Abstract
Morphology and cleavage rate remain the mainstay of embryo assessment. However, a number of additional technologies for this application are under investigation. These include the measurement of glucose, lactate, pyruvate or amino acid levels in the embryo culture media, assessment of oxygen consumption by the embryo, genomic and proteomic profiling, and most recently, analytical examination of the embryonic metabolome. As the number of assisted reproduction cycles increases worldwide, improvements in the ability to quickly and non-invasively identify the best embryos for transfer remain a critical goal for reproductive medicine. Recent studies suggest that metabolomic profiling of embryo culture media using optical and non-optical spectroscopies may provide a useful adjunct to the current embryo assessment strategies and provide insight into the phenotype of embryos with increasing reproductive potential.
Keywords: metabolomics, in vitro fertilization, non-invasive embryo assessment
Introduction
Soon after the report of the first successful pregnancy with in vitro fertilization (IVF) (Steptoe and Edwards, 1978), and development of controlled ovarian stimulation in order to generate more than one embryo in a given cycle (Trounson et al., 1981), it became apparent that morphology and cleavage rate of embryos correlate with their implantation potential (Edwards et al., 1984). Thereafter, grading systems based on embryo cleavage rate and morphology were developed (Scott and Smith, 1998; Gardner and Schoolcraft, 1999; Gerris et al., 1999; Tesarik and Greco, 1999; VanRoyan et al., 1999; Veeck, 1999) leading to significant improvements in implantation and pregnancy rates and reductions in multiple gestation rates (Toner, 2002).
Today, among the treatment modalities offered to infertile couples, those utilizing IVF are associated with the highest success rates. Consequently, IVF use has been increasing steadily, from 64 681 cycles reported in 1996 in the USA to 134 260 cycles in 2005 (SART, 1996, 2005). However, despite its widening application, IVF is currently associated with two important issues that derive, at least in part, from our inability to adequately assess the reproductive potential of individual embryos: (i) low implantation rates and (ii) high multiple pregnancy rates.
Approximately two of three IVF cycles fail to result in pregnancy, causing significant physical, emotional and financial distress for women undergoing infertility treatment (SART, 2006). Even more striking is the failure of more than 8 of 10 transferred embryos to implant (Kovalevsky and Patrizio, 2005; Bromer and Seli, 2008). Our inability to determine the embryos with highest reproductive potential is likely to contribute to the high rate of implantation and IVF cycle failures.
In order to prevent failures, centers have historically chosen to perform simultaneous transfer of multiple embryos, accepting the related risk of multiple pregnancies. In the USA, a mean number of 2.45 embryos were transferred in IVF cycles using fresh non-donor oocytes in 2006, resulting in a 34.3% live birth rate per transfer, of which 32% were multiple-infant live births (SART, 2006). Similarly, a mean number of 2.3 embryos were transferred in IVF cycles using fresh donor oocytes, achieving a 52.3% live birth rate per transfer, 40.8% of which were multiple-infant live births (SART, 2006). In total, while IVF treatment accounts for only 1% of all births in the USA, 18% of multiple births result from IVF. Moreover, 51% of all IVF neonates are the products of multiple gestations (Wright et al., 2006), a frequency of 15- to 20-fold greater than with spontaneous conceptions (Reddy et al., 2007).
The high multiple pregnancy rates associated with IVF have significant public health consequences (Bromer and Seli, 2008) as the increased rate of preterm delivery in multiple-infant pregnancies compromises the survival of neonates and increases their risk of lifelong disability. Indeed, while multiple births constitute approximately 1/33 of all births in the USA, they account for 1/8 of preterm births (<37 weeks) and 1/4 of very low birthweight infants (<2000 g) (Luke and Martin, 2004). Largely due to the increased incidence of prematurity, cerebral palsy is increased 8-fold in twins and 47-fold in triplets (Keith et al., 2000), while infant deaths (birth to 1 year) are increased 6-fold in twins, and 17-fold in triplets and higher order gestations (Oleszczuk et al., 2003). Recently, it has been estimated that preterm births that result from IVF-related multiple pregnancies, account for ∼$1 billion of cost to the society in the USA yearly (Bromer and Seli, 2008). These estimations do not include maternal complications associated with multiple gestations, including a 2- to 4-fold increase in pregnancy induced hypertension and post-partum hemorrhage (Luke and Brown, 2007).
The medical and financial complications associated with multiple pregnancies have now led a number of countries to impose legal restrictions on the number of embryos transferred in IVF cycles (Bromer and Seli, 2008). However, the majority of countries have not yet adopted strict regulations with respect to IVF practice; most probably due to high cost of treatment and patients’ desire to increase their chances to achieve pregnancy in a given cycle. Consequently, decreasing multiple gestations while maintaining or increasing overall pregnancy rates is an important goal of contemporary infertility treatment, and an improvement over the current methods of embryo assessment would be beneficial for this purpose.
Embryo metabolism as a predictor of reproductive potential
Given the limitations of morphologic evaluation, many investigators have pursued adjunctive technologies for the assessment of the reproductive potential of an embryo. Within this context, many metabolic parameters of developing embryos have been studied using a variety of non-invasive methods (Sakkas and Gardner, 2005). We will briefly review studies that investigated metabolomic parameters of in vitro grown human embryos as they relate to embryo growth and potential to result in a pregnancy. These studies demonstrate an underlying metabolic difference between embryos that result in a pregnancy and those that do not, and form the basis of a metabolomic approach in the assessment of embryo viability.
Pyruvate metabolism
There are two main pathways in ATP generation that are necessary for embryonic cellular metabolism: aeorobic glycolysis, or the tricarboxylic acid (Krebs) cycle, and anaerobic glycolysis, by the Embden–Meyerhof pathway. Carboxylic acid-based metabolism predominates in the early preimplantation (cleavage stage) development, where pyruvate and lactate are the embryo's main sources of energy, and glucose uptake is minimal (Biggers et al., 1967; Conaghan et al., 1993a). Consequently, pyruvate uptake by embryos in culture has been investigated as a possible marker of embryo viability and growth potential.
Hardy et al. (1989) and Gott et al. (1990) reported higher pyruvate uptake in embryos that develop to blastocyst stage (Table I). Conaghan et al. (1993a,b), however, later reported contradictory findings and demonstrated an inverse relationship between pyruvate uptake by 2–8-cell embryos with embryo viability and pregnancy. A subsequent report by Turner et al. (1994) suggested that embryos display a wide range of pyruvate uptake values, but the variation is reduced in those embryos that were capable of implantation.
Table I.
Pyruvate, lactate and glucose metabolism as a predictor of embryo development and viability—human studies.
| Study | Embryo stage examined | Altered metabolite associated with improved outcome | Technology used | Outcome |
|---|---|---|---|---|
| Hardy et al. 1989 | Day 2-4 | ↑ pyruvate uptake | Ultramicrofluorescence assay | Blastocyst development |
| No association with glucose uptake | ||||
| Day 5 | ↑ pyruvate uptake | Ultramicrofluorescence assay | Blastocyst development | |
| ↑ glucose uptake | ||||
| Gott et al. 1990 | Day 2-4 | ↑ pyruvate uptake | Ultramicrofluorescence assay | Blastocyst development |
| ↑ lactate production | ||||
| No association with glucose uptake | ||||
| Day 5 | ↑ pyruvate uptake | Ultramicrofluorescence assay | Blastocyst development | |
| ↑ glucose uptake | ||||
| ↑ lactate production | ||||
| Conaghan et al., 1993 | Day 2 – 3 | ↓ pyruvate uptake | Ultramicrofluorescence assay | Clinical pregnancy |
| Turner et al., 1994 | Day 2 | Intermediate pyruvate uptake | Ultramicrofluorescence assay | Clinical pregnancy |
| Gardner et al., 2001 | Day 4 | ↑ pyruvate uptake | Ultramicrofluorescence assay | Blastocyst development |
| ↑ glucose uptake | ||||
| Seli et al. | Day 2-3 | A trend toward | Proton NMR | Pregnancy and delivery |
| ↑ pyruvate uptake | ||||
| ↑ glucose uptake |
Adapted from Bromer and Seli (2008).
Most recently, Gardner et al. (2001) assessed pyruvate uptake on Day 4 embryos as it relates to development to blastocyst stage. They found that pyruvate uptake on Day 4 is significantly higher in embryos that go on to form blastocysts compared with embryos that fail to develop to the blastocyst stage, consistent with the initial reports. Overall, it appears inconclusive whether pyruvate uptake is predictive for embryo development and viability.
