Abstract
One of the most important factors influencing embryo viability is chromosome imbalance (aneuploidy). Embryos derived from aneuploid gametes have little potential for forming a viable pregnancy, but cannot be distinguished from normal embryos using standard morphological evaluation. For more than a decade, preimplantation genetic screening (PGS) has been used to assist in the identification of aneuploid embryos. However, current strategies, based upon cell biopsy followed by fluorescent in situhybridization, allow less than half of the chromosomes to be screened. In this review, we discuss methods that overcome the limitations of earlier PGS strategies and provide screening of the entire chromosome complement in oocytes and embryos. In recent months, there has been a rapid growth in the number of PGS cycles utilizing one such method, comparative genomic hybridization (CGH). Data from IVF cycles utilizing CGH must be considered preliminary, but appear to indicate a dramatic increase in embryo implantation following comprehensive chromosomal screening. It is expected that methods based upon microarrays will yield similar clinical results and may be sufficiently rapid to permit comprehensive screening without the need for embryo cryopreservation. Some microarray platforms also offer the advantage of embryo fingerprinting and the potential for combined aneuploidy and single gene disorder diagnosis. However, more data concerning accuracy and further reductions in the price of tests will be necessary before microarrays can be widely applied.
Keywords: comparative genomic hybridization, aneuploidy, preimplantation genetic screening, in vitro fertilization, preimplantation genetic diagnosis
The need for embryo viability assessment
The use of controlled ovarian stimulation in assisted reproductive treatment (ART) cycles generally results in the production of multiple mature oocytes. Fertilization rates are usually high and consequently it is typical for several embryos to be produced each treatment cycle. In order to maximize the probability of obtaining a pregnancy, most IVF cycles involve the transfer of more than one embryo. Although this approach increases the likelihood of obtaining a pregnancy, it also leads to an elevated risk of multiple gestation (twins, triplets, etc). Multiple pregnancies are associated with significantly elevated risks of serious complications for both the mother and the children. For example, mothers have an increased incidence of pre-eclampsia, post-partum hemorrhage, hysterectomy and gestational diabetes (Walker et al., 2004), whereas infants are at greater risk of low birthweight, preterm delivery, cerebral palsy and congenital malformations (Bergh et al., 1999; Stromberg et al., 2002; Pinborg et al., 2003). Concern over the high incidence of multiple pregnancies in ART cycles has led to increasing pressure on physicians to restrict the number of embryos transferred to the uterus.
Not surprisingly, decreasing the number of embryos transferred to the uterus is extremely effective at reducing multiple pregnancy rates (Gerris, 2007; Sunde, 2007). However, the transfer of fewer embryos is also likely to have an adverse impact on ART pregnancy rates. The problem stems from the fact that there is great heterogeneity in the viability of individual human embryos generated in vitro. In order to maintain satisfactory pregnancy rates, while reducing the number of embryos transferred, it is essential to identify the embryos having the greatest potential for pregnancy formation and insure that these embryos are given priority for transfer. The most common strategy for the identification of viable embryos is based upon assessment of morphological criteria, such as cell size and number, presence of multinucleation, percentage of fragmentation and cleavage rate (Cummins et al., 1986; Puissant et al., 1987; Van Royen et al., 1999; reviewed in Sakkas and Gardner, 2005). Morphological evaluation is utilized in all IVF laboratories and remains the mainstay of embryo assessment. However, some of the most important aspects of embryo viability remain invisible to such analyses.
