2 189 593 | 94.2 | |
| P.gingivalis / FLX reads+Sanger mates (F1+S1) | |||||
| CABOG | 6 | 1 507 760 | 1 507 760 | 2 268 548 | 96.6 |
| Newbler | 51 | 1 489 797 | 1 489 797 | 2 185 214 | 94.3 |
| P.gingivalis / FLX mates+Sanger mates (M2+S1) | |||||
| CABOG | 1 | 2 317 095 | 2 317 095 | 2 317 095 | 98.7 |
| Newbler | 6 | 1 550 861 | 1 550 861 | 2 184 352 | 93.9 |
The analysis included all scaffolds 2 kb or longer found in each assembler's FASTA output. Scaffold length statistics are shown in bases excluding the lengths of the gaps between contigs. Note that scaffold sum may not equal contig sum (Table 2) due to the 2 kb threshold being applied at the scaffold not contig level. Cov, bases of the reference covered by a sum over single best alignments of each full or partial scaffold sequence.
In one case, CABOG produced exactly 1 scaffold, and it covered 99% of the reference sequence. That dataset, M2+S1, included short-range FLX mates and long-range Sanger mates. It is possible that the high concentration of mate constraints, or the combination of mate distances, enabled CABOG to resolve a single-scaffold assembly.
Scaffold span is an alternate measure of scaffold size. Span includes the estimated lengths of the gaps between the contigs, as well as the contig lengths. On many datasets, Newbler scaffold span statistics exceeded those of CABOG.
4.3 Assembly correctness
Selected assemblies were tested for their coverage of the reference genome sequence. Table 4 indicates the genome coverage provided by CABOG and Newbler assemblies of hybrid sets of P.gingivalis reads. CABOG coverage was consistently above 96% and was always higher than Newbler coverage. This test considered best matches only, so collapsed assemblies of repeat copies would cover only one repeat copy.
The same assemblies were measured for consensus accuracy. The alignments to reference were parsed to count all inserted, deleted, substituted and unmapped bases. Accuracy was expressed as the fraction of assembled bases that did not fall into one of these categories. For the four datasets in Table 4, CABOG accuracy varied between 99.932% and 99.980%. Newbler accuracy varied between 99.995% and 99.998%.
Selected alignments were assessed by visual inspection to reveal assembly errors, such as mis-oriented or misordered contigs within scaffolds. No errors were found at the scaffold level. Some rearrangements within CABOG contigs were noticed; these were also revealed by the subsequent analysis.
Next, alignments of contigs were inspected in detail. The analysis relied on manual and scripted review of textual representations of alignments. It covered four CABOG assemblies of P.gingivalis, four CABOG assemblies of P. sp CNPT3, and the corresponding Newbler assemblies. Breaks in the alignments were counted and inspected. Results based on MUMmer were confirmed by analyses with other software.
Table 5 lists some minor problems found in CABOG contigs: bad ends, bad contigs and collapsed tandem repeats. Collapse of tandem repeats has been observed before in Celera Assembler (She et al., 2004) and other assemblers. Most CABOG collapses involved the omission of <100 bases. CABOG's bad end and bad contig problems also involved small (under 1 kb) bits of sequence. CABOG assemblies showed more serious problems: chimeric joins and chimeric ends. The analysis of Newbler assemblies (data not shown) revealed no serious problems and three minor problems. Note Newbler's rate of collapsed tandem repeats would have been underestimated because only contigs that spanned a repeat region could contribute to the alignment breaks that were counted here.
Table 5.
Errors in CABOG assemblies
| Genome | Dataset | Chimeric | Chimeric | Bad | Bad | Collapsed |
|---|---|---|---|---|---|---|
| join | end | end | contig | tandem | ||
| P.gingivalis | F1 | 0 | 0 | 0 | 0 | 4 |
| P.gingivalis | F1+M2 | 3 | 8 | 1 | 1 | 11 |
| P.gingivalis | F1+S1 | 0 | 0 | 1 | 0 | 7 |
| P.gingivalis | M2+S1 | 0 | 1 | 2 | 0 | 9 |
| P.sp CNPT3 | F1 | 0 | 1 | 0 | 0 | 1 |
| P.sp CNPT3 | F2 | 0 | 2 | 0 | 0 | 4 |
| P.sp CNPT3 | F1+S1 | 0 | 0 | 0 | 0 | 1 |
| P.sp CNPT3 | F2+S1 | 0 | 0 | 0 | 0 | 0 |
The analysis included contigs at least 2 kb long. Chimeric join, a concatenation of unrelated sequences of at least 1 kb. Chimeric End, concatenation of less than 1 kb to a contig end. Bad end, less than 1 kb of unaligned sequence at a contig end. Bad Contig, unaligned contig. Collapsed Tandem, multiple alignments between a contig and the reference, partially overlapping in either sequence. Errors were estimated by analysis of alignments to reference sequences. Estimates were confirmed by two other alignment-based methods.
