Abstract
Noncanonical DNA structures are postulated to be responsible for some breakpoint hotspots that occur frequently in cancers. We developed a novel mouse model system using the naturally occurring H-DNA structure that deviate from the familiar right-handed helical B form found at the breakage hotspot in the human c-MYC promoter and a Z-DNA–forming CG repeat to test this idea directly. Large-scale chromosomal deletions and/or translocations occurred in 5 (7.7%, 95% confidence interval [CI] = 3.7% to 12.8%) of the 65 mice carrying the H-DNA–forming sequences and in 7 (6.6%, 95% CI = 3.8% to 11.6%) of the 106 mice carrying the Z-DNA–forming sequences, but in 0 of the 63 control mice (P = .042 and P = .035, respectively, two-sided test). Thus, the DNA structure itself can introduce instability in a mammalian genome.
CONTEXT AND CAVEATS
Prior knowledge
Nonrandom chromosomal rearrangements that originate from double-strand DNA breaks at genomic “hotspots” are known to occur in some types of cancer; however, how DNA structure is involved is unclear.
Study design
A transgenic mouse model using DNA sequences that form H-DNA, Z-DNA, and normal helical B-DNA structures.
Contribution
Chromosome breakage and rearrangements occurred in regions of H-DNA and Z-DNA but not in control B-DNA.
Implications
DNA structure may have a role in chromosome breakage and rearrangements.
Limitations
How these DNA structures might induce the chromosome rearrangements observed in human cancer and how they form in premalignant cells are still unresolved.
From the Editors
Cytogenetic analyses of a variety of cancers, particularly hematological malignancies (1), reveal that many patients carry nonrandom chromosomal breakage and translocation events at genomic “hotspots.” However, because DNA double-strand breaks can be caused by various endogenous cellular processes and exogenous insults, and because many genetic and epigenetic changes can be found in a cancer cell, it is difficult to delineate the factors that initiate chromosome breakage and translocation in these diseases. V(D)J recombination and class switch recombination generate double-strand breaks in immune cells to stimulate rearrangements, and the RAG complex can create double-strand breaks adjacent to recombination break points, which may be responsible for some chromosomal translocation events in immune cell-related cancer (2). Many genomic break points in common chromosomal translocations in leukemia, lymphoma, and sarcomas have been cloned and sequenced. Of particular interest is the fact that such breakpoint hotspots often cluster at specific genomic elements (eg, repetitive sequences capable of adopting noncanonical DNA structures that deviate from the familiar right-handed helical B form), implicating DNA structure in chromosome breakage (3,4). We previously reported (5,6) that a naturally occurring sequence from the human c-MYC gene, which is capable of forming H-DNA, and a model Z-DNA–forming GC repeat sequence, when placed in shuttle vectors, are highly mutagenic and induce double-strand breaks in mammalian cells. These results suggest that helical distortions may result in spontaneous chromosome breakage and the translocation of genes in human disease (eg, c-MYC gene in Burkitt lymphoma and BCL-2 in follicular lymphomas).
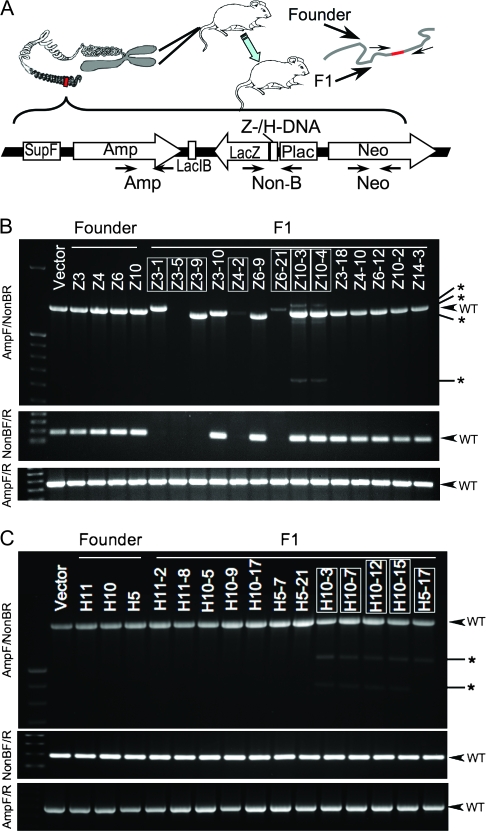
Prompted by these observations, we established a transgenic mouse model to assess the effect of DNA secondary structures on genetic instability in a chromosomal context. We constructed p2RT (Figure 1, A), which is a recoverable dual reporter vector, to generate transgenic mice for rapid analysis of mutations that are generated in the mouse genome. A 33-bp fragment in the lacZ′ mutation reporter gene in the recoverable vector, p2RT (Figure 1, A), was replaced with the H-DNA–forming sequence (5) from the human c-MYC promoter (p2RT-MYC) or with a Z-DNA–forming CG(14) repeat (6) (p2RT-CG14), for facile analysis of instability generated in the mouse genome (for insert and flanking DNA sequences, see Supplementary Figure 1, available online). A distal second reporter gene, supF, served as an internal control for the background mutations due to integration position effects. Two separated LacI-binding sequences were used to recover the reporter DNA from mouse chromosomes (7).