Aneuploidy in oocytes and embryos
Many morphologically normal embryos do not achieve implantation or spontaneously abort during pregnancy. In most cases, the underlying basis of this failure remains unknown. However, it is likely that many unsuccessful IVF attempts can be explained by the presence of numerical chromosomal abnormalities (aneuploidy). The high prevalence of aneuploidy in human oocytes and embryos has long been recognized (Angell et al., 1983; Veiga et al., 1987; Delhanty et al., 1993; Kamiguchi et al., 1993; Munné et al., 1993) and the developmental impact of these anomalies, at least those of meiotic origin, is well documented. Early data from Hassold and others demonstrated that at least 50% of first trimester spontaneous miscarriages are chromosomally abnormal (Hassold and Jacobs, 1984), whereas more recent studies employing molecular cytogenetic techniques suggest the true incidence of aneuploidy may be even higher, perhaps exceeding 65% (Menasha et al., 2005). Data from prenatal samples and spontaneous miscarriages have indicated a dramatic increase in chromosomally abnormal pregnancies with advancing maternal age. These data are mirrored by findings from the direct cytogenetic analysis of human oocytes, which demonstrates oocyte aneuploidy rates in excess of 50% for many women over 40 years of age (Sandalinas et al., 2002; Kuliev et al., 2003; Pellestor et al., 2003; Gutiérrez-Mateo et al., 2004; Fragouli et al., 2006; Hassold et al., 2007; Sher et al., 2007).
Given that chromosome imbalance of meiotic origin is common, and almost always lethal to the embryo or fetus, it has been suggested that screening oocytes or embryos for chromosome abnormalities could greatly assist the identification of the most viable embryos. Such an approach is expected to be particularly beneficial for patients predisposed to the production of large numbers of aneuploid gametes, such as those of advanced maternal age (AMA). Theoretically, aneuploidy screening and preferential transfer of euploid embryos should lead to improved pregnancy rate, decreased miscarriage rate and reduced risk of aneuploid syndromes, such as Down, Edwards, Patau, Klinefelter and Turner (Munné et al., 1993).
In recent years, several groups have initiated research aimed at identifying non-invasive markers of chromosome imbalance. Some investigators have detected characteristic transcriptional changes in the cumulus cells attached to aneuploid oocytes (Fragouli et al., 2007), whereas others have identified alterations in the molecules secreted by aneuploid embryos (i.e. the secretome) (Katz-Jaffe et al., 2006). Another study discovered several genes, producing cell surface or secretory proteins, which display aberrant expression in chromosomally abnormal oocytes (Wells et al., 2006). Although these studies have revealed some promising marker genes and proteins, further validation is necessary, and a widely available clinical test still seems some way off.
Currently, the only reliable methods for aneuploidy detection require biopsy of material from the oocyte or embryo. Information concerning the chromosomes of oocytes can be obtained by analysis of the associated polar bodies (Kuliev and Verlinsky, 2005), while embryos can be assessed by testing single blastomeres biopsied at the cleavage stage (Day 3 post-fertilization) (Gianaroli et al., 1997; Munné et al., 1999) or by analysis of several cells removed from the trophectoderm at the blastocyst stage (Day 5 post-fertilization) (McArthur et al., 2005, 2008).
The application of conventional techniques of chromosome preparation (karyotyping) to human preimplantation embryos has proven to be difficult and unsatisfactory. In most cases, only one cell is available for analysis, greatly reducing the likelihood of obtaining the high-quality metaphase chromosomes essential for chromosome banding studies. Indeed, biopsied blastomeres are almost always in interphase, with the chromosomes contained within the nucleus rather than visible as distinct entities. On the rare occasions when blastomeres in metaphase are obtained, the chromosomes tend to be highly contracted, a morphology unsuitable for traditional karyotyping methods. Morphology is similarly poor for chromosomes derived from polar bodies. For these reasons, molecular cytogenetic techniques, especially fluorescence in situ hybridization (FISH), have become the methods of choice for the analysis of chromosomes from embryos (Munné et al., 1993, 1997; Delhanty et al., 1993; Gianaroli et al., 1997).
The use of FISH for screening oocytes and embryos
FISH with chromosome-specific DNA probes can be applied to single cells and gives detectable signals in interphase nuclei as well as on metaphase chromosomes. The first application of this technology to human blastomeres was demonstrated by Griffin et al. (1992). Since that time, thousands of IVF patients have had their embryos screened for aneuploidy using FISH, a process variously termed preimplantation genetic screening (PGS), preimplantation genetic diagnosis for aneuploidy screening (PGD-AS) or PGD for infertility. Most of these patients are considered to be at elevated risk of producing aneuploid embryos due to AMA, repeated implantation failure or recurrent miscarriages. Several groups have reported a decrease in spontaneous miscarriages (Munné et al., 2005; Schoolcraft et al., 2008a) and/or an increase in the implantation and live birth rates per embryo transfer following these procedures (Munné et al., 1999, 2003, 2005; Gianaroli et al., 1999).