The chimeric joins in the CABOG assemblies corresponded to repetitive regions of the reference genome sequences. In no case did a Newbler contig span the corresponding region. Thus, it appears that CABOG was more aggressive than Newbler about including whole repeats inside larger contigs while committing false joins in a few repetitive regions.
On both genomes for which alignments were studied, the chimer rate dropped when the S1 (Sanger mates) set was included. This is consistent with CABOG's use of long-range mate constraints to correct mis-assembly errors within unitigs. Sanger sequencing provided the long-range mates here, but long-range mate constraints may be available soon from the pyrosequencing platform (Jarvie and Harkins, 2008). In summary, CABOG has a chimeric join rate that may be acceptably low for some genome projects, and that is diminished by inclusion of long-range mate data.
Assemblies of the C.muris genome were validated by EST mapping since no independent reference was available. All available ESTs were mapped to the CABOG and Newbler scaffolds with a threshold of 95% identity over 95% of EST length. No EST mapping spanned multiple scaffolds in either assembly. Of 27 498 ESTs, there were 14 148 unspliced and 2312 spliced alignments to CABOG's assembly. Thus, over half the ESTs confirmed CABOG scaffolds by full-length, high-stringency alignments. There were 13 214 unspliced and 1883 spliced alignments to Newbler's assembly. Thus, the CABOG assembly showed a higher rate of EST confirmation than the Newbler assembly.
4.4 Large genomes
Large eukaryotic genome projects present additional problems of scale and complexity. To test whether CABOG would scale up to such problems, it was applied to human genome data. It was run on a hybrid set consisting of 6X 454 FLX unmated reads from the Watson genome project (Wheeler et al., 2008) plus 3X in 10 kb and larger Sanger mate pairs from the Venter genome project (Levy et al., 2007). The computation consumed 5209 CPU hours over 5 days on our grid. The assembly's statistics included:
Contig count=145 971
Contig N50=36 460 bp
Contig max=310 470 bp
Contig sum=2 715 539 585 bp
Scaffold N50=10 913 700 bp
Correctness is more difficult to evaluate on larger genomes. Using the NCBI B36 human reference sequence, a whole-genome alignment was generated by the ATAC method (Istrail et al., 2004). Reference coverage by ungapped matches was 97%, indicating completeness and short-range agreement. A measure of long-range agreement was provided by the maximal one-to-one mappings between reference chromosomes and assembled scaffolds 2 kb or longer. These mappings span at most one chromosome and one scaffold. Ninety-three percent of mapped scaffolds were included in exactly one mapping. The 223 discontinuously mapped scaffolds could indicate incorrect assembly or other factors including reference errors, population differences or alignment artifacts. For comparison, the Venter assembly was reported to have 12 chimera (Levy et al., 2007) though it has 116 discontinuous mappings by this technique. Thus, CABOG produced a reasonable assembly of the human genome from this hybrid mixture of pyrosequencing reads plus mates. On the same dataset, Newbler reported overflow conditions and terminated.
5 DISCUSSION
The rapid recent emergence of new sequencing technologies has made it difficult for assembly software to keep pace. Especially challenging has been the problem of assembling heterogeneous mixtures of data so as to exploit the relative advantage of each data type. The hybrid assembly problem is new but it will retain importance as long as different platforms each offer different characteristics and compelling advantages. It is not surprising that assemblers, such as Newbler and Velvet, explicitly support hybrid datasets. Hybrid assembly software is critical even for some seemingly homogenous data. The 454 paired-end protocol produces a mixture of paired and non-paired reads, where the paired reads are less than half the length of the non-paired, on average. This phenomenon would persist even if the new 454 FLX ‘Titanium’ upgrade is able to deliver Sanger-length reads.
Here, we described improvements to the Celera Assembler that were embodied in a pipeline called CABOG. CABOG parses native 454 output and Sanger reads. It handles mate pairs of either type alone or in combination. These abilities make CABOG a versatile tool for modern assembly tasks.
CABOG assemblies of heterogeneous data compare favorably to those produced by other assembly software. CABOG assembles more bases into fewer and larger contigs and scaffolds. CABOG is more aggressive than Newbler at repeat resolution. Its large-contig and large-scaffold output would provide more substrate for manual review and automatic annotation. CABOG is a valuable tool for projects where repeat resolution is desirable.
