Abstract
Background
Glucocorticoids are used in the curative treatment of acute lymphoblastic leukemia (ALL). Resistance to glucocorticoids is an important adverse prognostic factor in newly diagnosed ALL patients but its mechanism is unknown. Because SWI/SNF complex–mediated chromatin remodeling is required for glucocorticoid transcriptional activity in vitro, we investigated whether expression of subunits of the SWI/SNF complex was related to glucocorticoid resistance in ALL.
Methods
Gene expression and in vitro sensitivity to prednisolone and dexamethasone were assessed in a training set of primary ALL cells from 177 children with newly diagnosed ALL and a validation set of cells from an independent cohort of 95 ALL patients. The global test method was used to select pathways whose genes were associated with drug sensitivity. Genes involved in chromatin remodeling were identified by use of the Gene Ontology database. Short hairpin RNA (shRNA) was used to knock down mRNA expression of SMARCA4 in glucocorticoid-sensitive Jurkat human ALL cells. Spearman rank correlation, multiple linear regression, and logistic regression were used to investigate associations between gene expression and glucocorticoid sensitivity. All statistical tests were two-sided.
Results
Statistically significant associations between decreased expression in ALL cells of genes for core subunits of the SWI/SNF complex—SMARCA4, ARID1A, and SMARCB1—and resistance to prednisolone and dexamethasone were identified in the training cohort. In the validation cohort, expression of SMARCA4 (P < .001 and r = −0.43), ARID1A (P = .016 and r = −0.29), and SMARCB1 (P = .019 and r = −0.29) in ALL cells was statistically significantly associated with dexamethasone sensitivity, and SMARCA4 expression (P = .018 and r = −0.28) was statistically significantly associated with prednisolone sensitivity. Prednisolone resistance was higher in SMARCA4 shRNA-transfected Jurkat cells (drug concentration lethal to 50% of the leukemia cells [LC50] = 277 μM) than in control shRNA-transfected cells (LC50 = 174 μM, difference = 103 μM, 95% confidence interval of the difference = 100 to 106 μM; P < .001, t test).
Conclusion
Decreased expression of as many as three subunits of the SWI/SNF complex appears to be associated with glucocorticoid resistance in primary ALL cells.
CONTEXT AND CAVEATS
Prior knowledge
Glucocorticoids are a curative treatment for acute lymphoblastic leukemia (ALL), and patients who develop resistance to glucocorticoid treatment have poor prognosis. SWI/SNF complex–mediated chromatin remodeling is required for glucocorticoid transcriptional activity in vitro.
Study design
Associations between expression of genes encoding components of the SWI/SNF complex and sensitivity to prednisolone and dexamethasone were studied in ALL cells from a training cohort and an independent validation cohort of children with newly diagnosed ALL. RNA interference was used to reduce mRNA expression of SMARCA4 in glucocorticoid-sensitive Jurkat human ALL cells.
Contribution
ALL cells from every patient studied expressed only SMARCA4 as the catalytic subunit of the SWI/SNF complex; none expressed SMARCA2, the alternate catalytic component. Expression of genes for three components of the SWI/SNF complex (SMARCA4, ARID1A, and SMARCB1) in ALL cells was statistically significantly associated with dexamethasone sensitivity, and expression of one component (SMARCA4) was statistically significantly associated with prednisolone sensitivity. Decreased SMARCA4 expression in Jurkat cells was associated with prednisolone resistance.
Implications
Future studies should further investigate the association between subunits of the SWI/SNF complex and glucocorticoid resistance in ALL patients.
Limitations
Glucocorticoid resistance is probably a multigenic phenomenon and may involve additional genes not investigated in this study. The sample size of ALL patients was relatively small, especially of the St. Jude validation cohort.
From the Editors
Synthetic glucocorticoids, such as dexamethasone and prednisolone, are widely used anti-inflammatory, antiproliferative, and immunomodulatory medications (1). In lymphoid malignancies, especially childhood acute lymphoblastic leukemia (ALL), glucocorticoids are an essential element of curative combination chemotherapy regimens (2,3). Glucocorticoids have clinically significant antileukemic activity in part because of their relatively specific cytotoxicity for cells in the lymphoid lineage (4,5). Also, glucocorticoids have relatively few acute side effects compared with other antileukemic agents (5). It is also important that, among patients with childhood ALL, the initial response to glucocorticoid treatment is a statistically significant predictor of long-term treatment outcome (6,7) and that in vitro cellular glucocorticoid resistance, as determined in primary ALL cells from ALL patients, identifies patients who are at high risk for a poor response to treatment (8,9).
Glucocorticoids enter cells by passive diffusion and bind to the intracellular glucocorticoid receptor, which is the product of the NR3C1 gene and which is a ubiquitously expressed member of the nuclear receptor superfamily (10). After a glucocorticoid binds to the NR3C1 glucocorticoid receptor, conformational changes in the receptor expose nuclear localization signal protein domains, so that the glucocorticoid–NR3C1 complex can be translocated to the nucleus. Once in the nucleus, the complex regulates the transactivation (via binding to glucocorticoid response elements located in the promoter regions of target genes) and transrepression (via interactions with transcription factors such as AP-1) of glucocorticoid-responsive genes (10). The transactivation and/or transrepression of downstream target genes then induce apoptosis in glucocorticoid-sensitive cells (5).
Although prognosis is poor for ALL patients who are resistant to glucocorticoid treatment under contemporary protocols (6–9,11), mechanisms underlying glucocorticoid resistance remain poorly understood. Studies of glucocorticoid resistance (5,12,13) have focused on mechanisms upstream of the binding of glucocorticoid to its receptor, including decreased extracellular or intracellular glucocorticoid concentrations or NR3C1 mutations, splice variants, or insufficient expression. These studies also reported resistance mechanisms downstream of the binding of glucocorticoid to its receptor that involve the glucocorticoid signaling pathway and result in the inhibition of apoptosis or the inappropriate activation of survival signals.
Using a genome-wide approach to assess gene expression in ALL cells that were isolated from patients with newly diagnosed childhood ALL, we have previously identified (14) 33 genes that were differentially expressed in prednisolone-sensitive cells compared with prednisolone-resistant cells. These 33 genes provided new insights into mechanisms of cellular resistance to prednisolone and revealed potentially new targets that were involved in this process. Specifically, expression of SMARCB1 (also known as SNF5, INI1, or BAF47), which encodes a core subunit of the SWI/SNF chromatin-remodeling complex, was strongly associated with prednisolone resistance. That is, lower SMARCB1 gene expression was associated with prednisolone-resistant ALL and higher SMARCB1 gene expression was associated with prednisolone-sensitive ALL (14,15).
Condensed chromatin structure inhibits gene transcription by blocking access of the transcriptional machinery to the DNA and by blocking the interaction between gene-specific transcription factors and their DNA recognition sequences (16). Transcriptional activation and efficient transcription of genes require dynamic structural changes in chromatin, and the ATP-dependent SWI/SNF complex is involved in chromatin restructuring, especially that associated with steroid hormone-activated transcription (16,17).
The SWI/SNF complex is a highly conserved multisubunit complex composed of nine to 12 subunits (18,19). In humans, the SWI/SNF complex contains either SMARCA4 protein (also known as BRG1) or SMARCA2 protein (also known as BRM) as the catalytic ATPase subunit. This subunit is bound to a multiprotein complex containing BAF proteins (ie, BRG1- or BRM-associated factors) and products of the following genes: SMARCC1 (also known as BAF155), SMARCC2 (also known as BAF170), SMARCD1-3 (also known as BAF60a-c), SMARCE1 (also known as BAF57), ACTL6A-B (also known as BAF53a-b), and SMARCB1 (also known as BAF47) (18,19). In vitro studies in yeast and in mammalian cells have shown that the SWI/SNF complex is necessary for glucocorticoid-dependent transcription because of its chromatin-remodeling and nucleosome-disruption activities (20–22) and that glucocorticoid activation of the promoter of the mouse mammary tumor virus requires SWI/SNF complexes that contain SMARCA4 protein as the catalytic subunit (23,24).
From these results, we hypothesized that decreased expression of not only SMARCB1 protein but also other subunits of the SWI/SNF complex is related to glucocorticoid resistance in ALL. To assess whether expression of genes for subunits of the SWI/SNF complex differs between primary ALL cells that are resistant to glucocorticoids and those that are sensitive to glucocorticoids, we measured glucocorticoid sensitivity and gene expression in primary ALL cells from a discovery cohort of ALL patients and from an independent validation cohort of ALL patients.