Glucose metabolism
The capacity to metabolize glucose increases significantly at the transition from the morula to blastocyst stage and also seems to reflect the embryo's developmental potential and viability (Devreker, 2007). Gardner and Leese (1987) measured glucose uptake by microfluorescence in Day 4 mouse blastocysts prior to embryo transfer. Embryos that resulted in term pregnancies had a significantly higher glucose uptake in culture than embryos that failed to progress. Gardner et al. (2001) also reported that the glucose consumption by Day 4 human embryos was higher in embryos that formed blastocysts and correlated with morphologic grade (Table I).
Meanwhile, several other studies failed to demonstrate a relationship between glucose uptake and blastocyst development in human embryos (Hardy et al., 1989; Gott et al., 1990; Jones et al., 2001). It is noteworthy that the media used to assess embryo metabolism in these studies lacked pyruvate, lactate, amino acids and vitamins. Thus, it is possible that there was significant stress on the embryos under these culture conditions (Lane and Gardner, 1998), and the conclusions drawn may consequently have been limited by this factor.
Amino acid metabolism
Using high-performance liquid chromatography (HPLC), Houghton et al. (2002) determined the amino acids secreted and taken up by human embryos, during different stages of preimplantation development, correlated to blastocyst development. Their findings are summarized in Table II. In a subsequent study, Brison et al. (2004) used the same approach to examine the changes in concentration of amino acids secreted by individually cultured human embryos. They reported an association between decreased glycine and leucine, and increased asparagine levels in the culture media with increased clinical pregnancy and live birth rates. They also reported that embryos with greater viability have a lower or quieter amino acid metabolism than those that arrest. Sturmey et al. (2008) reported similar findings in cryopreserved embryos, and Stokes et al. (2007) provided further support for the quiet embryo hypothesis by demonstrating that the metabolic activity of bovine, porcine and human embryos at the blastocyst stage positively correlates with existing DNA damage.
Table II.
Amino acid uptake and secretion by the embryo as a predictor of embryo development viability—human studies.
| Study | Embryo stage examined | Altered metabolite associated with outcome | Technology used | Outcome |
|---|---|---|---|---|
| Houghton et al., 2002 | Day 2 – 3 | ↓ amino acid turnover (sum of depletion and appearance) | HPLC | Blastocyst development |
| ↓ glutamine, arginine, methionine uptake | ||||
| ↓ alanine and asparagine release | ||||
| 8 cell-Morula | ↓ amino acid turnover (sum of depletion and appearance) | HPLC | Blastocyst development | |
| ↓ serine uptake | ||||
| ↓ alanine and glycine release | ||||
| Brison et al., 2004 | Day 2 | ↓ glycine and leucine in culture media | HPLC | Clinical pregnancy and live birth |
| ↑ asparagine levels in culture media | ||||
| Seli et al. 2008 | Day 3 | ↑ glutamate levels in culture media | Proton NMR | Clinical pregnancy and live birth |
Adapted from Bromer and Seli (2008).
Most recently, Seli et al. (2008a) have used proton nuclear magnetic resonance (1H NMR) and found an association between higher glutamate levels in the culture media and clinical pregnancy and live birth.
Implications
The studies described above utilized a multitude of technologies and have suggested that metabolic differences exist among embryos that differ in their ability to result in a pregnancy. However, the application of these technologies to a clinical setting has remained limited for a variety of reasons. Many of these technologies require complex equipment and dedicated technical staff, which would be cost prohibitive to most embryology laboratories, and some do not produce results quickly enough to allow the information to be used clinically in the limited window of time acceptable for embryo transfer in the fresh cycle. None of the technologies has ever been prospectively validated on a test set of culture media samples, demonstrating a correlation with the implantation potential of embryos that have been transferred. Thus, the need remains for a validated technology that predicts viability of embryos non-invasively, through a rapid, on-site evaluation of multiple samples.
Metabolomics: the essentials
Metabolomics in systems biology
Evolving from genomics, transcriptomics and proteomics, the emerging ‘omics’ science, metabolomics, is the systematic analysis of the inventory of metabolites—as small molecule biomarkers—that represent the functional phenotype at the cellular level (Appendix Table A1—Glossary) (Posillico and Technologies, 2007). In particular, metabolomics is used to explain the underlying change in metabolic regulation as a function of disease and abnormal health conditions in a biological system (cell, tissue or organism). Realization that obtaining the genome sequence of humans or other species does not in itself explain the fundamental nature of many disease processes has increased interest in approaches that relate gene expression to phenotypic outcome (Nicholson et al., 2002). Metabolomics offers a unique opportunity to investigate the relationships between an organism's genotype and its resulting phenotype as the metabolome is the end product of gene expression, and also marks a relationship between an organism's physiology and environmental conditions.
The metabolome refers to the complete inventory of small molecules (<1 kDa), non-proteinaceous compounds such as metabolic intermediates (amino acids, lipids and nucleotides), ATP, hormones, other signaling molecules and secondary metabolites that are found within a biological sample (Table III) (Posillico and Technologies, 2007; Pasikanti et al., 2008). These compounds are the ultimate products of cellular metabolism and are very diverse in their physical and chemical properties, occurring in a wide concentration range. As the number of metabolites is typically lower than the number of genes and proteins in a cell or organism, thorough metabolomic analysis can be achieved in a relatively faster period of time than genomically related experiments. It is generally agreed that there are more than 25 000 genes in the human genome, encoding for approximately 100 000–200 000 transcripts, and 1 million proteins, whereas there may be as few as 2500–3000 small molecule metabolites that make up the human metabolome. Due to high labor and cost associated with proteomics and transcriptomics, metabolomics has become a front running research tool in understanding the ‘global picture’ of systems biology.
Table III.
Examples of metabolites from the human metabolome and their physical (molecular weight) and chemical (chemical class, chemical formula) properties.
| Metabolite | Chemical class | Chemical formula | Molecular weight (Da) |
|---|---|---|---|
| Urea | Organic compound—amino ketone | CH4N2O | 60.06 |
| Pyruvic acid | Organic compound—keto-acid | C3H4O3 | 88.06 |
| Alanine | Amino acid | C3H7NO2 | 89.09 |
| Glucose | Carbohydrate—monosaccharide | C6H12O6 | 180.16 |
| Oleic acid | Lipid—fatty acid | C18H34O2 | 282.46 |
| Lactose | Carbohydrate—disaccharide | C12H22O11 | 342.3 |
| Cholesterol | Lipid—steroid | C27H46O | 386.65 |
| Adenosine triphosphate | Nucleotide—co-enzyme | C10H16N5O13P3 | 507.18 |
| Glycogen | Carbohydrate—polysaccharide | C24H42O21 | 666.58 |
| Oxytocin | Peptide | C43H66N12O12S2 | 1007.19 |
The metabolomics experiment is managed by a multidisciplinary team of scientists in a workflow, from sample collection and preparation through analytical operations to processing of raw data and analysis of the processed data. Each step of the pipeline requires meticulous planning in order to extract the optimal amount of reproducible information and interpret its biological significance. As metabolomics is a data-driven, inductive science, the proper methodology of analysis, including key analytical technologies, must be selected in order to remove any bias. Within the metabolomics platform, each analytical approach offers an array of strengths and pitfalls, and are discussed here and summarized in Appendix Table A2.
Analytical technologies
To investigate complex metabolic profiles of a biological system, non-selective, but specific analytical technologies are required. Many spectroscopic/spectrometric and chromatographic techniques are excellent candidates and provide automated, high-throughput methodologies with information-rich profiles of target biological fluids. Investigators have coined the term ‘biospectroscopy’ to designate the analysis of biological fluids by spectroscopic technologies. The scientific platform that incorporates both metabolic profiling and biospectroscopy can be referred to as ‘biospectroscopy-based metabolomics’ or ‘BSM’. Common techniques applied in BSM studies include NMR spectroscopy, mass spectrometry (MS), which can be coupled with separation methods like gas chromatography (GC–MS), liquid chromatography (LC–MS) or HPLC–MS, and capillary electrophoresis (CE–MS). Optical spectroscopies, like Fourier transform infrared (FT-IR), near infrared (NIR) and Raman spectroscopies all provide complementary profiles of the various components within biological fluids due to the similar physical mechanisms involved in each technique.