Figure 1.
DNA secondary structure-induced genetic instability in mouse chromosomes. A) Schematic structure of the linearized mutation reporter shuttle plasmid p2RT (constructed based on plasmid pUC19, Invitrogen, Carlsbad, CA), which was integrated in the FVB/N mouse genome by microinjection into fertilized oocytes. Primers used for genotyping are also shown in the figure. Amp = ampicillin resistance; Neo = neomycin resistance; Non-B = inserted H-DNA, Z-DNA, or control sequence. For sequences of primers, H-DNA, Z-DNA, or control sequence and the context sequence in the vector, see Supplementary Figure and Table (available online). B) Polymerase chain reaction (PCR) analysis of the p2RT-CG14 (Z-DNA) mouse strain. C) PCR analysis of the p2RT-MYC (H-DNA) mouse strain. Three primer pairs shown in panel (A) were used to amplify the genomic DNA from tail samples of each F1 mouse. Top: AmpF/Non-BR primers; middle: NonBF/R primers; bottom: AmpF/R primers. Samples from mice demonstrating DNA secondary structure-induced genetic instability are boxed. The expected PCR products from wild-type (WT) transgenes are indicated by arrows; abnormal products are indicated by asterisks.
The linearized p2RT-MYC, p2RT-CG14, or p2RT (as a negative control) plasmids were microinjected into fertilized FVB/N mouse oocytes (NCI Frederick, Frederick, MD) to generate transgenic mice. Typically 150–300 oocytes were injected for each construct and were implanted into 5–10 pseudopregnant mice. Transgenic founder mice were identified by polymerase chain reaction (PCR) analysis of genomic DNA from tail biopsies using three primer pairs (four combinations) that are specific to various regions of the p2RT plasmid. AmpF/R primers spanned the ampicillin resistance gene; NonBF/R primers spanned the regions containing H-DNA, Z-DNA, or control sequences; and NeoF/R primers spanned the neomycin resistance gene (Figure 1, A; for primer information, see Supplementary Table 1, available online). Six Z-DNA founder mice (Z1, Z3, Z4, Z6, Z10, and Z14), three H-DNA founder mice (H5, H10, and H11), and three control founder mice (C1, C2, and C3) that tested positive in all four PCRs were crossed with wild-type FVB/N mice to obtain F1 offspring from each line, which were then genotyped. DNA from transgenic mouse tail samples was also subjected to quantitative PCR and real-time PCR (comparing reporter vector to the endogenous Gapdh gene, Table 1) to estimate the copy number of constructs per genome. Mice were housed and experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee.
Table 1.
Mutation frequency as a measure of chromosome instability in mice carrying H-DNA (H), Z-DNA (Z), or control (C) sequences
| Mouse strain | Copy no.* | No. positive† | No. of mutants | Mutation frequency, (95% CI), %‡ |
| H5 | 40–80 | 27 | 1 | |
| H10 | 10–20 | 19 | 4 | |
| H11 | 20–40 | 19 | 0 | |
| H-DNA total§ | 65 | 5 | 7.7 (3.7 to 12.8) | |
| Z3 | 1–2 | 14 | 3 | |
| Z4 | 2–4 | 9 | 1 | |
| Z14 | 1–2 | 20 | 0 | |
| Z10 | 15–20 | 22 | 2 | |
| Z1 | 5–10 | 19 | 0 | |
| Z6 | 10–15 | 22 | 1 | |
| Z-DNA total§ | 106 | 7 | 6.6 (3.8 to 11.6) | |
| C1 | 20–40 | 31 | 0 | |
| C2 | 40–60 | 9 | 0 | |
| C3 | 3–6 | 23 | 0 | |
| Control total | 63 | 0 | ND‖ |
Copy numbers were estimated by combining the results of quantitative polymerase chain reaction (PCR) and real-time PCR by comparing the signal intensity or ΔΔCt of integrated fragments to the Gapdh gene.
Mouse tail DNA showing a positive signal in any one of the PCRs using AmpF/R, NonBF/R, or NeoF/R primers was considered as “positive”.
The 95% confidence intervals (CIs) of mutation frequency in Z-DNA and H-DNA mice were estimated using Tukey’s Honest Significant Difference method.
Because differences among the subgroups within each strain (ie, Z-DNA, H-DNA, or control) were not statistically significant using the Pearson chi square test, we summed the mutant number for each group and analyzed the data using the Fisher exact test (Z-DNA or H-DNA vs control; P =.035 and P = .042, respectively). All statistical tests were two-sided.
ND = None detected.