Patients and Methods
Patients With ALL
Bone marrow and peripheral blood samples were obtained at diagnosis from patients with childhood ALL who enrolled in the German Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia protocol COALL-92/97 (between August 1, 1992, and August 1, 2003) or the ALL-IX Dutch Childhood Oncology Group (DCOG) protocol at the Erasmus MC/Sophia Children's Hospital (between January 1997 and October 2004) (14,25) (hereafter referred to as the COALL-DCOG training or discovery cohort). The training cohort, thus, included the 177 consecutive ALL patients who had sufficient ALL cells for successful gene expression analysis and in vitro drug sensitivity testing (Table 1), as described previously (14). The independent validation cohort included 95 patients with childhood ALL who were enrolled in the St. Jude Children's Research Hospital protocol, Total Therapy 15 (Table 1) (ie, the St. Jude validation cohort). Prednisolone sensitivity was measured in 71 of these 95 patients, and dexamethasone sensitivity was measured in 66 patients (with 42 of the 95 patients being tested for both drugs) (26). We similarly collected data on consecutive patients with sufficient ALL cells for successful gene expression analysis and in vitro drug sensitivity testing in the St. Jude validation cohort. Enrollment in the St. Jude Total Therapy 15 trial started on June 29, 2000, and we routinely measured in vitro sensitivity after March 1, 2002. The gene expression analysis of these patients has not been published previously. Patients were enrolled in either of these studies if they had a diagnosis of ALL, were younger than 18 years of age, and had not been previously treated for ALL. The investigation was approved either by the institutional review boards at St. Jude Children's Research Hospital or by the central study office of the COALL and at Sophia Children's Hospital. Signed informed consent was obtained from parents or legal guardians before enrollment of all patients participating in these studies.
Table 1.
Characteristics of patients in the German Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia COALL-92/97 and the ALL-IX Dutch Childhood Oncology Group discovery cohort and in the St. Jude validation cohort*
| No. of patients |
|||
| Variable | COALL-DCOG (n = 177) | St. Jude (n = 95) | P value† |
| Age, y | .13 | ||
| 1–10 | 130 | 78 | |
| >10 | 47 | 17 | |
| Subtype‡ | .97 | ||
| B-lineage other | 47 | 22 | |
| BCR-ABL | 5 | 3 | |
| E2A-PBX1 | 8 | 5 | |
| MLL-AF4 | 3 | 1 | |
| TEL-AML1 | 44 | 22 | |
| Hyperdiploid | 43 | 28 | |
| T cell | 28 | 14 | |
| WBCs, No. × 109 per liter | .66 | ||
| <10 | 42 | 28 | |
| 10–49 | 67 | 36 | |
| 50–100 | 28 | 15 | |
| >100 | 39 | 16 | |
COALL-DCOG = German Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia COALL-92/97 and the ALL-IX Dutch Childhood Oncology Group (the discovery cohort); St. Jude = validation cohort; WBCs = white blood cells.
Fisher exact test. All statistical tests were two-sided.
The BCR-ABL gene fusion is a translocation of parts of chromosomes 9 and 22, t(9;22), and involves the gene break point cluster region protein and Abelson murine leukemia viral (v-abl) oncogene homolog 1.The t(1;19) translocation creates E2A-PBX1 fusion gene and involves pre-B-cell leukemia homeobox 1 and E2A immunoglobulin enhancer-binding factor E12/E47. The t(4;11) translocation creates the MLL-AF4 fusion gene involving the mixed-lineage leukemia and the AF4–FMR2 family member 1 gene. The t(12;21) translocation creates the TEL-AML1 fusion gene, involving the ets variant gene 6 (TEL oncogene) and the runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene). Hyperdiploid cells were identified by cytogenetics as having more than 51 chromosomes.
Isolation of Leukemia Cells
Mononuclear cells were isolated from bone marrow or peripheral blood by sucrose density gradient centrifugation (sucrose density = 1.077 g/mL; Lymphoprep, Nycomed Pharma, Oslo, Norway) within 24 hours of collection. For in vitro sensitivity testing, cells were resuspended in modified RPMI-1640 medium supplemented with 20% fetal calf serum (Integro, Zaandam, the Netherlands), 2 mM L-glutamine, gentamycin (Gibco BRL, Breda, the Netherlands; 200 μg/mL), penicillin (100 IU/mL), streptomycin (100 μg/mL), and Fungizone (Gibco BRL, Rockville, MD; 0.125 μg/mL), as well as ITS medium supplement containing insulin (5 μg/mL), transferrin (5 μg/mL), and sodium selenite (5 ng/mL) (Sigma-Aldrich Chemie B.V., Zwijndrecht, the Netherlands), as described previously (14). If necessary, cells purified by density gradient centrifugation were further enriched to a cell population that contained more than 90% blasts by removing nonmalignant cells with immunomagnetic beads (Dynal, Breda, the Netherlands). To remove normal T lymphocytes, we used 10-fold more anti-CD3 immunomagnetic beads than the number of normal T lymphocytes. These normal T lymphocytes were depleted according to the manufacturer's recommendation. To remove other contaminating normal cells, we used a mixture of mouse anti-human monoclonal antibodies, including anti-CD3 for T lymphocytes, IgM for B lymphocytes, anti-CD13 for immature myeloid cells, anti-CD14 for monocytes, anti-CD15 for granulocytes and bands (ie, less mature neutrophils), anti-CD13–15 for metamyelocytes, anti-CD33 for myelocytes, and anti-H1 for normocytes and erythroblasts. Briefly, we added 2 μL of undiluted antibody per 100 μL containing 10 × 106 cells, incubated the mixture for 30 minutes at 37°C in a water bath, washed the cells three times with culture medium, and centrifuged the mixture at 300 g for 5 minutes at 4°C. Subsequently, we incubated the cell–antibody mixture with sheep anti-mouse immunobeads, in accordance with the manufacturer's recommendation. Enrichment was checked by microscopy in a cytospin preparation to determine the fraction of leukemic cells by use of Wright–Giemsa staining (product 23-036507, Fisher Scientific, Landsmeer, the Netherlands). For the St. Jude validation cohort, the procedures were the same except that no antibiotics were used in the culture medium.
Drug-Resistance Assay and Determination of the Drug Concentration Lethal to 50% of the Leukemia Cells (LC50 Value)
Prednisolone sensitivity was determined for leukemia cells isolated from all 177 patients in the training set. Asparaginase and vincristine sensitivity was determined for leukemia cells isolated from 176 of the 177 patients. Sensitivity to prednsiolone (Bufa Pharmaceutical Products, Uitgeest, the Netherlands), vincristine (TEVA Pharma, Mijdrecht, the Netherlands), and asparaginase (Paronal, Christiaens, Breda, the Netherlands) was determined in tumor cells from patients in the training set by use of a 4-day in vitro 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide (MTT) drug resistance assay, as described elsewhere (14). Tumor cells were cultured in a 96-well microculture plate with the indicated drugs, and viable cells reduced MTT (yellow) to formazan (purple). Formazan crystals were dissolved in acidified isopropanol and the concentration of formazan was determined spectrophotometrically. At each drug concentration, leukemic cell survival was calculated. The LC50 value (as detailed below) was then determined from a dose–response curve and used as the most reliable estimate for comparing drug resistance or sensitivity, respectively, across the population of patients.
Prednisolone and dexamethasone sensitivities of the leukemia cells that were isolated from 95 patients in the St. Jude validation cohort were determined by use of bone marrow aspirates that were prepared as described above, except that no antibiotics were used in the culture medium. Dexamethasone was from the American Reagent Laboratories, Inc. (Shirley, NY) and prednisolone (Solu-Medrol) was from Pharmacia & Upjohn Company (New York, NY). LC50 values were determined as for the training set.
The LC50 value was determined by first fitting the following model to the data:
where Ymax is the maximum viability, KM is the half saturation parameter, Ymin is the minimum viability, n is the Hill coefficient (which regulates the steepness of the viability curve), and C is the concentration of the drug. From this equation, we estimated the LC50 by solving for the concentration, C, at which the effect was 50%. For this estimation, we used the nonlinear parameter estimation maximum likelihood method, which is available in ADAPT II (27). The variability was estimated for the LC50 and its 95% confidence interval (CI) (by assuming that a normally distributed error was made by the maximum likelihood method). These measures of variability were also used to determine the statistical significance of the difference between the SMARCA4 knockdown and control cell LC50 values for prednisolone and asparaginase (27).