Although several FISH studies have yielded positive clinical data, other reports have suggested that PGS using this method does not lead to improved IVF outcomes, at least in terms of pregnancy/birth rate per treatment cycle (Staessen et al., 2004; Mastenbroek et al., 2007). Some of the disparities in the published data may be a consequence of differences in patient selection, methodology or limitations of the FISH technique itself (Munné et al., 2007a,b). The number of chromosomes screened and the choice of which chromosomes to assess are both vital aspects of the method, yet show significant variation between the studies. Additionally, it is essential that the accuracy rate and the proportion of cells producing a diagnostic result are high, which has not always been the case for published FISH studies (Mastenbroek et al., 2007).
Although embryo screening using well-optimized FISH methods may lead to improved outcomes for appropriately indicated patients, it is acknowledged that this approach does have some technical limitations. First, relatively few chromosomes can be assessed, as only a limited number of spectrally distinct fluorochromes (colors) are available for the labeling of DNA probes. To get around this limitation, multiple rounds of FISH can be employed, analyzing one set of probes then washing them off and recycling the same colors in order to assess a different set of chromosomes. However, the accuracy of the FISH analysis decreases with each additional round of hybridization and for this reason it is inadvisable to perform more than two or three sequential rounds of FISH. The most comprehensive FISH methods currently used for routine embryo screening assess approximately half of the chromosomes and it is therefore inevitable that some abnormal embryos remain undetected. Another limitation of the FISH procedure is that it is dependent on fixation of a single cell onto a microscope slide, a procedure that requires skill and experience. The spreading of a cell on a slide can lead to overlapping or split signals, which are difficult to score correctly, necessitating an additional round of confirmatory FISH analysis using an alternative probe for the questionable chromosome, a process that has been termed no result rescue (Colls et al., 2007).
Comparative genomic hybridization
In 1996, a molecular cytogenetic method allowing the simultaneous enumeration of all of the chromosomes in a single cell was developed and applied to blastomeres for the first time (Wells and Delhanty, 1996). The method was based upon comparative genomic hybridization (CGH), a technique originally developed for the evaluation of chromosomal losses and gains in tumors (Kallioniemi et al., 1992). CGH is a DNA-based method, which is applicable to cells in any phase of the cell cycle and avoids fixation and spreading. The technique employs a competitive hybridization of differentially labeled DNA samples (DNA from the sample: green; chromosomally normal reference DNA: red) to normal metaphase chromosomes on a microscope slide. Fragments of the red (reference) and green (sample) DNAs anneal to their complementary sequences on the chromosomes, such that each chromosome becomes coated with thousands of red and green DNA fragments (Fig. 1). The ratio of green:red fluorescence along the length of each chromosome reveals the relative number of chromosome copies in the test sample compared with the reference (Kallioniemi et al., 1992). An excess of green fluorescence on a specific chromosome is indicative of a chromosomal gain (e.g. a trisomy), whereas an excess of red fluorescence is indicative of chromosome loss. The CGH method requires ∼1 µg of DNA whereas a single cell contains only 5–10 pg, and for this reason it is necessary to amplify the entire genome of cells prior to CGH analysis. Although various methods can be employed for this purpose, the most widely used is a technique based upon the annealing of semi-degenerate primers followed by polymerase chain reaction (PCR) mediated amplification, known as degenerate oligonucleotide primed (DOP) PCR (Telenius et al., 1992; Wells et al., 1999).