Non-optical spectroscopy
Nuclear magnetic resonance (NMR) spectroscopy
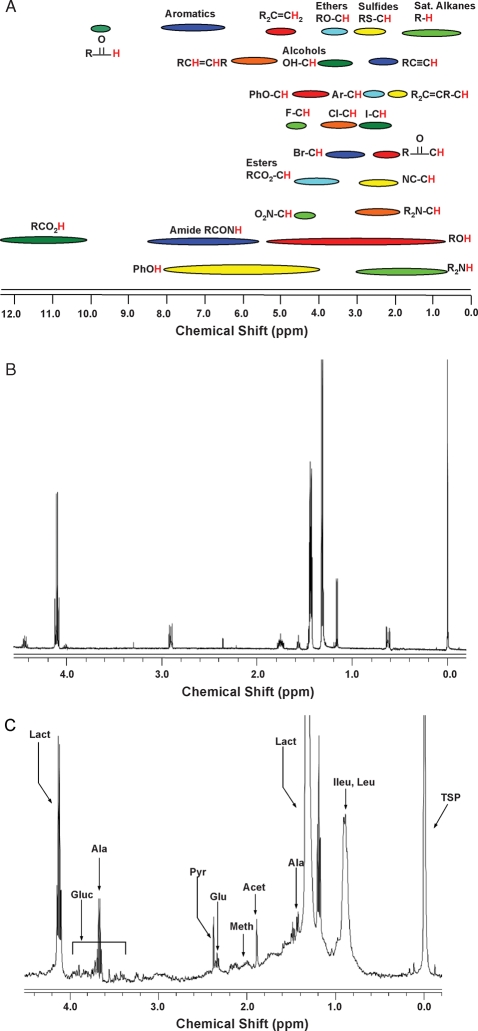
One of the most common and widely used spectroscopic techniques for metabolomics is NMR spectroscopy (Fig. 1). Based on magnetic properties of atomic nuclei, this technique provides a multivariate analysis approach by non-destructively examining intact tissue and biological fluids (Nicholson et al., 2002). NMR is intrinsically rich in information, offering structural and quantitative information simultaneously, with metabolite identification capabilities of even the most complex of mixtures, without the need of separation or sample preparation (Lindon, 2004). An important aspect of NMR spectroscopy is that the fundamental physicochemical mechanism is completely different from other analytical techniques and provides a different scientific perspective (Pauli et al., 2005). However, NMR spectroscopy is expensive, as highly advanced spectrometers with increasing magnetic field strengths are required to make biological measurements discernable. NMR experiments can also be time-costly, ranging from less than a minute to a few hours per measurement. In addition, the method requires highly trained personnel to execute experiments and to interpret the resulting spectra (Fig. 1). Although the NMR signal is intrinsically quantitative, advanced software is required to extract such information. Yet with each of these pitfalls, NMR remains a spectroscopic method of choice for metabolomics.
Figure 1:
NMR spectroscopy examples of pure component and biological fluid metabolomic analyses.
(A) Known chemical shift regions for proton NMR spectroscopy (0–12 ppm). (B) Fourier transform proton NMR spectrum (0–5 ppm) of 6 mM lactic acid in water obtained using a 500 MHz Varian spectrometer. (C) Fourier transform proton NMR spectrum (0–5 ppm) of 18 mL spent IVF culture media in 600 mL 10% D2O/90% H2O obtained using a 500 MHz Varian spectrometer. Ileu, isoleucine; Leu, leucine; Lact, lactate; Ala, alanine; Acet, acetate; Meth, methionine; Glu, glutamate; Pyr, pyruvate; Gluc, glucose.
Mass spectrometry
A counterpart to NMR spectroscopy that is also used extensively in BSM is MS (Dettmer et al., 2007). Mass spectrometers operate by ionization of metabolites, separation of ionic fragments according to their mass-to-charge ratio and then detection of these separated ions. As only medium-high abundant chemicals can be detected by NMR spectroscopy (detection limit ∼0.1 mM (Bock, 1994)), MS-based metabolomics provides a more analytically sensitive alternative with the ability to detect metabolites at the micromolar concentration level, typical of physiological concentrations (Dunn, 2008). MS is also capable of quantitative analyses, and metabolite and structure identification through molecular weight determination. In a recent study, Katz-Jaffe et al. (2006) applied MS to the analysis of embryo culture medium and reported differences among the secretome of embryos at different stages of development.
When MS is coupled with separation techniques, the complexity of the mass spectrum is reduced and provides additional information on the physiochemical properties of the metabolites. Chromatography is the collective term for a family of laboratory techniques for the separation of mixtures. It involves passing a mixture in a ‘mobile phase’ through a ‘stationary phase’ (bound to columns and capillaries), which separates the analytes to be measured into its comprising components. Separation occurs as a result of the analytes affinity for the stationary phase, which is typically dependent on the physical (size) or chemical (polarity) properties of the analyte. Chromatographic-coupled MS usually requires thorough a priori knowledge of the biological sample and may necessitate extensive sample preparation. Consequently, chromatographic techniques can cause metabolite losses by discrimination between metabolite classes (Pasikanti et al., 2008).
Gas chromatography–mass spectrometry (GC-MS)
GC–MS is biased towards volatile, thermally stable, low molecular weight metabolites (<350 Da). As most of the metabolites are polar and non-volatile in nature, they cannot be readily analyzed by GC–MS. Therefore, this technique requires chemical derivatization at the polar functional groups to reduce polarity and to increase thermal stability and volatility (Pasikanti et al., 2008). Although these processes allow the detection of many metabolite classes (including amino acids, fatty acids and some lipids, sugars, sugar alcohols and phosphates, amines, amides and thiol containing metabolites), this extra processing step can introduce greater technical variability and complexity to the data as a single metabolite can produce multiple derivatized metabolite peaks (Dunn, 2008). Derivatization, extensive sample preparation and long chromatographic analysis (∼10–60 min per run) render GC–MS a relatively low-throughput technique for metabolomic applications.
Liquid chromatography–mass spectrometry (LC-MS)
A related chromatographic technique, LC–MS, differs from GC–MS in distinct ways as it not only operates in a different state of matter (with a solid stationary phase and a liquid mobile phase), but can function at both high and low temperatures and does not require sample volatility. These conditions simplify sample preparation and make LC–MS a better selection for the metabolomic analysis of biological fluids. Fluids like urine can be directly injected into an LC–MS system, whereas samples such as plasma need minimal pretreatment (protein precipitation) (Lu et al., 2008). As a result, LC–MS, and HPLC–MS alike, are capable of moderate- to high-throughput sampling. It is also applicable to a reasonable chemical dynamic range, with molecular weights extending from low molecular weight species that are detectable by GC–MS to molecular weights >600 Da. This chemical inventory includes the analysis of phospholipids, proteins, amino acids, glycosides and sugars (Dunn, 2008).
Capillary electrophoresis–mass spectrometry (CE-MS)
A final separation method coupled to MS, which focuses on the separation of charged metabolites is CE. Although CE–MS is used to a lesser extent in metabolomics, it offers significant potential with its capability of analyzing a wide range of analytes from inorganic ions to large proteins (Dunn and Ellis, 2005). As a major portion of metabolites are polar and ionic, CE–MS can typically resolve samples that may be more difficult with conventional techniques like GC or LC, without requiring rigorous sample preparation (Montona and Soga, 2007). Yet the most attractive feature of CE is its small sample requirement (few µL), making it useful for samples that are limited by volume (Montona and Soga, 2007). However, capillary and nanocolumn advances in HPLC may offer the same low-volume requirement and offer greater advantages as an analytical technology over CE.
The choice between NMR and MS approaches is ultimately matrix or problem dependent. While chromatographic-coupled MS provides an increase in sensitivity, the need to select and adjust experimental conditions may result in an unintended bias to specific analytes. Whereas the use of NMR spectroscopy is less selective and not chemically restricted, but continues to be challenged by its detection limits. Combining both NMR spectroscopy and MS can yield complementary information and prove to be useful in metabolomics studies (Lenz et al., 2004; Williams et al., 2005; Wilsona et al., 2005). Still, both technologies require further development, especially of high-throughput and data-processing methods, to optimize their use in complex metabolomic studies.
Optical spectroscopy
Ultimately, what limits the use of NMR and chromatographic MS technologies in clinical applications are the cost, reproducibility and practicality as a commercial, ‘bench-top’ product. Although both technologies are excellent candidates in research applications, they are limited by their size and complexity of operation. An alternative is the relatively inexpensive and simple platforms of optical spectroscopies.