To validate the prednisolone sensitivity phenotype from the training set, we used the LC50 values for two different glucocorticoids, prednisolone or dexamethasone, in the validation set. From the distribution of the LC50 data in the training and validation sets, we defined those with an LC50 value for prednisolone of more than 150 μg/mL as being resistant to prednisolone and those with an LC50 value of less than 0.1 μg/mL as sensitive to prednisolone. These definitions were from previous reports (14,28–30) in which these criteria were associated with treatment outcome. We defined the patients with an LC50 value for dexamethasone of more than 5.8 μg/mL as being resistant to dexamethasone and those with an LC50 value of less than 0.02 μg/mL as being sensitive to dexamethasone. We chose these cutoffs so that, for both glucocorticoids, 20% of the population was defined as glucocorticoid resistant and 33% was defined as being glucocorticoid sensitive. These percentages are exactly the same as those in the training set (20% and 33%, respectively).
Gene Expression
Total RNA was extracted from mononuclear cells isolated from every ALL patient in both cohorts (which contained >85% leukemia cells) with Trizol (Gibco BRL) according to the manufacturer's recommendation. High-quality total RNA (5 μg) was used to determine the gene expression profile of leukemia cells from each patient at diagnosis. Generation of cDNA and complementary RNA from the total RNA and hybridization of the complementary RNA to the U133A GeneChip oligonucleotide microarray (Affymetrix, Santa Clara, CA) were performed in accordance with to the manufacturer's protocol. This microarray contains 22 215 gene probe sets, representing approximately 12 357 human genes, plus approximately 3820 expressed sequence tag clones with unknown function. Default settings of GCOS software version 1.2 (Affymetrix) were used to calculate scaled gene expression values.
Bioinformatics
A total of 224 candidate gene probe sets related to the SWI/SNF complex (representing 123 unique genes and six cDNA clones) were selected for the analysis. Among these probe sets, 204 gene probe sets (representing 119 unique genes and six cDNA clones) were found in the Gene Ontology database entry “chromatin modification,” and 46 gene probe sets (representing 16 unique genes and one cDNA clone) were related to the SWI/SNF complex. An overlap of 26 gene probe sets was found in both categories. The Gene Ontology database (located at http://www.geneontology.org/) uses predefined and authorized terms to organize information about gene products in terms of their association with three categories (ie, Biological Process, Cellular Component, and Molecular Function) (31). Expression signals were log transformed to normalize their distribution. The candidate probe sets were defined as expressed if they were absent by the Affymetrix Call algorithm in fewer than 5% of the 177 cell samples obtained from the 177 patients in the training cohort or if the probe sets had an average scaled expression value of greater than 10.5.
To assess any association between the expression of a gene and sensitivity to antileukemic agents, we used two public global pathway databases, Gene Ontology (see above) and GenMAPP (located at http://www.genmapp.org/). The GenMAPP database is a free computer application that maps genomic data from groups of biologically related genes and maintains a collection of well-curated metabolic and signaling pathways.
Pathways listed in the GenMAPP database (n = 311 entries) and in the Biological Process category of Gene Ontology database (n = 119 entries) were tested for associations with prednisolone, asparaginase, and vincristine sensitivity by use of LC50 data and the global test method (32), which was implemented with the R Bioconductor package (33). This test was used to infer overrepresentation of specific biological pathways by comparing the prediction accuracy of the phenotype by the use of the expression profile of genes in the pathway that were not associated with the phenotype. The “geneplot” function was used to plot the association between selected genes and default parameters.
Three of the 119 Biological Process categories in the Gene Ontology database or three of the 311 GenMAPP entries included the word chromatin (ie, “chromatin assembly,” “chromatin assembly or disassembly,” and ”chromatin modification”). Because the chromatin-related pathways in both databases had the same names and contained most of the same genes, we selected the Gene Ontology database for this analysis. The Gene Ontology Biological Process and GenMAPP databases contained nine proteins encoded by the SWI/SNF chromatin-remodeling genes, including ARID1A, SMARCB1, SMARCC1, SMARCC2, SMARCA1, SMARCA2, SMARCA5, SMARCD1, and SMARCD2, but were both missing SMARCA4. The “Cellular Component” entry “SWI/SNF complex” (accession number in Gene Ontology = GO:0016514) in the Gene Ontology database contained 11 gene products and most of the crucial core subunits (Supplementary Table 1, available online) of the SWI/SNF complex, including ACTL6B, ARID1A, ARID1B, NCTR1, SMARCB1, SMARCC1, SMARCC2, SMARCA2, SMARCA4, SMARCD2, and SMARCD3 (34). The core subunits form the minimal basic structure of the SWI/SNF protein complex. This entry in the Gene Ontology database also included SMARCA4. These 11 gene products in the Gene Ontology database corresponded to 27 gene probe sets on the U133A GeneChip.
Cell Culture, Lentiviral Transduction, and RNA Interference
Jurkat human ALL cells (American Type Culture Collection, Rockville, MD) were maintained in RPMI-1640 medium (BioWhittaker, Walkersville, MD), supplemented with 10% fetal bovine serum (HyClone, Logan, UT) and 1% L-glutamine (200 mM, BioWhittaker). The cells were cultured at 37°C in an atmosphere of 5% CO2 and air with a humidity of 95%.
Knockdown of SMARCA4 protein expression was achieved by use of RNA interference with a lentiviral vector-based short hairpin RNA (shRNA) approach developed by The RNAi Consortium and distributed commercially as the MISSION TRC-Hs 1.0 library (Sigma, St Louis, MO). We used lentiviral particles corresponding to the MISSION shRNA SHVRS-NM_003072 target set (including five different shRNA constructs) and the MISSION Non-Target shRNA control. We transduced all five shRNA constructs and determined the knockdown efficiency by western blot analysis. Briefly, for the RNA interference experiments, 1 × 104 Jurkat cells were transduced with lentiviruses carrying shRNA at a multiplicity of infection of 20 in wells of a 96-well plate (Fisher Scientific) containing RPMI-1640 medium. The wells had been precoated with retronectin at the concentration recommended by the manufacturer to enhance virus-mediated gene transduction. Three days after transfection, cells were transferred into medium containing puromycin (Sigma; 2.5 μg/mL) to select transduced cells and maintained under selective pressure at this concentration of puromycin. MTT drug resistance assays and western blot analysis were performed within a week after selection. MTT assays were performed in cells transduced with the most efficient shRNA (clone NM_003072.2-5350s1c1); the most efficient shRNA was defined as the shRNA that resulted in the lowest expression of SMARCA4 protein as assessed by western blot analysis.
Western Blot Analysis
We homogenized 200 000 Jurkat cells that had been transduced with SMARCA4 shRNA or control shRNA in microsome storage buffer (100 mM potassium phosphate at pH 7.4, 1 mM EDTA, and 20% glycerol) containing a protease inhibitor mixture (Roche, Indianapolis, IN) and subjected the homogenate to further disruption by sonication (Cole Palmer Ultrasonic Homogenizer 4710 Series, Cole Palmer Instruments, Chicago, IL; with the output control at a setting of 20 for 10 seconds). Protein concentrations of the lysates were measured with the Bio-Rad Protein Assay (Bio-Rad Laboratory, Hercules, CA). Proteins (25 μg per lane) were separated in a 3%–8% gel by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (NuPAGE Tris-acetate gel, Invitrogen, Carlsbad, CA) and then electrotransferred to nitrocellulose membranes (Invitrogen). Filters were blocked for 2 hours in Tris-buffered saline (50 mM Tris–HCl at pH 7.4 and 150 mM NaCl) containing 1% Tween-20 (TBST) and 5% nonfat dry milk and then incubated with the primary antibody (goat anti-SMARCA4, diluted 1:200, or anti-glyceraldehyde-3-phosphate dehydrogenase [anti-GAPDH], diluted 1:500; both from Santa Cruz Biotechnology Inc., Santa Cruz, CA) in TBST containing 5% nonfat dry milk overnight at 4°C on a shaker. Filters were washed for two 15-minute periods in TBST and then incubated in TBST containing 5% nonfat dry milk for 1 hour with horseradish peroxidase–conjugated anti-goat IgG secondary antibody (diluted 1:5000; Santa Cruz Biotechnology). After three washes as described above, filters were activated by use of the enhanced chemiluminescence western blotting detection reagent as described by the manufacturer (ECL Plus, Amersham Biosciences, Piscataway, NJ) and exposed to autoradiographic film (Hyperfilm ECL, Amersham Biosciences). The gels were also scanned with the Storm 860 PhosphoImager (Molecular Dynamics, Sunnyvale, CA) and analyzed by the ImageQuant software.
Statistical Analysis
Highly correlated probe sets (defined as those with a Pearson correlation coefficient r of >0.6) from the same gene were considered to be measuring a single transcript and were combined into one expression score variable that was represented by the first component of the principal component analysis of the standardized microarray signals of each probe set. The Spearman rank correlation test was used to investigate the relationship between drug resistance and the expression level of each transcript selected. Differences in gene expression were evaluated by use of the Kruskal–Wallis test.