F1 mice carrying H-DNA– or Z-DNA–forming sequences showed genetic instability near the noncanonical DNA sequences, whereas the vector backbone remained largely intact within the chromosomes, suggesting a DNA structure–specific mutagenesis rather than general instability events at unstable integration sites. For example, although all mice included in this study tested positive for AmpF/R by PCR (Figure 1, B and C, bottom panel), indicating the existence of vector backbone in their genomes, 3 of 14 Z3 F1 mice showed either absence of PCR amplification when NonBF/R primers were used (Figure 1, B, middle panel) or heterogeneity of PCR products when AmpF-NonBR primers were used (Figure 1, B, top panel). Similarly, 4 of 19 H10 F1 mice showed multiple PCR products of both normal and abnormal sizes, indicating that multiple mutations occurred in different copies (Figure 1, C, top panel). In contrast, we did not detect any variation in the sizes of the PCR products that were generated from the 63 control F1 mice. The mutation frequencies in mice carrying H-DNA and Z-DNA sequences were statistically significantly higher than those in control mice, as assessed by Fisher exact test (7.7%, 95% CI = 3.7% to 12.8% and 6.6%, 95% CI = 3.8% to 11.6%, respectively, vs 0%; P = .042 and P = .035; Table 1), suggesting that H-DNA and Z-DNA structures are unstable in mouse chromosomes.
The H-DNA– and Z-DNA–forming sequences induced a high level of genetic instability compared with the control sequence in this mouse model. It is particularly notable that the instability events (large-scale deletions and/or rearrangements) occurred in the mouse tail tissue (ie, devoid of immune cells). Double-strand breaks in immunoglobulin genes or T-cell receptor genes can result from RAG recombinase activity during V(D)J recombination, or the deamination of cytidines, which is catalyzed by activation-induced cytidine deaminase (AID) in class switch recombination, and these proteins might also contribute to the double-strand breaks in translocated oncogenes, such as c-MYC and BLC-2 (8,9). However, the data presented here provide direct evidence that non-B DNA-induced genetic instability can occur in V(D)J and class switch recombination-deficient tissues.
The fact that we could easily detect instability at regions capable of adopting H-DNA or Z-DNA structures in integrated reporter vectors suggests that chromosome breakage and translocation occur more frequently than previously thought (10). Perhaps the majority of chromosome breakage events go undetected because they do not result in noticeable phenotypes, or when instability occurs in critical genes, it may result in cytotoxicity such that those cells are eliminated from the population (11). Because the dual reporter vector used in this study does not contain a gene that has an important cellular function in mammalian cells, the deletion or translocation of this fragment per se will not result in an altered phenotype or in a growth advantage or disadvantage; thus, it enabled the sensitive detection of deletion and translocation events.
We noticed that the different strains of H-DNA or Z-DNA F1 mice exhibited different levels of instability, suggesting that context sequences and/or the local chromosome metabolism at the integration sites of the reporter vectors might also be important for the observed instability events. Further experiments to map the integration sites and to sequence the break points could provide useful information on the mechanisms involved in the DNA structure-induced genetic instability.
In summary, we have constructed a novel transgenic mouse model to detect DNA structure-induced chromosome breakage, deletion, and rearrangement events from sequences that map to break points in leukemia and lymphoma patients. This model should prove useful to further our understanding of the mechanisms that are involved in cancer etiology. These data provide the first evidence that H-DNA– and Z-DNA–forming sequences can lead to genetic instability in a chromosomal context in living organisms. In addition, these results suggest that helical distortions can result in spontaneous chromosome breakage and the translocation of genes involved in human diseases, including cancer.
Funding
National Institutes of Health/National Cancer Institute grant (CA093729 to K.M.V.); a National Institute of Environmental Health Sciences grant (ES015707 to K.M.V.) and a center grant for core services (ES007784); and an Odyssey Fellowship (to G.W.) from University of Texas M. D. Anderson Cancer Center.
Supplementary Material
Footnotes
We thank Ms Sarah Henninger for manuscript preparation and Ms Xiuixa Wu, Dr Howard Thames, and Mr Kevin Lin for assistance with statistical analyses.
The sponsors had no role in the design of the study, data collection and analysis, interpretation of the results, the preparation of the manuscript, or the decision to submit the manuscript for publication.
References
- 1.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372(6502):143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5(9–10):1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Bacolla A, Jaworski A, Larson JE, et al. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc Natl Acad Sci USA. 2004;101(39):14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst) 2006;5(9–10):1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 5.Wang G, Vasquez KM. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc Natl Acad Sci USA. 2004;101(37):13448–13453. doi: 10.1073/pnas.0405116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc Natl Acad Sci USA. 2006;103(8):2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boerrigter ME, Dolle ME, Martus HJ, Gossen JA, Vijg J. Plasmid-based transgenic mouse model for studying in vivo mutations. Nature. 1995;377(6550):657–659. doi: 10.1038/377657a0. [DOI] [PubMed] [Google Scholar]
- 8.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428(6978):88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 9.Duquette ML, Pham P, Goodman MF, Maizels N. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24(38):5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 10.Kato T, Inagaki H, Yamada K, et al. Genetic variation affects de novo translocation frequency. Science. 2006;311(5763):971. doi: 10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Libura J, Slater DJ, Felix CA, Richardson C. Therapy-related acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion. Blood. 2005;105(5):2124–2131. doi: 10.1182/blood-2004-07-2683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.