Optical spectroscopy measures the interaction of a species with electromagnetic radiation—the electromagnetic radiation absorbed, emitted or scattered by the sample analyzed. The term ‘optical spectroscopy’ in fact encompasses a broad range of analytical techniques which includes ultraviolet/visible absorption, luminescence and circular dichroism spectroscopies (of which are not discussed herein), as well as a subset of techniques that are more formally referred to as ‘vibrational spectroscopies’: IR, NIR and Raman spectroscopies. Optical spectroscopic instrumentation is relatively simple compared with that required for NMR or MS—it incorporates basic optical components like light sources, mirrors, lenses, gratings and detectors. In fact, some dispersive optical spectrometers incorporate no moving parts at all, which results in very fast, highly reproducible measurements. Additionally, these instruments are extremely stable over time and can be easily maintained and operated by minimally trained users. Optical spectroscopic technologies typically enable the rapid (common measurement times range from less than a second to a few minutes) and non-destructive analysis of a diverse range of sample types (Dunn and Ellis, 2005).
Fourier transformation infrared (FT-IR) spectroscopy
FT-IR is a well-established vibrational spectroscopic method that is extensively used to analyze molecular species in biological samples. The principle of FT-IR is based on molecular vibrational modes, like stretching and bending of chemical bonds in a molecule. Upon exposure to light (electromagnetic radiation), molecules will increase in vibrational energy due to light absorbance and vibrate at characteristic frequencies depending on their chemical structure. Spectra of vibrational frequencies absorbed (between 400–4000 cm−1 or 2500–25 000 nm) can then be correlated to single bonds or functional groups of a molecule (–OH, –CH, –CH2, –CH3, –NH, –NH2, –C=O etc.). Multivariate analysis of several spectral peaks generally allows for identification of the various components within a sample by matching the peak pattern(s) against a library of previously recorded reference spectra of individual components. FT-IR may also be used as a quantitative method for determining concentrations of components within a sample. Generally, FT-IR spectroscopy is able to analyze carbohydrates, amino acids, lipids and fatty acids as well as proteins and polysaccharides simultaneously (Dunn and Ellis, 2005). While sensitivity and selectivity are not as high as methods like MS, the fact that FT-IR is a high-throughput method, capable of measuring thousands of spectra in a single day, should not be overlooked. One drawback to FT-IR is that water absorbs IR radiation very strongly, making analysis of aqueous samples challenging. However, short path-length transmission cells and attenuated total reflectance sampling accessories are both feasible ways of recording FT-IR spectra from aqueous samples. In most cases, water will still contribute the strongest features in the spectrum and potentially interfere with the detection of some metabolites.
Near infrared (NIR) spectroscopy
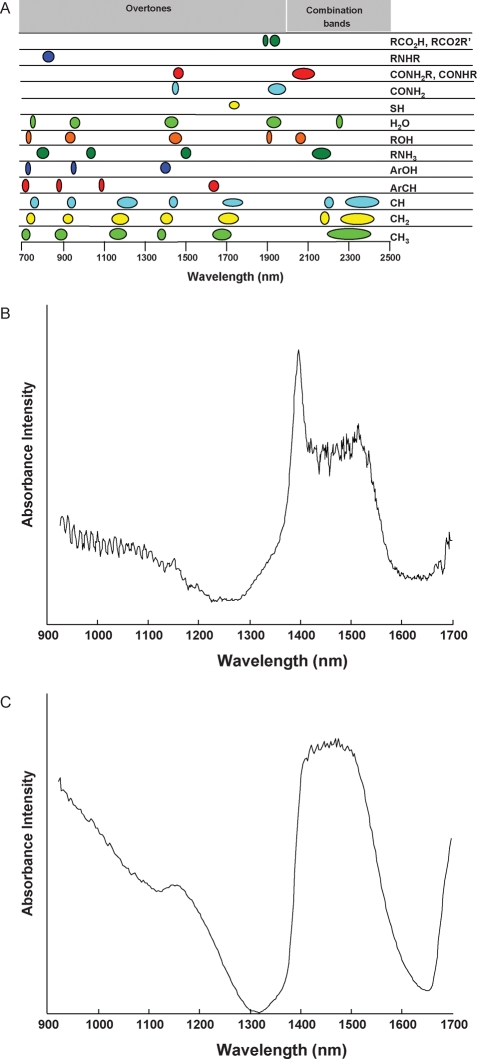
Although FT-IR is used in the vast majority of metabolomic studies, other classes of vibrational spectroscopy are making headway in these applications. Some work has been carried out using NIR spectroscopy (Fig. 2), having significant potential for metabolomics studies sharing many of the same advantages as FT-IR. In particular, NIR spectroscopy has been used to monitor lactate in the blood (Lafrance et al., 2004), and metabolite content of culture media in IVF (Seli et al., 2007). NIR differs from FT-IR in that it predominately measures overtone and combination vibrational frequencies of functional groups (R–H, N–H, C–H, S–H and O–H) (Fig. 2) between 700–2500 nm (or 4000–14 300 cm−1). The vibrational transitions corresponding to overtone and combination bands, as a result of quantum mechanics, are at least an order of magnitude weaker in comparison to fundamental transitions in the FT-IR spectral range. However, relatively low NIR absorbance is advantageous in many respects as many different types of materials (solids, aqueous samples, tissues etc.) may be analyzed without sample preparation. Also, instrumental cost and complexity is reduced compared with IR spectrometers. The main drawback to NIR spectroscopy is that the overtone and combination bands tend to be very broad and overlapping relative to the sharp, well-separated fundamental features found in the IR region (Fig. 2). As a result, multivariate algorithms are required for analysis of NIR spectra to extract chemically relevant information. One such method is to measure NIR spectra of differing sample populations, followed by identification of spectral features that distinguish between them. As such, NIR analysis is not typically used for target metabolite identification, but is used for overall spectral profile comparisons. However, due to the low absorbance and correspondingly longer path length in NIR, multivariate models built from NIR data tend to be more accurate for quantitative purposes than those derived from IR data.
Figure 2:
NIR spectroscopy examples of pure component and biological fluid metabolomic analyses.
(A) Known combination and overtone NIR vibrational absorption bands of organic molecules visible between 700 and 2500 nm. (B) NIR spectrum (900–1700 nm) of 200 mM lactic acid in phosphate buffered saline (PBS). Distinct absorption bands are visible for water (1390–1590 nm) and lactic acid (1650–1700 nm). (C) NIR spectrum (900–1700 nm) of spent IVF culture media (G1; VitroLife AB, Goteborg, Sweden) supplemented with 5% human serum albumin (HSA; Irvine Scientific, Santa Ana, CA, USA). Broad combination and overtone vibrational bands are characteristic of these spectra.
Raman spectroscopy
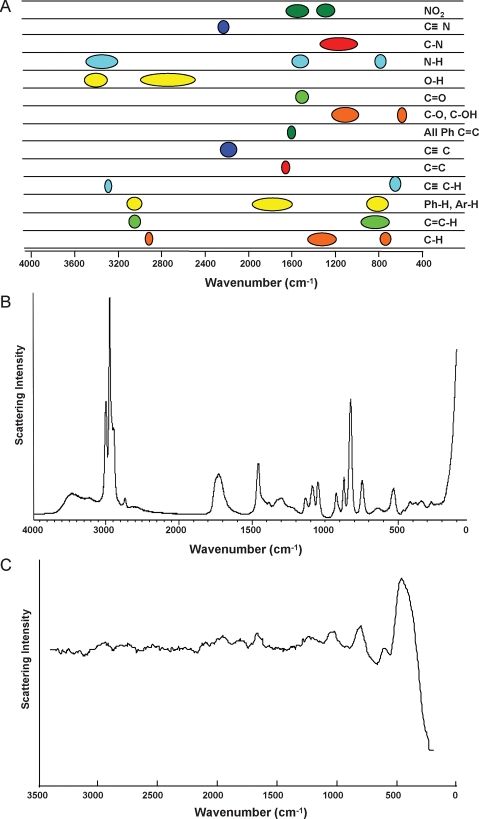
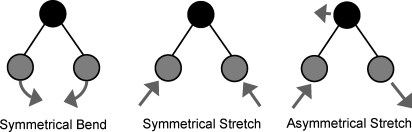
Raman spectroscopy (Fig. 3) is another vibrational technique which finds application in the field of metabolomics. In a Raman experiment, a laser is utilized to promote a vibrational excitation (Stokes) or relaxation (anti-Stokes) along a molecular vibrational mode. By analyzing the scattered light from the laser-illuminated sample, the amount of light absorbed at a number of vibrational frequencies can be deduced. The resulting spectrum contains vibrational fundamental information similar to that seen in FT-IR/IR spectroscopy (400–4000 cm−1, Fig. 3) but with a key difference. IR spectra tend to contain information from asymmetric vibrational modes, while Raman spectra preferentially reflect symmetric vibrational modes (Fig. 4). Thus, IR and Raman are often considered to be perfectly complementary techniques: when combined, they provide information on all of the fundamental vibrational modes of a molecule. An advantage of Raman for metabolomics is that water provides only very weak Raman signals, making analysis of aqueous samples relatively straightforward. However, unlike IR absorbance, Raman scattering is an intrinsically insensitive spectroscopic technique. Some approximations put the odds of Raman scattering in the order of one photon out of every billion hitting the sample (Harris and Bertolucci, 1994). The newest commercial Raman systems contain no moving parts and contain compact, stable laser diode sources, resulting in affordable, robust and portable instruments.