The relationship between the in vitro prednisolone sensitivity of 177 patients and the expression of ARID1A, SMARCA4, SMARCB1, and SMARCC2 genes (as mRNAs) was investigated by use of a multiple linear regression model. We also included expression of the glucocorticoid receptor gene NR3C1 because the SWI/SNF chromatin-remodeling complex is recruited for the transcriptional regulation of glucocorticoid-responsive genes by a complex containing glucocorticoid, NR3C1 glucocorticoid receptor, and DNA (18,19). We used logistic regression to determine whether this model could predict which of the 177 patients in the COALL-DCOG training cohort were resistant to prednisolone and which were sensitive. We built the logistic regression model with the discovery data from the COALL-DCOG training cohort and predicted the glucocorticoid sensitivity of patients in the validation cohort on the basis of their gene expression. We then cross-tabulated the predicted sensitivity and the observed drug sensitivity by use of the aforementioned cut points for groups that were sensitive, intermediate, or resistant to a glucocorticoid. For prednisolone, sensitive was defined as an LC50 of 0.1 μg/mL or less, intermediate was defined as an LC50 of more than 0.1 μg/mL but less than 150 μg/mL, and resistant was defined as an LC50 of 150 μg/mL or more. For dexamethasone, sensitive was defined as an LC50 of 0.02 μg/mL or less, intermediate was defined as an LC50 of more than 0.02 μg/mL but less than 5.8 μg/mL, and resistant was defined as an LC50 of 5.8 μg/mL or more. Dexamethasone is a more potent glucocorticoid than prednisolone and, therefore, its LC50 concentration ranges were lower than those for prednisolone. Additionally, sensitivity and resistance to dexamethasone were defined as having the same distribution as prednisolone resistance in the discovery set.
A power calculation was not done. We used the t test to compare the LC50 values for prednisolone and asparaginase of SMARCA4 knockdown vs control cells. All statistical analyses were performed with R version 2.6.1 software. All statistical tests were two-sided.
Results
Prednisolone Sensitivity and Biological Pathway Analysis
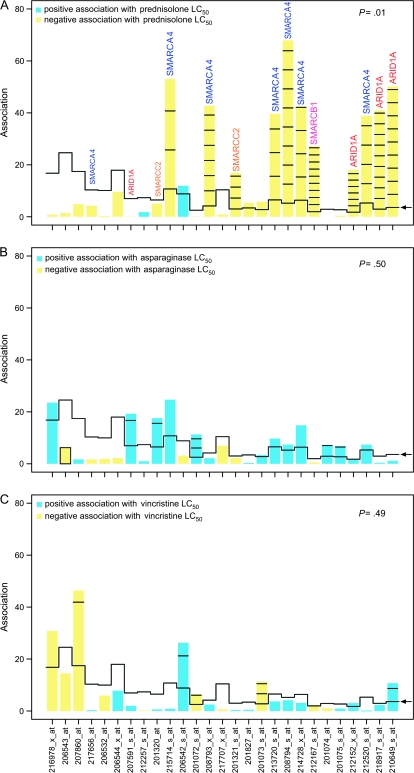
This investigation was undertaken to determine whether the expression of chromatin-remodeling genes differed between primary ALL cells that are resistant to glucocorticoids and those that were sensitive. We measured glucocorticoid sensitivity in primary ALL cells from 177 newly diagnosed patients with ALL and used microarrays to assess differences in expression of genes encoding proteins that are related to chromatin remodeling in sensitive and resistant leukemia cells. Using the Gene Ontology Biological Process and GenMapp databases and the global test method, we found a statistically significant association between prednisolone sensitivity in primary ALL cells and the expression of genes for various subunits in specific biological pathways that are related to chromatin regulation. Although the Gene Ontology Biological Process entries did not include the SMARCA4 protein, the Gene Ontology Cellular Component entry “SWI/SNF complex” did include SMARCA4 protein, along with another eight proteins that can be found in the SWI/SNF complex, including ACTL6B, ARID1A, SMARCA2, SMARCB1, SMARCC1, SMARCC2, SMARCD2, and SMARCD3. The expression of these genes was statistically significantly associated with prednisolone sensitivity (P = .010, global test) but not with sensitivity to the mechanistically distinct antileukemic agents asparaginase or vincristine (Figure 1).
Figure 1.
Associations of each gene in the Gene Ontology accession set for the SWI/SNF complex with sensitivity of primary acute lymphoblastic leukemia cells to antileukemic drugs. Drug sensitivity was measured in vitro as the drug concentration lethal to 50% of the leukemia cells (LC50). A) Prednisolone. B) Asparaginase. C) Vincristine. For each gene probe set (x-axis) in this accession set in each panel, a bar and reference line (which gives the expected height of the bar for each probe set under the null hypothesis that the gene was not statistically associated with drug sensitivity) are shown. The height of the bars indicates the strength of the association of each probe set with sensitivity to the antileukemic drug. If the height of the bar exceeds the reference line, the probe set was statistically significantly related to drug sensitivity. The number of standard deviations by which the bar exceeds the reference line is shown by horizontal lines within the bars. In panel A, for example, the SD values for SMARCA4 are 2.82, 6.62, 3.64, 8.55, 4.05, and 4.51; for ARID1A are 6.59, 9.26, 9.25; for SMARCB1 are 9.20; and for SMARCC2 are 3.30. The SDs were determined for each gene probe set; multiple gene probe sets may represent the same gene. Blue bars = positive association with drug sensitivity; yellow = negative association with drug sensitivity; arrow = reference line.
Further, the statistically significant association between this pathway and prednisolone sensitivity was explained by the expression of only four genes in the SWI/SNF complex—ARID1A, SMARCA4, SMARCB1, and SMARCC2. That is, the expression of ARID1A, SMARCA4, and SMARCB1 was most strongly associated with prednisolone sensitivity, with the expression of each gene exceeding that of the reference value expected under the null hypothesis by 6–9 SDs. Expression of SMARCC2 was less strongly associated with prednisolone sensitivity, with its expression exceeding the reference by more than 3 SDs (Figure 1, A).
Expression of SWI/SNF-Related Genes in ALL Cells
To determine which SWI/SNF-related genes are expressed in ALL cells, we used the expression values of 43 probe sets that corresponded to 17 SWI/SNF-related genes in the COALL-DCOG cohort. Transcripts of 11 genes whose products can be found in the SWI/SNF complex (ie, ARID1A, HLTF, SMARCA4, SMARCA5, SMARCB1, SMARCC1, SMARCC2, SMARCD1, SMARCD2, SMARCE1, and PB1) were expressed in primary ALL cells (Supplementary Table 1, available online). We found no evidence that SMARCA2 was expressed in leukemia cells. Therefore, because the catalytic subunit of SWI/SNF chromatin-remodeling complexes can be either SMARCA4 or SMARCA2 protein, the ALL cells examined in our study must contain SWI/SNF complexes with only the SMARCA4 protein.
Expression of Gene Products in the SWI/SNF Complex and Drug Sensitivity
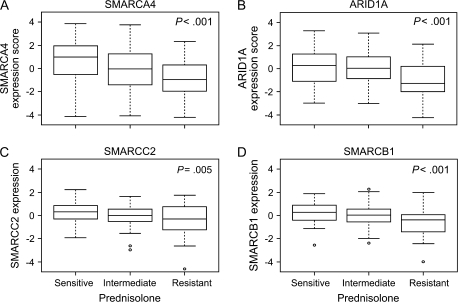
By use of data from the COALL-DCOG discovery cohort and the Spearman rank correlation test, we assessed whether the expression of genes whose products can bind to the SWI/SNF complex was statistically significantly associated with the sensitivity of primary ALL cells to prednisolone. We found that increased expression of SMARCA4 (P < .001 and r = −0.28), ARID1A (P = .002 and r = −0.23), SMARCB1 (P < .001 and r = −0.28), and SMARCC2 (P = .016 and r = −0.18) mRNAs was statistically significantly associated with prednisolone sensitivity, as measured by the LC50 for prednisolone (Supplementary Table 2, available online). No association was found between expression of these genes and sensitivity to either asparaginase or vincristine (data not shown). The expression of SMARCA4, ARID1A, SMARCC2, and SMARCB1 was investigated in primary ALL cells from subsets of the 177 patients with ALL who were sensitive to prednisolone (n = 58), neither sensitive nor resistant to prednisolone (intermediate; n = 83), or resistant to prednisolone (n = 36). Prednisolone-resistant cells had statistically significantly lower expression of all four genes than cells from the other subgroups, as shown by linear regression analysis (for SMARCA4, estimate = −0.71, 95% CI = −1.07 to –0.34, P < .001; for ARID1A, estimate = –0.56, 95% CI = –0.86 to –0.26, P < .001; for SMARCC2, estimate = –0.30, 95% CI = –0.51 to –0.1, P = .005; and for SMARCB1, estimate = –0.42, 95% CI = –0.62 to –0.23, P < .001) (Figure 2).