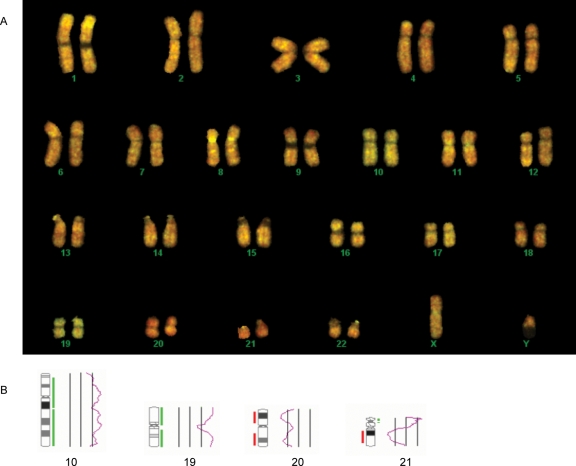
Figure 1:
Clinical screening of a human blastocyst using CGH.
(A) Normal metaphase chromosomes hybridized with test and reference DNAs. The test DNA (green) was composed of amplified material derived from biopsied trophectoderm cells. The reference DNA (red) was derived from a chromosomally normal male. (B) Ratio profiles for chromosomes 10, 19, 20 and 21, revealing additional copies of chromosomes 10 and 19 and loss of chromosomes 20 and 21. The most likely karyotype for this embryo is 46,XY,+10,+19,−20,−21.
Studies applying CGH to the analysis of embryos provided fascinating data on the variety and frequency of chromosome abnormalities in human embryos and confirmed that aneuploidy can affect any chromosome during human preimplantation development, including the largest chromosomes. Some of the aneuploidies detected are of types never observed in prenatal samples or material from spontaneous miscarriages and are presumed to cause developmental arrest during the preimplantation phase, implantation failure or extremely early miscarriage. CGH was also capable of detecting some abnormalities not readily detectable using FISH, including chromosome breakage (Wells and Delhanty, 2000; Voullaire et al., 2000).
Analysis of the published CGH data reveals that 20–40% of embryos carry chromosome abnormalities that would not be detectable using commercially available FISH screens used for PGS (Wells and Delhanty, 2000; Voullaire et al., 2002; Wilton et al., 2003). The fact that current aneuploidy screening protocols are not 100% successful at preventing the transfer of aneuploid embryos potentially reduces their effectiveness as a method for embryo evaluation. However, CGH has confirmed that not all chromosomes have an equal risk of aneuploidy. To date, FISH panels have tended to focus on screening of the chromosomes most often aneuploid in prenatal samples and miscarriages, but it is clear from CGH results that these chromosomes are not necessarily the most important in terms of preimplantation aneuploidy. Recently, a new panel of FISH probes has been introduced for embryo screening, based upon the 10 chromosomes most often shown to be aneuploid in embryos using CGH. It is estimated that this strategy could allow the detection of over 85% of aneuploid embryos (Reprogenetics, unpublished).
Although it is possible to achieve high detection rates for aneuploid embryos without screening the entire chromosome complement, it is inevitable that maximum accuracy will only be achieved when all the chromosomes are evaluated. For this reason, the application of CGH to the screening of oocytes and embryos is an attractive possibility. The principal difficulties in using CGH clinically are the length of time required for the method (∼4 days), which is incompatible with the restricted timeframe available for preimplantation testing, and the high complexity of the technique. One early attempt at clinical application focused on the use of an accelerated CGH protocol applied to first polar bodies biopsied on the day of fertilization (Day 0). This strategy allowed results to be obtained by Day 3 post-fertilization and was therefore compatible with a fresh embryo transfer (Wells et al., 2002). However, aneuploidies arising in meiosis II, as well as those of paternal origin, could not be detected using this approach. More recently, pregnancies have been obtained after comprehensive chromosome screening of polar bodies using CGH, combined with diagnosis of single gene disorders. For this purpose, the first polar body was biopsied and analyzed using CGH and a single blastomere was biopsied on Day 3 and used for single gene testing (Obradors et al., 2008).
An alternative approach to polar body analysis utilized Day 3 blastomere biopsy coupled with cryopreservation (Wilton et al., 2001, 2003; Voullaire et al., 2002). In this case, embryos remained frozen while CGH analysis was carried out, with those diagnosed normal thawed and transferred in a subsequent cycle. This strategy was successfully clinically applied, leading to multiple births for a variety of infertile patients. The main drawback of this approach is that conventional freezing and thawing methods lead to a reduction in embryo implantation potential, a problem exacerbated by embryo biopsy.