Figure 3:
Raman spectroscopy examples of pure component and biological fluid metabolomic analyses.
(A) Known fundamental IR vibrational absorption bands of organic molecules visible between 400 and 4000 cm−1. (B) Raman spectrum (0–4000 cm−1) of lactic acid. (C) Raman spectrum (0–3400 cm−1) of spent IVF culture media (G1, VitroLife AB) supplemented with 5% human serum albumin (HSA; Irvine Scientific).
Figure 4:
Asymmetrical and symmetrical vibrational modes of water.
Non-invasive metabolomic profiling of embryo culture media by vibrational spectroscopy to determine the viability of individual embryos
Proof-of-concept studies
In 2007, Seli et al. (2007) reported the results of a proof-of-concept study where Day 3 spent culture media of individually cultured embryos with known pregnancy outcome were analyzed using NIR and Raman spectroscopies (Fig. 5, Table IV). Using a multivariate analysis approach, spectral profiles were compared between embryos that resulted in live birth and embryos that failed to implant. Spectral regions that discriminated between the two study populations and were most predictive of pregnancy outcome were identified as markers of oxidative stress, including vibrational modes of –CH, –NH and –OH groups. These spectral regions were then quantified in a multi-linear regression algorithm and expressed as a viability score of each individual embryo's potential to result in a pregnancy and delivery.
Figure 5:
Viability scores calculated using (A) NIR and (B) Raman spectra of culture media are shown for embryos that implanted and lead to delivery (empty), and those that did not implant (shaded).
The implantation index values were significantly different between the two groups assessed by both NIR and Raman spectroscopy (P < 0.001). Modified from Seli et al. (2007).
Table IV.
Studies of non-invasive metabolomic profiling of embryo culture media to assess embryo viability in IVF.
| Study | Study design | n | Day of transfer | Number of embryos transferred | Analytical technique | Center | Findings |
|---|---|---|---|---|---|---|---|
| Seli et al. (2007) | Algorithm development | 36 | Day 3 | MET | Raman | YFC | A |
| Scott et al. (2008) | Blinded analysis | 41 | Day 3 and 5 | MET | Raman | RMANJ | B |
| Seli et al. (2007) | Algorithm development | 33 | Day 3 | MET | NIR | RMANJ | A |
| Seli et al. (2007) | Blinded analysis | 16 | Day 3 | MET | NIR | YFC | B |
| Seli et al. (2008b) | Algorithm development | 121 | Day 2 | SET | NIR | KLC | A, C |
| Seli et al. (2008b) | Blinded analysis | 60 | Day 2 | SET | NIR | KLC | B, D |
| Vergouw et al. (2008) | Algorithm development | 29 | Day 2 | SET | NIR | VUMC | A, C |
| Vergouw et al. (2008) Seli et al. (2008b) | Algorithm development | 304 | Day 3 | SET | NIR | VUMC | A, C |
| Hardarson et al. (2008) | Algorithm development | 137 | Day 5 | SET | NIR | FCG, SG | A, C, D |
SET, single embryo transfer; MET, multiple embryo transfer; YFC, Yale Fertility Center, New Haven, CT, USA; RMANJ, Reproductive Medicine Associates, Morristown, New Jersey, USA; VUMC, Vrije Universiteit Medical Center, Amsterdam, The Netherlands; KLC, Kato Ladies Clinic, Tokyo, Japan; FCG, Fertilitets centrum, Göteborg, Sweden; SG, Shady Grove Fertility Reproductive Science Center, Rockville, Maryland, USA.
A, the mean viability score of embryos that implanted and resulted in fetal cardiac activity or live birth was significantly higher compared with the mean viability score of embryos that failed to implant; B, spectroscopic analysis by an observer blinded to pregnancy outcome, using a previously established regression algorithm demonstrated that the mean viability score of embryos that resulted in a pregnancy was higher compared with embryos that failed to implant; C, study showed the metabolomic profile of embryo culture media to be independent of morphology; D, a positive correlation was detected between increasing viability scores and the potential of individual embryos to result in a pregnancy.
In this initial study (Seli et al., 2007), using both NIR and Raman spectroscopy, the mean viability score (also called viability index) of embryos that implanted and resulted in a live birth was significantly higher (P < 0.01) compared with the mean viability score of embryos that failed to implant (Fig. 5). Raman and NIR spectroscopies achieved a sensitivity of 76.5% and 83.3%, and a specificity of 86% and 75%, respectively. Moreover, the test was rapid (<1 min per sample) and required a very small sample volume (<15 µl).
Subsequently, the regression algorithm for Raman spectra developed by Seli et al. (2007) described above was tested in a blinded trial (Scott et al., 2008) analyzing Day 3 and Day 5 spent media collected at a different IVF center, where embryos were cultured in a different volume (50 µl instead of 30 µl) and type [Quinn's Advantage (SAGE) versus Scandinavian G1 (VitroLife)] of culture medium. In this study, the previously developed algorithm was applied to Raman spectra of embryo culture media and successfully predicted the pregnancy outcome for embryos transferred on Day 3 and Day 5 (P < 0.05).
Validation studies using metabolomic profiling of embryo culture media collected in single embryo transfer cycles
The initial studies described above were followed by studies with of larger sample size from IVF centers that routinely perform single embryo transfer (SET) (Kato et al., 2007; Hardarson et al., 2008; Seli et al., 2008b; Vergouw et al., 2008). In these studies, spent culture media were analyzed by NIR spectroscopy as described previously (Seli et al., 2007), and independent regression algorithms for NIR spectra were developed for embryos that underwent SET on Day 2 (n = 121, Seli et al., 2008b; and n = 29, Vergouw et al., 2008), Day 3 (n = 304, Vergouw et al., 2008) and Day 5 (n = 137, Hardarson et al., 2008). Consistent with the findings of the initial studies (Seli et al., 2007; Scott et al., 2008), the more recent studies using larger sample size and SET model, consistently showed higher mean viability scores for embryos that resulted in a pregnancy with fetal heart activity, compared with those that did not (Kato et al., 2007; Hardarson et al., 2008; Seli et al., 2008b; Vergouw et al., 2008). They also showed that metabolomic profiling of embryo culture media was independent of morphology (Hardarson et al., 2008; Seli et al., 2008b; Vergouw et al., 2008), providing an independent parameter in embryo viability assessment. In addition, retrospective analysis suggested that combining morphology with metabolomic profiling may improve identification of embryos with higher implantation potential (Seli et al., 2008b).
Most recently, spent culture media of 60 embryos collected following SET on Day 2 were evaluated with NIR spectroscopy by an observer blinded to pregnancy outcome, and viability scores were determined using the regression algorithm previously established for Day 2 samples (Seli et al., 2008b). Viability scores of embryos that implanted and resulted in fetal cardiac activity were higher compared with embryos that failed to implant (P < 0.05). In addition, a positive correlation existed between increasing viability score values of individual embryos and their potential to result in a pregnancy (P < 0.001).
Conclusions and future directions
The data summarized above strongly suggest that in vitro cultured embryos that have a high implantation potential alter their environment differently compared to embryos that do not result in a pregnancy, and that the difference is detectable using non-invasive metabolomic profiling of embryo culture media using biospectroscopy. Further investigation is necessary to validate the proposed regression algorithms in different types and volumes of embryo culture media and to determine whether an on-site testing will be clinically valuable in aiding morphologic assessment.
A multitude of advantages may ensue from the use of a rapid, non-invasive and reliable technology as an adjunct for embryo assessment. An improved understanding of embryo viability may help in identifying the embryos that are most likely to result in a pregnancy and allow more accurate decisions to be made about the number of embryos to be transferred. This in turn may reduce the likelihood of multiple gestations while maintaining or even increasing implantation rates. Additional studies will be necessary to determine the value and limitations of the use of metabolomics in IVF practice.
Conflict of interest: L.B. and D.S. are employed by the company developing this technology for clinical application (Molecular Biometrics, LLC). E.S. and D.S. serve on the scientific advisory board of Molecular Biometrics.