CABOG can generate mis-assemblies, but the problem appears to be mitigated by inclusion of long-range mate data. Indeed, CABOG makes broad use of mate constraints to build larger contigs, to span repeats, and to avoid mis-assemblies. CABOG should be valuable to sequencing projects that include mate pairs, whether those are derived from Sanger sequencing or pyrosequencing. With the expected availability of long-range, as well as short-range, mates from the 454 GS FLX platform, CABOG could become the preferred assembler for projects with 100% FLX data.
CABOG used Celera Assembler's consensus module without any modification specific to pyrosequencing reads. CABOG's consensus accuracy, though high, is less than Newbler's. Thus, the consensus module may need to be tuned for pyrosequencing reads.
To our knowledge, CABOG is the only software capable of calculating a de novo assembly of the human genome from pyrosequencing and Sanger whole-genome shotgun reads. CABOG is a modification to Celera Assembler, which had previously assembled Sanger-only data from human (Levy et al., 2007). The 454 technology had previously been applied to sequencing an individual human (Wheeler et al., 2008) and to comparing individual humans (Korbel et al., 2007), though neither experiment employed de novo whole-genome shotgun assembly. Our test on human data showed that CABOG is able to run on a large-eukaryotic dataset within the memory limitations of modern computers. Our attention has shifted toward the testing and tuning of CABOG for hybrid datasets from large genomes. Other proportions of mated reads and mate distances, or further adjustment to the software, may refine CABOG's large-genome capabilities.
Supplementary Material
ACKNOWLEDGEMENTS
Gennady Denisov, Aaron Halpern, Saul Kravitz, Laura Sheahan, Tim Stockwell, Shibu Yooseph and an anonymous reviewer provided helpful feedback. Swapna Annavarapu, Les Foster, Hernan Lorenzi, Diana Radune, Joana Da Silva and Indresh Singh assisted with the data preparation.
Funding: NIAID (contract No. HHSN266200400038C), ‘Bioinfor-matics Resource Centers for Biodefense and EmergingRe-emerging Infectious Diseases’; NIAID (contract No. N01-AI-30071), ‘Microbial Genome Centers’; NIGMS (grant R01-GM077117); the J. Craig Venter Institute.
Conflict of Interest: none declared.
REFERENCES
- Bentley DR. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Denisov G, et al. Consensus generation and variant detection by Celera Assembler. Bioinformatics. 2008;24:1035–1040. doi: 10.1093/bioinformatics/btn074. [DOI] [PubMed] [Google Scholar]
- Florea L, et al. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg SM, et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc. Natl Acad. Sci. USA. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusfield D. Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. Cambridge, UK: Cambridge University Press; 1997. [Google Scholar]
- Hall N. Advanced sequencing technologies and their wider impact in microbiology. J. Exp. Biol. 2007;210:1518–1525. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- Huang X, Yang SP. Generating a genome assembly with PCAP. Curr. Protoc. Bioinformatics. 2005 doi: 10.1002/0471250953.bi1103s11. Chap. 11, Unit11 3. [DOI] [PubMed] [Google Scholar]
- Istrail S, et al. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc. Natl Acad. Sci. USA. 2004;101:1916–1921. doi: 10.1073/pnas.0307971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe DB, et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvie T, Harkins T. De novo assembly and genomic structural variation analysis with genome sequencer FLX 3K long-tag paired end reads. Biotechniques. 2008;44:829–831. doi: 10.2144/000112894. [DOI] [PubMed] [Google Scholar]
- Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, et al. The diploid genome sequence of anindividual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, et al. A whole-genome assembly of Drosophila. Scienc. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Nelson KE, et al. Complete genome sequence of the oral pathogenic Bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner PA, et al. An Eulerian path approach to DNA fragment assembly. Proc. Natl Acad. Sci. USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M, et al. A preprocessor for shotgun assembly of large genomes. J. Comput. Biol. 2004;11:734–752. doi: 10.1089/cmb.2004.11.734. [DOI] [PubMed] [Google Scholar]
- Roche . Genome Sequencer FLX Data Analysis Software Manual. Mannheim, Germany: Roche Applied Science; 2007. [Google Scholar]
- She X, et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- Sutton GG, et al. TIGR Assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci. Technol. 1995;1:9–19. [Google Scholar]
- Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- White JR, et al. Figaro: a novel statistical method for vector sequence removal. Bioinformatics. 2008;24:462–467. doi: 10.1093/bioinformatics/btm632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7:275. doi: 10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.