Figure 2.
Expression of genes for the subunits of the SWI/SNF complex in acute lymphoblastic leukemia (ALL) cells from 177 ALL patients and prednisolone sensitivity. A) SMARCA4. B) ARID1A. C) SMARCC2. D) SMARCB1. Pednisolone sensitivity was measured in primary leukemia cells by use of the 4-day in vitro 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide drug resistance assay. Expression score is the median mRNA level or the median mRNA expression score if multiple gene probe sets exist for the same gene. Box plots show the medians (horizontal line), interquartile ranges (box), and ranges (whiskers) of the expression level or scores, excluding outliers, for prednisolone-sensitive, intermediate, and prednisolone-resistant ALL cells. Sensitive cells were defined as having a drug concentration lethal to 50% of cells (LC50) for prednisolone of 0.1 μg/mL or less, cells with intermediate sensitivity were defined as having an LC50 of more than 0.1 μg/mL but less than 150 μg/mL, and resistant cells were defined as having an LC50 of 150 μg/mL or more (14,27–29). Among the 177 ALL patients, 58 were prednisolone sensitive, 83 were intermediate, and 36 were prednisolone resistant. P values and estimates were determined by linear regression that compared gene expression with sensitivity. Estimates were as follows: for SMARCA4, –0.71 (95% confidence interval [CI] = –1.07 to –0.34); for ARID1A, –0.56 (95% CI = –0.86 to –0.26); for SMARCC2, –0.30 (95% CI = –0.51 to –0.1); and for SMARCB1, –0.42 (95% CI = –0.62 to –0.23). All statistical tests were two-sided. Circles = outliers.
The multiple linear regression model with the expression of all five genes (ARID1A, SMARCA4, SMARCB1, SMARCC2, and NR3C1) as covariates appeared to account for 22% of total variation in prednisolone sensitivity (P < .001, Supplementary Table 3, available online). We included expression of the NR3C1 glucocorticoid receptor gene in the model because, after the complex containing glucocorticoid, NR3C1 protein, and DNA is formed, the SWI/SNF chromatin-remodeling complex is recruited, via protein–protein interaction of ARID1A and SMARCD1 with NR3C1, for the transcriptional regulation of glucocorticoid-responsive genes (18,19). Furthermore, the expression of SMARCC2 and particularly of SMARCA4 was correlated with the expression of ARID1A and of SMARCB1 (Table 2), explaining why they were not independently statistically significant in the multiple linear regression model. By using expression of these five genes (ARID1A, SMARCA4, SMARCB1, SMARCC2, and NR3C1), we could correctly identify patients in the COALL-DCOG training cohort as being sensitive (n = 58) or resistant (n = 36) to prednisolone, with a 76% (95% CI = 65% to 85%) prediction accuracy (P < .001), a sensitivity of correctly identifying prednisolone-resistant patients of 72% (95% CI = 54% to 87%), and a specificity of 87% (95% CI = 75% to 94%, five- and three-gene model, Supplementary Table 4, available online). Seven patients in the training set who were sensitive and 15 who were resistant to prednisolone were misclassified by our model.
Table 2.
Correlations among mRNA expression of genes implicated in prednisolone sensitivity*
| Gene |
||||
| Gene | ARID1A | SMARCB1 | SMARCA4 | SMARCC2 |
| NR3C1 | –0.16 (.02) | –0.11 (.14) | –0.29 (<.001) | –0.23 (.002) |
| ARID1A | 0.29 (<.001) | 0.49 (<.001) | 0.28 (<.001) | |
| SMARCB1 | 0.46 (<.001) | 0.28 (<.001) | ||
| SMARCA4 | 0.32 (<.001) | |||
Estimates from the discovery cohort using a Pearson correlation test are reported, with P values in parentheses. All statistical tests were two-sided.
Finally, we used an independent validation cohort of 95 patients with childhood ALL who were treated according to the St. Jude Children's Research Hospital Total 15 protocol to validate these findings. Using leukemia cells from these 95 patients, we determined gene expression levels (ie, as mRNA) for the five genes in our model (ARID1A, SMARCA4, SMARCB1, SMARCC2, and NR3C1) and their sensitivity to prednisolone (n = 71 patients) and dexamethasone (n = 66 patients). Consistent with the findings with our model and data from the COALL-DCOG discovery cohort (Supplementary Table 2, available online), low gene expression of SMARCA4 (P = .018 and r = –0.28 for prednisolone, P < .001 and r = –0.43 for dexamethasone) was statistically significantly associated with resistance to prednisolone and to dexamethasone in primary ALL cells from patients in the St. Jude validation cohort. With ARID1A (P = .2 and r = –0.15 for prednisolone; and P = .016 and r = –0.29 for dexamethasone) and SMARCB1 (P = .29 and r = –0.13 for prednisolone; and P = .019 and r = –0.29 for dexamethasone), we found a statistically significant association only for dexamethasone resistance (Table 3). In the validation cohort, gene expression of NR3C1 was statistically significantly associated with prednisolone sensitivity (P = .036 and r = –0.25) but not with dexamethasone sensitivity (P = .07 and r = –0.22). In the COALL-DCOG training cohort, gene expression of NR3C1 was not associated with prednisolone sensitivity (P = .13 and r = –0.11). Using the five-gene model in the validation cohort, we achieved a prediction accuracy of 71% (95% CI = 58% to 81%) for prednisolone and 77% (95% CI = 63% to 91%) for dexamethasone. The sensitivity of predicting which patients would have glucocorticoid resistance was 67% for both glucocorticoids (with 95% CI = 50% to 83% for prednisolone and 95% CI = 39% to 89% for dexamethasone) and a specificity of 76% (95% CI = 58% to 91%) for prednisolone and 91% (95% CI = 72% to 100%) for dexamethasone. Thus, results in the validation cohort with two different glucocorticoids support our five-gene model (Supplementary Table 4, available online).
Table 3.
Gene probes associated with prednisolone and dexamethasone sensitivity, as measured by the concentration lethal to 50% of cells, in primary acute lymphoblastic leukemia cells: the St. Jude validation cohort*
| Dexamethasone |
Prednisolone |
||||||
| Gene symbol | Probe set ID | P† | r† | R/S ratio‡ | P† | r† | R/S ratio‡ |
| ARID1A | 212152_x_at | .023 | –0.27 | 0.79 (.03) | .4 | –0.1 | 0.94 (.27) |
| ARID1A | 218917_s_at | .005 | –0.34 | 0.83 (.01) | .088 | –0.2 | 0.84 (.07) |
| ARID1A | 210649_s_at | .035 | –0.26 | 0.71 (.04) | .18 | –0.16 | 0.89 (.11) |
| ARID1A‡ | .016 | –0.29 | 0.26 (.02) | .2 | –0.15 | 0.48 (.11) | |
| SMARCA4 | 208793_x_at | .002 | –0.37 | 0.53 (.002) | .11 | –0.19 | 0.69 (.05) |
| SMARCA4 | 208794_s_at | <.001 | –0.42 | 0.54 (.001) | .01 | –0.3 | 0.78 (.001) |
| SMARCA4 | 214728_x_at | <.001 | –0.43 | 0.50 (<.001) | .003 | –0.34 | 0.79 (.002) |
| SMARCA4 | 212520_s_at | <.001 | –0.43 | 0.44 (<.001) | .27 | –0.13 | 0.71 (.11) |
| SMARCA4§ | <.001 | –0.43 | 0.15 (<.001) | .018 | –0.28 | 0.49 (.002) | |
| SMARCC2 | 201321_s_at | .46 | –0.09 | 1.01 (.66) | .055 | –0.23 | 0.77 (.02) |
| SMARCB1 | 212167_s_at | .019 | –0.29 | 0.79 (.007) | .29 | –0.13 | 0.84 (.16) |
LC50 = concentration lethal to 50% of cells; R/S = resistant to sensitive.
P value and correlation coefficient value (r) were from a Spearman rank correlation test for the concentration of prednisolone that is lethal to 50% of cells vs gene expression. All statistical tests were two-sided.
R/S ratios and their P value (Wilcoxon rank sum test, in parentheses) compared the median gene expression of resistant group with the median gene expression of sensitive group. For prednisolone, resistance = LC50 ≥ 150 μg/mL; sensitivity = LC50 ≤ 0.1 μg/mL. For dexamethasone, resistance = LC50 ≥ 5.8 μg/mL; sensitivity = LC50 ≤ 0.02 μg/mL.