Appendix
Table A1.
Glossary of terms.
| Biospectroscopy | The analysis of biological fluids by spectroscopic technologies |
| Capillary electrophoresis | Analytical method of separation in which charged species migrate in a viscous medium under the influence of an electric field. Separation occurs as a result of varying mass-to-charge ratios |
| Chromatography | Family of analytical methodologies for the separation of mixtures |
| Endometabolome | Endogenous (intracellular) metabolites. These include amino acids, amines, sugars, steroids, nucleic acid bases and other substances that are intermediates in cellular metabolism |
| Exometabolome | Exogenous (extracellular) metabolites. This metabolome reflects the influence of the intracellular metabolic network on its external environment, by the uptake of extracellular metabolites and secretion of intracellular metabolites |
| Gas chromatography | Analytical method of separation in which a volatile sample is transported through a column by a flow of an inert, gaseous mobile phase. Separation occurs as a result of components varying affinity for the stationary (liquid) phase in the column |
| Genomics | A systematic study of an organism's genome, including structural genes, regulatory sequences and non-coding DNA segments, and its influence on biological pathways, networks and function |
| Liquid chromatography | Analytical method of separation in which a liquid sample is transported through a column by a flow of a liquid mobile phase. Separation occurs as a result of components varying affinity for the stationary (solid) phase in the column |
| Mass spectrometry | Analytical method of separation and molecular weight determination of sample components. Components are ionized and separated according to their mass-to-charge ratio |
| Metabolite | Low molecular weight molecules (metabolites, typically <1 kDa) present in a biologic environment that are participants in general metabolic reactions and that are required for the maintenance, growth and normal function of a cell |
| Metabolome | Qualitative and quantitative collection of all low molecular weight molecules (metabolites, typically <1 kDa) present in a biologic environment that are participants in general metabolic reactions and that are required for the maintenance, growth and normal function of a cell |
| Metabolomics | A systematic study of the inventory of metabolites—as small molecule biomarkers—that represent the functional phenotype |
| Metabonomics | A quantitative measurement of multi-parametric metabolic responses of multi-cellular systems to pathophysiological stimuli or genetic signaling. Can be interchanged with the term ‘metabolomics’ |
| Non-optical spectroscopy | Analytical spectroscopic/spectrometric technologies that do not measure the interaction of a species with electromagnetic radiation and are based on alternative chemical and physical phenomena |
| Nuclear magnetic resonance spectroscopy | Analytical spectroscopic technique that exploits the physical phenomenon based upon the quantum mechanical magnetic properties of an atom's nucleus. A sample is placed in an external magnetic field and nuclear spin are excited by a resonant radio-frequency. When the radio-frequency is turned off, the nuclei will relax and emit a detectable amount of energy |
| Optical spectroscopy | Optical spectroscopy measures the interaction of a species with electromagnetic radiation—the electromagnetic radiation absorbed, emitted or scattered by the sample analyzed |
| Proteomics | The study encompassing the identification and quantification of proteins, and the effect of their modifications, interactions, activities in a biological system |
| Systems biology | The study and understanding of the interplays of different hierarchies of biological information (DNA, RNA, proteins, macromolecular complexes, signaling networks, cells, organs, metabolic pathways) in a biological system, and how these interactions give rise to the function and behavior of that system |
| Transcriptomics | A systematic study of an organism's transcriptome, the expression level of a complete set of messenger RNA molecules, in understanding the involvement of genes and pathways in biological processes |
| Vibrational spectroscopy | A subclass of optical spectroscopy that measures the absorbance and/or scattering of IR light as a result of molecular vibrational transitions |
Table A2.
Analytical technologies applicable to metabolomics studies with their limitations, advantages and chemical ranges of detection.
| Analytical technology | Description | Chemical range | Advantages | Limitations |
|---|---|---|---|---|
| NMR spectroscopy | Multivariate analysis based on the quantum mechanical magnetic properties of specific atomic nuclei. A sample is placed in an external magnetic field and nuclear spin are excited by a resonant radio-frequency. When the radio-frequency is turned off, the nuclei will relax and emit a detectable amount of energy | Aqueous-based solutions Organic solvent solutions Complex biological mixtures of small molecules and protein-rich samples Detection limits of ∼0.1 mM Most metabolites Examples: carbohydrates (sugars—polysaccharide) Amino acids Lipids, fatty acids Proteins |
Non-destructive analysis of biological fluids and tissue Provides quantitative and structural information simultaneously Capable of extracting chemical information of complex mixtures without the need of separation Increasing sensitivity with increasing magnetic field strength |
Expensive system as high magnetic field strength spectrometers are required Low throughput of samples (depending on experimental conditions) Requires highly trained personnel to execute experiments and to interpret resulting spectra Requires computational software to extract spectral and quantitative information |
| GC–MS | Chromatographic-coupled mass spectrometry. A volatile sample is transported through a column by a flow of an inert, gaseous mobile phase. Separation occurs as a result of components' varying affinity for the stationary (liquid) phase in the column. Mass spectrometry operates by ionization of metabolites, separation of ionic fragments according to their mass-to-charge ratio, and then detection of these separated ions |
Low molecular weight compounds (<350 Da) Volatile and thermally stable molecules Detection limits of ∼µM Examples: some lipids (fatty acids) Amino acids Sugars and sugar alcohols Organic compounds (phosphates, amines, amides, thiols) |
Analytically sensitive (detection limits <µM) Molecular structure determination Metabolite identification through molecular weight determination Analysis of many metabolite classes after derivatization |
Cannot readily analyze polar, non-volatile analytes without derivatization Low throughput (10–60 minutes per analysis) Sample preparation can introduce greater technical variability and data complexity A priori sample knowledge required Requires maintenance and highly trained personnel to execute experiments |
| LC–MS | Chromatographic-coupled mass spectrometry. A liquid sample is transported through a column by a flow of a liquid mobile phase. Separation occurs as a result of components varying affinity for the stationary (solid) phase in the column. Mass spectrometry operates by ionization of metabolites, separation of ionic fragments according to their mass-to-charge ratio, and then detection of these separated ions |
Molecular weights extending from those detectable by GC–MS to molecular weights >600 Da Detection limits of ∼µM Examples: amino acids Peptides Sugars and Glycosides Organic compounds Lipids (fatty acids, phospholipids) Proteins |
Analytically sensitive Can operate at both high and low temperatures Does not require sample volatility Larger chemical dynamic range Limited sample preparation required without the need of derivatization Direct injection possible |
Expensive system Fragmentation data complex and requires computational software to extract structural information Requires maintenance and highly trained personnel to execute experiments |
| CE–MS | Capillary electrophoresis coupled mass spectrometry where charged metabolites are separated by charge-to-mass ratio, in a conductive liquid medium, under the influence of an electric field. Mass spectrometry operates by ionization of metabolites, separation of ionic fragments according to their mass-to-charge ratio, and then detection of these separated ions |
Large proteins Ionic metabolites (organic and inorganic) |
Can resolve polar and ionic samples that may be more difficult with GC–MS or LC–MS Limited sample preparation required Small sample requirement (few µL) |