Representative P value and correlation coefficient value that are based on principal component analysis.
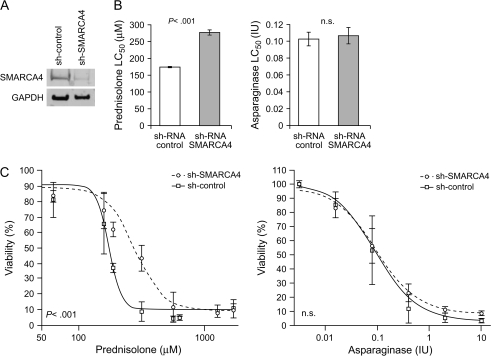
Knockdown of SMARCA4 Expression and Glucocorticoid Resistance
To determine whether modulating SMARCA4 expression affects glucocorticoid sensitivity, Jurkat cells were transduced with a shRNA targeting SMARCA4 or a control shRNA and knockdown of SMARCA4 protein expression was evaluated by western blot analysis (Figure 3, A). The expression of SMARCA4 protein was 86% and 93% (as determined in two independent transductions) lower in SMARCA4 shRNA-transfected cells than that in control shRNA-transfected cells. After treatment with either prednisolone or asparaginase for 4 days, prednisolone resistance was higher in SMARCA4 shRNA-transfected cells than in control shRNA-transfected cells (59% increase in LC50) in each of two independent transductions (LC50 for prednisolone in control cells = 174 μM and LC50 for prednisolone in SMARCA4 shRNA-transfected cells = 277 μM, difference = 103 μM, 95% CI = 100 to 106 μM; P < .001, t test, 32 data points). In contrast, decreased expression of SMARCA4 in SMARCA4 shRNA-transfected cells did not alter the LC50 for the mechanistically distinct antileukemic agent asparaginase (LC50 for asparaginase in control cells = 0.103 IU and LC50 for asparaginase in SMARCA4 shRNA-transfected cells = 0.107 IU) (Figure 3, B and C).
Figure 3.
Short hairpin RNA (shRNA)–mediated SMARCA4 depletion and glucocorticoid resistance. Human Jurkat leukemia cells were stably transduced with control shRNA or SMARCA4 shRNA. A) SMARCA4 protein expression. Cells that were transduced with SMARCA4 shRNA or control shRNA, and SMARCA4 protein expression were evaluated by western blot analysis with anti-SMARCA4 antibodies. The loading control was glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. B) Drug concentration lethal to 50% of cells (LC50) values derived from the in vitro drug sensitivity analysis (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide [MTT] assays). Bars show the mean of two independent experiments, with duplicate samples. Error bars are 95% confidence intervals (CIs). C) Viability of Jurkat cells stably transduced with control shRNA or SMARCA4 shRNA after a 4-day exposure to either prednisolone or asparaginase. Drug concentrations are indicated. Viability was determined by a MTT assay. Each point represents the mean of two transductions performed in duplicate. Error bars are 95% confidence intervals. Statistical significance in the difference of the parameter estimates (specifically LC50 values) was assessed by a two-sided t test. n.s. = not statistically significant (P > .05).
Discussion
Pathway analysis by use of the global test method with the GenMAPP and Gene Ontology database applied on the COALL-DCOG cohort detected statistically significant associations between expression of SWI/SNF complex–related genes—in particular, SMARCA4, ARID1A, SMARCB1, and SMARCC2—and prednisolone resistance in ALL cells. We showed that primary ALL cells from all patients in our study expressed only the SMARCA4 gene, not the SMARCA2 gene, as the catalytic subunit for the SWI/SNF complex. The expression of SMARCA4 gene which translates into the catalytic subunit of the SWI/SNF complex was related to the sensitivity of primary ALL to prednisolone in the training cohort and in two independent validation cohorts of patients (in the training cohort with prednisolone, P < .001 and r = –0.28; in the validation cohort with prednisolone, P = .018 and r = –0.28; and in the validation cohort with dexamethasone, P < .001 and r = –0.43). Finally, we demonstrated the functional importance of the SWI/SNF complex for glucocorticoid resistance by knocking down the expression of SMARCA4 with RNA interference and showing that glucocorticoid resistance was induced in the cells with decreased expression of SMARCA4 protein.
Drug resistance is the major cause of treatment failure in patients with ALL, yet the mechanisms responsible for resistance remain largely unknown (35). Glucocorticoids are one of the most important classes of drugs used in the curative treatment of ALL (3). Glucocorticoid resistance, as determined either in vitro or in vivo, has been strongly linked to treatment outcome (6–9). Thus, elucidating mechanisms of glucocorticoid resistance should provide important new insights to improve the efficacy of chemotherapy for ALL.
We have shown previously (14,15) that the SMARCB1 gene, which encodes a core component of the SWI/SNF complex, is expressed at lower levels in primary ALL cells that are resistant to predisolone than in cells that are sensitive to predisolone. Because SMARCB1 protein is a subunit of the SWI/SNF chromatin-remodeling complex, the current work was undertaken to determine whether expression of other subunits of the SWI/SNF complex influence glucocorticoid resistance in ALL.
Previous studies (23,24) have shown that the SWI/SNF complex containing SMARCA4 protein as the catalytic subunit is required for glucocorticoid-induced transcriptional activation in human SW-13 cells and in yeast. Cytoplasmic rather than nuclear localization of SMARCA4 protein has also been observed in pituitary corticotroph adenomas that exhibit loss of feedback inhibition after high-dose dexamethasone treatment (36). However, the relation of SMARCA4 expression to glucocorticoid resistance in human leukemia cells has not, to our knowledge, been investigated previously. Glucocorticoid transcriptional activation has been widely studied in vitro by use of the mouse mammary tumor virus promoter model (37–40). This promoter, when stably integrated into chromatin, adopts an organized chromatin structure containing steroid response elements and restricts transcription factor access (41,42). With this model, it has been shown that glucocorticoid-mediated activation of the mouse mammary tumor virus promoter is dependent solely on the SWI/SNF complex containing SMARCA4 protein (23,24,43). After glucocorticoid binding, the glucocorticoid–NR3C1–receptor complex binds to its recognition site on the nucleosome and then recruits the SWI/SNF complex containing SMARCA4 protein, which remodels chromatin by disrupting nucleosome structure so that transcription factors and other regulatory elements have access to their DNA binding sites (43). In particular, other SWI/SNF complex–related proteins, such as SMARCD1 and ARID1A, have been shown to interact with the NR3C1 or to potentiate glucocorticoid transactivation and chromatin remodeling (24). The NR3C1 glucocorticoid receptor has been shown (44,45) to bind directly to SMARCD1 protein in a ligand-independent manner and to bind to ARID1A protein in a ligand-dependent manner. These results are consistent with our finding that expression of ARID1A, and not SMARCD1, is statistically significantly and specifically associated with prednisolone sensitivity and not with sensitivity to asparaginase or vincristine (in the discovery cohort, P = .002 and r = –0.23; Supplementary Table 3, available online), indicating ligand dependence.
In this study, we also found that, among genes for subunits of the SWI/SNF complex, the expression levels of SMARCA4, SMARCC2, SMARCB1, and ARID1A before glucocorticoid treatment were statistically significantly lower in glucocorticoid-resistant leukemia cells than in glucocorticoid-sensitive leukemia cells. Expression of these genes, however, was not related to resistance of leukemia cells to the either of the two other antileukemic agent tested, asparaginase and vincristine. In addition, a global test analysis identified the “chromatin modification” pathway (which includes nine of the subunits of the SWI/SNF complex) among the most discriminating biological pathways that were related to prednisolone sensitivity, which also supports the gene expression data. Furthermore, the “SWI/SNF complex” entry of the Gene Ontology “Cellular Component” category, which named 11 subunits of the SWI/SNF complex, was statistically significantly related to prednisolone sensitivity but not to asparaginase or vincristine sensitivity (Figure 1). This analysis also revealed that prednisolone sensitivity was most strongly related to the expression of ARID1A, SMARCA4, and SMARCB1 and less strongly to the expression of SMARCC2. Furthermore, prednisolone sensitivity was associated statistically significantly and independently with the expression of SMARCB1, ARID1A, and NR3C1 glucocorticoid receptor gene (11), and the expression of SMARCC2 and SMARCA4 were strongly correlated with NR3C1, ARID1A, and SMARCB1 (for the SMARCC2–NR3C1 comparison, r = −0.23 and P = .002; for SMARCC2–ARID1A, r = 0.28 and P < .001; for SMARCC2–SMARCB1, r = 0.28 and P < .001; for SMARCA4–NR3C1, r = –0.29 and P < .001; for SMARCA4–ARID1A, r = 0.49 and P < .001; for SMARCA4–SMARCB1, r = 0.46 and P < .001) (Table 2). These data indicate that there is a common regulatory mechanism for these three genes. Also, low expression of SMARCB1, ARID1A, and NR3C1 was associated with prednisolone resistance, with the strongest associations being for ARID1A and SMARCB1 (Supplementary Table 3, available online).