Low reproducibility Requires conductive liquid medium that may not be compatible with MS Requires maintenance and highly trained personnel to execute experiments |
| FT-IR | Based on the asymmetrical vibrational modes of chemical bonds in a molecule. Electromagnetic radiation absorption causes an increase in characteristic vibrational energy states depending on their chemical structure. Spectra reflect fundamental vibrational transitions |
Most metabolites Examples: Carbohydrates (sugars—polysaccharides) Fatty acids Amino acids Organic compounds Lipids Proteins |
Unknown identification using peak pattern matching against a database Quantitative capabilities Simultaneous analysis of multiple analytes High-throughput method Resolved spectral features (in comparison to NIR) Higher signal intensity (in comparison to NIR) Minimal user training required |
Relatively insensitive and unselective in comparison to MS Challenging aqueous sample analysis. Requires short path length transmission cells and attenuated total reflectance sampling accessories Interferometer systems comprise of moving parts (moving mirror). Requires maintenance |
| NIR | Based on the asymmetrical vibrational modes of chemical bonds in a molecule. Electromagnetic radiation absorption causes an increase in characteristic vibrational energy states depending on their chemical structure. Spectra reflect overtone and combination vibrational bands |
Most metabolites Metabolites with asymmetrical vibrational modes Examples: Carbohydrates (sugars—polysaccharides) Fatty acids Amino acids Organic compounds Lipids Proteins |
Can analyze many types of materials (solids, aqueous samples, tissues etc.) without sample preparation Reduced instrumental complexity and cost NIR data more accurate and quantitative as a result of multivariate algorithm analysis Minimal user training required |
Absorptions are at weaker in magnitude compared with FT-IR Broad, overlapping spectral features that require multivariate algorithm analysis Requires thorough chemical knowledge to interpret spectral results |
| Raman | Analysis of scattered light from laser-illuminated sample, promoting vibrational excitation (Stokes) or relaxation (anti-Stokes) along a symmetrical vibrational mode. Spectra reflect fundamental vibrational transitions (similar to FT-IR) | Most metabolites Symmetrical molecules (that are IR inactive) Examples: Carbohydrates (sugars—polysaccharides) Fatty acids Amino acids Organic compounds Lipids Proteins |
Complementary to IR spectral information, representing additional fundamental vibrational modes Simple analysis of aqueous samples as a result of weak water signal No sample preparation required Raman systems with no moving parts, compact, portable, and inexpensive systems Minimal user training required Useful for symmetric molecule analysis (not IR active) |
Insensitive technique as a result of rare Raman scattering Weak signals require advanced detector amplification Requires laser source, may alter sample properties as a result of Requires thorough chemical knowledge to interpret spectral results |
References
- Biggers JD, Whittingham DG, Donahue RP. The pattern of energy metabolism in the mouse oocyte and zygote. Proc Natl Acad Sci USA. 1967;58:560–567. doi: 10.1073/pnas.58.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock JL. Metabolic profiling of amniotic fluid by proton nuclear magnetic resonance spectroscopy: correlation with fetal maturation and other clinical variables. Clin Chem. 1994;40:56–61. [PubMed] [Google Scholar]
- Brison DR, Houghton FD, Falconer D, Roberts SA, Hawkhead J, Humpherson PG, Lieberman BA, Leese HJ. Identification of viable embryos in IVF by non-invasive measurement of amino acid trunover. Hum Reprod. 2004;19:2319–2324. doi: 10.1093/humrep/deh409. [DOI] [PubMed] [Google Scholar]
- Bromer JG, Seli E. Assessment of embryo viability in assisted reproductive technologies: shortcomings of current approaches and the emerging role of metabolomics. Curr Opin Obstet Gynecol. 2008;20:234–241. doi: 10.1097/GCO.0b013e3282fe723d. [DOI] [PubMed] [Google Scholar]
- Conaghan J, Handyside AH, Winston RM, Leese HJ. Effects of pyruvate and glucose on the development of human preimplantation embryos in vitro. J Reprod Fertil. 1993;a 99:87–95. doi: 10.1530/jrf.0.0990087. [DOI] [PubMed] [Google Scholar]
- Conaghan J, Hardy K, Handyside A, Winston RML, Leese HJ. Selection criteria for human embryo transfer: a comparison of pyruvate uptake and morphology. J Assist Reprod Genet. 1993;b 10:21–30. doi: 10.1007/BF01204436. [DOI] [PubMed] [Google Scholar]
- Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreker F. Uptake and release of metabolites in human preimplantation embryos. In: Cohen J, Elder K, editors. Human Preimplantation Embryo Selection. London, UK: Taylor and Francis; 2007. pp. 325–336. [Google Scholar]
- Dunn WB. Current trends and future requirements for the mass spectrometric investigation of microbial, mammalian and plant metabolomes. Phys Biol. 2008;5:1–24. doi: 10.1088/1478-3975/5/1/011001. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Ellis DI. Metabolomics: Current analytical platforms and methodologies. Trends Anal Chem. 2005;24:285–294. [Google Scholar]
- Edwards R, Fishel S, Cohen J. Factors influencing the success of in vitro fertilization for alleviating human infertility. J In Vitro Fert Embryo Transf. 1984:3–23. doi: 10.1007/BF01129615. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Leese HJ. Assessment of embryo viability prior to transfer by the noninvasive measurement of glucose uptake. J Exp Zool. 1987;242:103–105. doi: 10.1002/jez.1402420115. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Schoolcraft WB. In vitro culture of human blastocysts. In: Mortimer D, Jansen R, editors. Towards Reprodcutive Certainty: Fertility and Genetics Beyond. Carnforth: Parthenon publishing; 1999. pp. 378–388. [Google Scholar]
- Gardner DK, Lane M, Stevens J, Schoolcraft WB. Noninvasive assessment of human embryo nutrient consumption as a measure of developmental potential. Fertil Steril. 2001;76:1175–1180. doi: 10.1016/s0015-0282(01)02888-6. [DOI] [PubMed] [Google Scholar]
- Gerris J, De Neubourg D, Mangelschots K, et al. Prevention of twin pregnancy after in-vitro fertilization or intracytoplasmic sperm injection based on strict embryo criteria: a prospective randomized clinical trial. Hum Reprod. 1999;14:2581–2587. doi: 10.1093/humrep/14.10.2581. [DOI] [PubMed] [Google Scholar]
- Gott AL, Hardy K, Winston RML, Leese HJ. Non-invasive measurement of pyruvate and glucose uptake and lactate production by single human preimplantation embryos. Hum Reprod. 1990;5:104–108. doi: 10.1093/oxfordjournals.humrep.a137028. [DOI] [PubMed] [Google Scholar]
- Hardarson T, Tucker M, Seli E, Botros L, Roos P, Sakkas D. San Francisco, CA: American Society for Reproductive Medicine; 2008. Non-invasive metabolic profiling of day 5 embryo culture media adds to the discriminatory power of blastocyst culture for single embryo transfer (SET) [Google Scholar]
- Hardy K, Hooper MAK, Handyside AH, Rutherford AJ, Winston RML, Leese HJ. Non-invasive measurement of glucose and pyruvate uptake by individual human oocytes and preimplantation embryos. Hum Reprod. 1989;4:188–191. doi: 10.1093/oxfordjournals.humrep.a136869. [DOI] [PubMed] [Google Scholar]
- Harris DC, Bertolucci MD. Symmetry and spectroscopy: an introduction to vibrational and electronic spectroscopy. In: Harris DC, Bertolucci MD, editors. Mineola, NY: Dover Publications; 1989. pp. 90–99. Vibrational spectroscopy. [Google Scholar]
- Houghton FD, Hawkhead JA, Humpherson PG, Hogg JE, Balen AH, Rutherford AJ, Leese HJ. Non-invasive amino acid turnover predicts human embryo developmental capacity. Hum Reprod. 2002;17:999–1005. doi: 10.1093/humrep/17.4.999. [DOI] [PubMed] [Google Scholar]
- Jones GM, Trounson AO, Vella PJ, Thouas GA, Lolatgis N, Wood C. Glucose metabolism of human morula and blastocyst-stage embryos and its relationship to viability after transfer. Reprod Biomed Online. 2001;3:124–132. doi: 10.1016/s1472-6483(10)61980-3. [DOI] [PubMed] [Google Scholar]
- Kato O, Terramoto S, Morita H, Botros L, Roos P, Burns DH. American Society for Reproductive Medicine; 2007. Metabolomic assessment of day 2 embryos based on pregnancy outcome after single embryo transfer (SET) [Google Scholar]
- Katz-Jaffe MG, Schoolcraft WB, Gardner DK. Analysis of protein expression (secretome) by human and mouse preimplantation embryos. Fertil Steril. 2006;86:678–685. doi: 10.1016/j.fertnstert.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Keith LG, Oleszczuk JJ, Keith DM. Multiple gestation: reflections on epidemiology, causes, and consequences. Int J Fertil Womens Med. 2000;45:206–214. [PubMed] [Google Scholar]