Our findings that the SWI/SNF pathway might be able to explain approximately 22% of the total variation in prednisolone resistance in ALL patients (correlation coefficients for these genes and prednisolone resistance ranged from 20% to 30% in the training set of 177 ALL patients; Supplementary Tables 2 and 3, available online) indicate that glucocorticoid resistance may be multifactorial. Nevertheless, the SWI/SNF complex is a biological system with a previously unrecognized association with the glucocorticoid sensitivity of leukemia cells. To validate these findings, we used an independent cohort of 95 patients for whom we obtained in vitro sensitivity data for two glucocorticoids, prednisolone and dexamethasone. Statistical significance of the association between glucocorticoid sensitivity and gene expression was confirmed for all genes except SMARCC2 by use of the more potent glucocorticoid dexamethasone, and expression of SMARCA4, the gene with the catalytic subunit of the SWI/SNF complex, was strongly associated with both prednisolone and dexamethasone sensitivity (Table 3). In the COALL-DCOG training cohort, the accuracy of predicting whether ALL cells were resistant or sensitive to prednisolone treatment by use of the expression of five genes encoding subunits of the SWI/SNF complex was 76% and, in the St. Jude validation cohort, accuracy was 71% for predicting prednisolone sensitivity or resistance and 77% for predicting dexamethasone sensitivity or resistance (Supplementary Table 4, available online).
We further documented the important role of the SWI/SNF complex in glucocorticoid resistance by showing that shRNA-mediated inhibition of SMARCA4 expression in human Jurkat leukemia cells reduced the sensitivity of leukemia cells to glucocorticoid treatment. Thus, SMARCA4 expression appears to be involved in the antileukemic mechanism of glucocorticoids in that decreased SMARCA4 expression reduced SWI/SNF complex–related chromatin-remodeling activity, which reduced glucocorticoid-induced transcriptional activity, which in turn then reduced glucocorticoid-induced cytotoxicity.
Glucocorticoid resistance in leukemia cell lines has been associated previously with defects in the NR3C1 glucocorticoid receptor (46–48), but such defects have rarely been observed in primary leukemia cells (11). These observations have led to the hypothesis that clinical resistance occurs downstream in the glucocorticoid signaling pathway, with loss of the death response or inappropriate activation of survival signals (13,49–55). However, we found a new mechanism for glucocorticoid resistance in primary leukemia cells, whereby a defect in the chromatin-remodeling process appears to reduce the cytotoxic effects of glucocorticoid on leukemia cells.
Our study had several limitations. Glucocorticoid resistance is most likely a multigenic phenomenon, and genes or gene groups other than those investigated in this report might cause glucocorticoid resistance. Furthermore, genetic mutations that do not change the mRNA level but do alter gene function might also contribute to resistance. These possibilities are supported by our finding that we were able to explain only approximately 20% of the variation in glucocorticoid resistance in patients. Finally, future studies are thus warranted to investigate the levels of proteins that are encoded by SMARCA4, SMARCB1, and ARID1A, the genes that expressed low mRNA levels, to determine whether protein expression is also associated with glucocorticoid resistance in ALL cells. Another limitation of our study is the relatively small sample size, especially of the St. Jude validation cohort.
In conclusion, to our knowledge for the first time, we found an association between the expression of as many as three genes encoding key subunits of the SWI/SNF complex—SMARCA4, SMARCB1, and ARID1A—and resistance of ALL cells to glucocorticoid treatment. These findings provide a basis for future studies to investigate the association between genetic or epigenetic determinants of subunits of the SWI/SNF complex and glucocorticoid resistance of ALL.
Funding
This work was supported in part by the following National Institutes of Health grants (R37 CA36401 to W.E.E., M.V.R.), (R01 CA78224 to W.E.E., M.V.R.), (U01 GM61393 to M.V.R., W.E.E.), Cancer Center Support Grant (CA21765), and by the American Lebanese Syrian Associated Charities; and M.L.D.B., R.P. supported by the Dutch Cancer Society and Pediatric Oncology Foundation Rotterdam.
Supplementary Material
Footnotes
Present address: University of Iowa, Iowa City, IA (M. Assem).
The authors had full responsibility for the design of the study, the collection of the data, the analysis and interpretation of the data, the decision to submit the manuscript for publication, and the writing of the manuscript.
We thank the patients and their parents for their participation in this study and our clinical staff for facilitating protocol-based patient care. We also thank Yan Wang for outstanding technical assistance and Dr Suraj Mukatira and Mark Wilkinson for mathematical and bioinformatics support.
References
- 1.Miner JN, Hong MH, Negro-Vilar A. New and improved glucocorticoid receptor ligands. Expert Opin Investig Drugs. 2005;14(12):1527–1545. doi: 10.1517/13543784.14.12.1527. [DOI] [PubMed] [Google Scholar]
- 2.Irving JA, Minto L, Bailey S, Hall AG. Loss of heterozygosity and somatic mutations of the glucocorticoid receptor gene are rarely found at relapse in pediatric acute lymphoblastic leukemia but may occur in a subpopulation early in the disease course. Cancer Res. 2005;65(21):9712–9718. doi: 10.1158/0008-5472.CAN-05-1227. [DOI] [PubMed] [Google Scholar]
- 3.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354(2):166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 4.Clarke AR, Purdie CA, Harrison DJ, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362(6423):849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 2004;11(suppl 1):S45–S55. doi: 10.1038/sj.cdd.4401456. [DOI] [PubMed] [Google Scholar]
- 6.Dordelmann M, Reiter A, Borkhardt A, et al. Prednisone response is the strongest predictor of treatment outcome in infant acute lymphoblastic leukemia. Blood. 1999;94(4):1209–1217. [PubMed] [Google Scholar]
- 7.Lauten M, Matthias T, Stanulla M, Beger C, Welte K, Schrappe M. Association of initial response to prednisone treatment in childhood acute lymphoblastic leukaemia and polymorphisms within the tumour necrosis factor and the interleukin-10 genes. Leukemia. 2002;16(8):1437–1442. doi: 10.1038/sj.leu.2402545. [DOI] [PubMed] [Google Scholar]
- 8.Kaspers GJ, Pieters R, Van Zantwijk CH, van Wering ER, van der Does-van den Berg A, Veerman AJ. Prednisolone resistance in childhood acute lymphoblastic leukemia: vitro-vivo correlations and cross-resistance to other drugs. Blood. 1998;92(1):259–266. [PubMed] [Google Scholar]
- 9.Hongo T, Yajima S, Sakurai M, Horikoshi Y, Hanada R. In vitro drug sensitivity testing can predict induction failure and early relapse of childhood acute lymphoblastic leukemia. Blood. 1997;89(8):2959–2965. [PubMed] [Google Scholar]
- 10.Zhou J, Cidlowski JA. The human glucocorticoid receptor: one gene, multiple proteins and diverse responses. Steroids. 2005;70(5–7):407–417. doi: 10.1016/j.steroids.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Tissing WJ, Lauten M, Meijerink JP, et al. Expression of the glucocorticoid receptor and its isoforms in relation to glucocorticoid resistance in childhood acute lymphocytic leukemia. Haematologica. 2005;90(9):1279–1281. [PubMed] [Google Scholar]
- 12.Holleman A, den Boer ML, Kazemier KM, et al. Decreased PARP and procaspase-2 protein levels are associated with cellular drug resistance in childhood acute lymphoblastic leukemia. Blood. 2005;106(5):1817–1823. doi: 10.1182/blood-2004-11-4296. [DOI] [PubMed] [Google Scholar]
- 13.Tissing WJ, Meijerink JP, den Boer ML, Pieters R. Molecular determinants of glucocorticoid sensitivity and resistance in acute lymphoblastic leukemia. Leukemia. 2003;17(1):17–25. doi: 10.1038/sj.leu.2402733. [DOI] [PubMed] [Google Scholar]
- 14.Holleman A, Cheok MH, den Boer ML, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351(6):533–542. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 15.Pottier N, Cheok MH, Yang W, et al. Expression of SMARCB1 modulates steroid sensitivity in human lymphoblastoid cells: identification of a promoter SNP that alters PARP1 binding and SMARCB1 expression. Hum Mol Genet. 2007;16(19):2261–2271. doi: 10.1093/hmg/ddm178. [DOI] [PubMed] [Google Scholar]
- 16.Percipalle P, Farrants AK. Chromatin remodelling and transcription: be-WICHed by nuclear myosin 1. Curr Opin Cell Biol. 2006;18(3):267–274. doi: 10.1016/j.ceb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Trotter KW, Archer TK. Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol. 2007;265–266:162–167. doi: 10.1016/j.mce.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts CW, Orkin SH. The SWI/SNF complex—chromatin and cancer. Nat Rev Cancer. 2004;4(2):133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 19.Wade PA, Wolffe AP. Transcriptional regulation: SWItching circuitry. Curr Biol. 1999;9(6):R221–R224. doi: 10.1016/s0960-9822(99)80134-1. [DOI] [PubMed] [Google Scholar]
- 20.Wallberg AE, Neely KE, Hassan AH, Gustafsson JA, Workman JL, Wright AP. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor tau1 activation domain. Mol Cell Biol. 2000;20(6):2004–2013. doi: 10.1128/mcb.20.6.2004-2013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258(5088):1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 22.Ostlund Farrants AK, Blomquist P, Kwon H, Wrange O. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol Cell Biol. 1997;17(2):895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393(6680):88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 24.Trotter KW, Archer TK. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol Cell Biol. 2004;24(8):3347–3358. doi: 10.1128/MCB.24.8.3347-3358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harms DO, Janka-Schaub CE. Co-operative study group for childhood acute lymphoblastic leukemia(COALL): long-term follow-up of trials 82, 85, 89 and 92. Leukemia. 2000;14(12):2234–2239. doi: 10.1038/sj.leu.2401974. [DOI] [PubMed] [Google Scholar]
- 26.Ross ME, Zhou X, Song G, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102(8):2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- 27.D’Argenio DZ, Schumitzky A. ADAPT II User's Guide: Pharmacokinetic/Pharmacodynamic Systems Analysis Software. Los Angeles, CA: Biomedical Simulations Resource; 1997. [Google Scholar]
- 28.den Boer ML, Harms DO, Pieters R, et al. Patient stratification based on prednisolone-vincristine-asparaginase resistance profiles in children with acute lymphoblastic leukemia. J Clin Oncol. 2003;21(17):3262–3268. doi: 10.1200/JCO.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 29.Kaspers GJ, Veerman AJ, Pieters R, et al. In vitro cellular drug resistance and prognosis in newly diagnosed childhood acute lymphoblastic leukemia. Blood. 1997;90(7):2723–2729. [PubMed] [Google Scholar]
- 30.Pieters R, Huismans DR, Loonen AH, et al. Relation of cellular drug resistance to long-term clinical outcome in childhood acute lymphoblastic leukaemia. Lancet. 1991;338(8764):399–403. doi: 10.1016/0140-6736(91)91029-t. [DOI] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goeman JJ, van de Geer SA, de KF, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20(1):93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 33.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13(2):136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 35.Cheok MH, Evans WE. Acute lymphoblastic leukaemia: a model for the pharmacogenomics of cancer therapy. Nat Rev Cancer. 2006;6(2):117–129. doi: 10.1038/nrc1800. [DOI] [PubMed] [Google Scholar]
- 36.Bilodeau S, Vallette-Kasic S, Gauthier Y, et al. Role of Brg1 and HDAC2 in GR trans-repression of the pituitary POMC gene and misexpression in Cushing disease. Genes Dev. 2006;20(20):2871–2886. doi: 10.1101/gad.1444606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deroo BJ, Archer TK. Glucocorticoid receptor activation of the I kappa B alpha promoter within chromatin. Mol Biol Cell. 2001;12(11):3365–3374. doi: 10.1091/mbc.12.11.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denolet E, Gendt KD, Swinnen JV, et al. Transfection with steroid-responsive reporter constructs shows glucocorticoid rather than androgen responsiveness in cultured Sertoli cells. J Steroid Biochem Mol Biol. 2006;98(2–3):164–173. doi: 10.1016/j.jsbmb.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Fruchter O, Kino T, Zoumakis E, et al. The human glucocorticoid receptor (GR) isoform {beta} differentially suppresses GR{alpha}-induced transactivation stimulated by synthetic glucocorticoids. J Clin Endocrinol Metab. 2005;90(6):3505–3509. doi: 10.1210/jc.2004-1646. [DOI] [PubMed] [Google Scholar]
- 40.Szatmary Z, Garabedian MJ, Vilcek J. Inhibition of glucocorticoid receptor-mediated transcriptional activation by p38 mitogen-activated protein (MAP) kinase. J Biol Chem. 2004;279(42):43708–43715. doi: 10.1074/jbc.M406568200. [DOI] [PubMed] [Google Scholar]
- 41.Richard-Foy H, Hager GL. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6(8):2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Archer TK, Cordingley MG, Wolford RG, Hager GL. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11(2):688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Kinyamu HK, Archer TK. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol Endocrinol. 2006;20(1):1–13. doi: 10.1210/me.2005-0192. [DOI] [PubMed] [Google Scholar]
- 44.Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23(17):6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inoue H, Furukawa T, Giannakopoulos S, Zhou S, King DS, Tanese N. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J Biol Chem. 2002;277(44):41674–41685. doi: 10.1074/jbc.M205961200. [DOI] [PubMed] [Google Scholar]
- 46.Strasser-Wozak EM, Hattmannstorfer R, Hala M, et al. Splice site mutation in the glucocorticoid receptor gene causes resistance to glucocorticoid-induced apoptosis in a human acute leukemic cell line. Cancer Res. 1995;55(2):348–353. [PubMed] [Google Scholar]
- 47.Powers JH, Hillmann AG, Tang DC, Harmon JM. Cloning and expression of mutant glucocorticoid receptors from glucocorticoid-sensitive and -resistant human leukemic cells. Cancer Res. 1993;53(17):4059–4065. [PubMed] [Google Scholar]
- 48.Hala M, Hartmann BL, Bock G, Geley S, Kofler R. Glucocorticoid-receptor-gene defects and resistance to glucocorticoid-induced apoptosis in human leukemic cell lines. Int J Cancer. 1996;68(5):663–668. doi: 10.1002/(SICI)1097-0215(19961127)68:5<663::AID-IJC17>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 49.Haarman EG, Kaspers GJ, Pieters R, Rottier MM, Veerman AJ. Glucocorticoid receptor alpha, beta and gamma expression vs in vitro glucocorticod resistance in childhood leukemia. Leukemia. 2004;18(3):530–537. doi: 10.1038/sj.leu.2403225. [DOI] [PubMed] [Google Scholar]
- 50.Bachmann PS, Gorman R, Mackenzie KL, Lutze-Mann L, Lock RB. Dexamethasone resistance in B-cell precursor childhood acute lymphoblastic leukemia occurs downstream of ligand-induced nuclear translocation of the glucocorticoid receptor. Blood. 2005;105(6):2519–2526. doi: 10.1182/blood-2004-05-2023. [DOI] [PubMed] [Google Scholar]
- 51.Quddus FF, Leventhal BG, Boyett JM, Pullen DJ, Crist WM, Borowitz MJ. Glucocorticoid receptors in immunological subtypes of childhood acute lymphocytic leukemia cells: a Pediatric Oncology Group Study. Cancer Res. 1985;45(12 pt 1):6482–6486. [PubMed] [Google Scholar]
- 52.Tissing WJ, Meijerink JP, Brinkhof B, et al. Glucocorticoid-induced glucocorticoid-receptor expression and promoter usage is not linked to glucocorticoid resistance in childhood ALL. Blood. 2006;108(3):1045–1049. doi: 10.1182/blood-2006-01-0261. [DOI] [PubMed] [Google Scholar]
- 53.Hillmann AG, Ramdas J, Multanen K, Norman MR, Harmon JM. Glucocorticoid receptor gene mutations in leukemic cells acquired in vitro and in vivo. Cancer Res. 2000;60(7):2056–2062. [PubMed] [Google Scholar]
- 54.Holleman A, den Boer ML, Kazemier KM, Janka-Schaub GE, Pieters R. Resistance to different classes of drugs is associated with impaired apoptosis in childhood acute lymphoblastic leukemia. Blood. 2003;102(13):4541–4546. doi: 10.1182/blood-2002-11-3612. [DOI] [PubMed] [Google Scholar]
- 55.Holleman A, den Boer ML, Cheok MH, et al. Expression of the outcome predictor in acute leukemia 1 (OPAL1) gene is not an independent prognostic factor in patients treated according to COALL or St Jude protocols. Blood. 2006;108(6):1984–1990. doi: 10.1182/blood-2006-04-015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.