- Kovalevsky G, Patrizio P. High rates of embryo wastage with the use of assisted reproductive technology: a look at the trends between 1995 and 2001 in the United States. Fertil Steril. 2005;84:325–330. doi: 10.1016/j.fertnstert.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Lafrance D, Lands LC, Burns DH. In vivo lactate measurement in human tissue by near-infrared diffuse reflectance spectroscopy. Vib Spectrosc. 2004;36:195–202. [Google Scholar]
- Lane M, Gardner D. Amino acids and vitamins prevent culture-induced metabolic perturbations and associated loss of viability of mouse blastocysts. Hum Reprod. 1998;13:991–997. doi: 10.1093/humrep/13.4.991. [DOI] [PubMed] [Google Scholar]
- Lenz EM, Bright J, Knight R, Wilson ID, Major H. Cyclosporin A-induced changes in endogenous metabolites in rat urine: a metabonomic investigation using high field 1H NMR spectroscopy, HPLC-TOF/MS and chemometrics. J Pharm Biomed Anal. 2004;35:599–608. doi: 10.1016/j.jpba.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Lindon J. Metabonomics—Techniques and Applications. Metabometrix Ltd. Business Briefing: Future Drug Discovery. 2004:1–6. [Google Scholar]
- Lu X, Zhao X, Bai C, Zhao C, Lu G, Xu G. LC–MS-based metabonomics analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;866:64–76. doi: 10.1016/j.jchromb.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Luke B, Brown MB. Contemporary risks of maternal morbidity and adverse outcomes with increasing maternal age and plurality. Fertil Steril. 2007;88:283–293. doi: 10.1016/j.fertnstert.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Martin J. The rise in multiple births in the United States: who, what, when, where, and why. Clin Obstet Gynecol. 2004;47:118–133. doi: 10.1097/00003081-200403000-00016. [DOI] [PubMed] [Google Scholar]
- Montona MRN, Soga T. Metabolome analysis by capillary electrophoresis–mass spectrometry. J Chromatogr A. 2007;1168:237–246. doi: 10.1016/j.chroma.2007.02.065. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- Oleszczuk JJ, Oleszczuk AK, Keith DM, Keith LG. Twin and triplet births: facts, figures, and costs. Female Patient. 2003;28:11–16. [Google Scholar]
- Pasikanti KK, Ho PC, Chan EC. Gas chromatography/mass spectrometry in metabolic profiling of biological fluids. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:202–211. doi: 10.1016/j.jchromb.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Pauli GF, Jaki BU, Lankin DC. Quantitative 1H NMR: development and potential of a method for natural products analysis. J Nat Prod. 2005;68:133–149. doi: 10.1021/np0497301. [DOI] [PubMed] [Google Scholar]
- Posillico JT. and the Metabolomics Study Group for Reproductive Technologies. Selection of viable embryos and gametes by rapid, non-invasive metabolomic profiling of oxidative stress biomarkers. In: Cohen J, Elder K, editors. Human Preimplantation Embryo Selection. London, UK: Taylor and Francis; 2007. pp. 325–336. [Google Scholar]
- Reddy UM, Wapner RJ, Rebar RW, Tasca RJ. Infertility, assisted reproductive technology, and adverse pregnancy outcomes. Obstet Gynecol. 2007;109:967–977. doi: 10.1097/01.AOG.0000259316.04136.30. [DOI] [PubMed] [Google Scholar]
- Sakkas D, Gardner DK. Noninvasive methods to assess embryo quality. Curr Opin Obstet Gynecol. 2005;17:283–288. doi: 10.1097/01.gco.0000169106.69881.3e. [DOI] [PubMed] [Google Scholar]
- SART. National summary and fertility clinic reports. USA: Centers for disease control; 1996. Assisted reproductive technology success rates. [DOI] [PubMed] [Google Scholar]
- SART. National summary and fertility clinic reports. USA: Centers for disease control; 2005. Assisted reproductive technology success rates. [DOI] [PubMed] [Google Scholar]
- SART. National summary and fertility clinic reports. USA: Centers for disease control; 2006. Assisted reproductive technology success rates. [DOI] [PubMed] [Google Scholar]
- Scott LA, Smith S. The successful use of pronuclear embryo transfer the day following oocyte retrieval. Hum Reprod. 1998;13:1003–1013. doi: 10.1093/humrep/13.4.1003. [DOI] [PubMed] [Google Scholar]
- Scott RT, Seli E, Miller K, Sakkas D, Scott K, Burns DH. Non-Invasive metabolomic profiling of human embryo culture media using Raman spectroscopy predicts embryonic reproductive potential: A prospective blinded pilot study. Fertil Steril. 2008;90:77–83. doi: 10.1016/j.fertnstert.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Seli E, Sakkas D, Scott R, Kwok JS, Rosendahl S, Burns DH. Non-Invasive metabolomic profiling of human embryo culture media using Raman and near infrared spectroscopy correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil Steril. 2007;88:1350–1357. doi: 10.1016/j.fertnstert.2007.07.1390. [DOI] [PubMed] [Google Scholar]
- Seli E, Botros L, Sakkas D, Burns DH. Non-invasive metabolomic profiling of embryo culture media using proton NMR correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil Steril. 2008;a 90:2183–2189. doi: 10.1016/j.fertnstert.2008.07.1739. [DOI] [PubMed] [Google Scholar]
- Seli E, Vergouw CG, Kato O, Botros L, Roos P, Morita H, Lambalk CB, Yamashita N, Sakkas D. A viability index determined by non-invasive metabolomic profiling of embryo culture media correlates with ART outcome. 2008 European Society for Human Reproduction & Embryology, Barcelona, Spain. [Google Scholar]
- Steptoe PC, Edwards RG. Birth after the reimplantation of a human embryo. Lancet. 1978;2:366. doi: 10.1016/s0140-6736(78)92957-4. [DOI] [PubMed] [Google Scholar]
- Stokes PJ, Hawkhead JA, Fawthrop RK, Picton HM, Sharma V, Leese HJ, Houghton FD. Metabolism of human embryos following cryopreservation: implications for the safety and selection of embryos for transfer in clinical IVF. Hum Reprod. 2007;22:829–835. doi: 10.1093/humrep/del447. (Erratum in: Hum Reprod 2007;22:1196.) [DOI] [PubMed] [Google Scholar]
- Sturmey RG, Hawkhead JA, Barker EA, Leese HJ. DNA damage and metabolic activity in the preimplantation embryo. Hum Reprod. 2008 doi: 10.1093/humrep/den346. Epub ahead of print October 3. [DOI] [PubMed] [Google Scholar]
- Tesarik J, Greco E. The probability of abnormal preimplantation development can be predicted by a single static observation on pronuclear state morphology. Hum Reprod. 1999;14:1318–1323. doi: 10.1093/humrep/14.5.1318. [DOI] [PubMed] [Google Scholar]
- Toner JP. Progress we can be proud of: U.S. trends in assisted reproduction over the first 20 years. Fertil Steril. 2002;78:943–950. doi: 10.1016/s0015-0282(02)04197-3. [DOI] [PubMed] [Google Scholar]
- Trounson A, Leeton J, Wood C, Webb J, Wood J. Pregnancies in humans by fertilization in vitro and embryo transfer in the controlled ovulatory cycle. Science. 1981;216:681–682. doi: 10.1126/science.7221557. [DOI] [PubMed] [Google Scholar]
- Turner K, Martin KL, Woodward BJ, Lenton EA, Leese HJ. Comparison of pyruvate uptake by embryos derived from conception and non-conception natural cycles. Hum Reprod. 1994:2362–2366. doi: 10.1093/oxfordjournals.humrep.a138453. [DOI] [PubMed] [Google Scholar]
- VanRoyan E, Mangelschots K, De Neubourg D, Valkenburg M, Van de Meerssche M, Ryckaert G, Eestermens W, Gerris J. Characterization of a top quality embryo, a step towards single-embryo transfer. Hum Reprod. 1999;14:2345–2349. doi: 10.1093/humrep/14.9.2345. [DOI] [PubMed] [Google Scholar]
- Veeck L. An Atlas of Human Gametes and Conceptuses: An Illustrated Reference for Assisted Reproductive Technology. New York, USA: Parthenon Publishing; 1999. [Google Scholar]
- Vergouw CG, Botros LL, Roos P, Lens JW, Schats R, Hompes PGA, Burns DH, Lambalk CB. Metabolomic profiling by near-infrared spectroscopy as a tool to assess embryo viability: a novel, non-invasive method for embryo selection. Hum Reprod. 2008;23:1499–1504. doi: 10.1093/humrep/den111. [DOI] [PubMed] [Google Scholar]
- Williams RE, Lenz EM, Lowden JS, Rantalainen M, Wilson ID. The metabonomics of aging and development in the rat: An investigation into the effect of age on the profile of endogenous metabolites in the urine of male rats using 1H NMR and HPLC-TOF MS. Mol Biosyst. 2005;1:166–175. doi: 10.1039/b500852b. [DOI] [PubMed] [Google Scholar]
- Wilsona ID, Plumbb R, Grangerb J, Majorc H, Williamsa R, Lenza EM. HPLC-MS-based methods for the study of metabonomics. J Chromatogr B. 2005;817:67–76. doi: 10.1016/j.jchromb.2004.07.045. [DOI] [PubMed] [Google Scholar]
- Wright VJC, Jeng G, Macaluso M. Assisted reproductive technology surveillance—United States, 2003. MMWR Surveill Summ. 2006;55:1–22. [PubMed] [Google Scholar]