Although initial results from clinical CGH studies showed promise, relatively few cases were performed and interest in this approach appeared to be on the wane. However, the recent development of vitrification techniques has provided a means to cryopreserve biopsied oocytes and embryos without a significant decline in survival rates, removing the principal bar to the clinical application of CGH (Sher et al., 2007, Fragouli et al., unpublished). This advance has resulted in resurgence in cases of PGS employing CGH. New CGH strategies, utilizing vitrification, are now being clinically applied for oocyte, cleavage stage and blastocyst screening on a large scale. Our most recent blastocyst screening data are suggestive of a remarkable improvement in embryo implantation and pregnancy rates (Wells et al., 2008; Schoolcraft, Fragouli et al., 2008b). In a prospective trial, involving patients of mean maternal age 37.5, with at least one previous failed IVF attempt (mean 1.8), the proportion of CGH cycles resulting in a live birth was 80%. This compared with 60% for patients without aneuploidy screening. The probability of an individual transferred embryo forming a pregnancy was 66.7% for the CGH group compared with 27.9% without screening (P = 0.00027). These data suggest that the technical difficulties affecting earlier chromosome screening methods are successfully overcome using blastocyst CGH, allowing preimplantation aneuploidy screening to finally achieve the clinical potential predicted by theory. The highly significant improvement in the probability of a transferred embryo forming a pregnancy indicates that comprehensive chromosomal screening will be extremely useful in supporting efforts to maintain high pregnancy rates while reducing the number of embryos transferred per cycle.
Although the use of vitrification has removed the most significant obstacle to the clinical application of CGH, the complexity of the method remains a challenge for the laboratory responsible for the genetic analysis. The CGH technique requires expertise in both molecular genetic and cytogenetic methods that are not generally available to fertility clinics. Thus, widespread clinical application is dependent on the use of highly specialized reference centers. Furthermore, the need for cryopreservation may reduce the clinical options available to patients and physicians. A more rapid technique, requiring less molecular genetic and cytogenetic know-how, could be advantageous.
Comparative genomic hybridization using microarrays
Currently, the best hope for a simplified, rapid method of comprehensive chromosomal screening is the microarray. As with conventional CGH, most microarray methods involve the competitive hybridization of differentially labeled test and reference DNA samples. However, in this case, the labeled DNAs are hybridized to DNA probes affixed to a microscope slide rather than metaphase chromosomes. Each probe is specific to a different chromosomal region and occupies a discrete spot on the slide. Chromosomal loss or gain is revealed by the color adopted by each spot after hybridization (i.e. ratio of fluorescence intensity for the two colors). Microarrays have an advantage over conventional CGH in that the evaluation of fluorescence ratios is simple and easily automated and that the time required for hybridization is generally less.
Microarray CGH has been successfully applied for the detection of aneuploidies in single cells after whole genome amplification (WGA) using DOP-PCR or an alternative method known as multiple displacement amplification (MDA) (Hu et al., 2004; Wells et al., 2004; Le Caignec et al., 2006). These approaches have permitted comprehensive chromosome analysis to be achieved in <48 h, within the timeframe necessary for oocyte or cleavage stage embryo screening without cryopreservation. It is very likely that microarray methods can be further accelerated, possibly allowing screening of embryos at even later stages (i.e. blastocysts), when the window available for analysis is even narrower. In preliminary experiments, we have managed to obtain array-CGH results within 10 h of receiving samples. However, further research is needed to establish whether accuracy rates are maintained at acceptable levels when hybridization times are reduced.
Microarray CGH using bacterial artificial chromosome probes
There are a variety of alternative microarray-CGH platforms available for chromosomal screening and several different methods of WGA that could be used in conjunction with them. Some researchers have focused on the use of bacterial artificial chromosome (BAC) arrays, consisting of thousands of spots, each of which comprises DNA fragments covering relatively large fragments of chromosome (typically in the region of 150–200 kb). Most, BAC-arrays contain a few thousand individual probes (BAC clones), fewer than some alternative microarray platforms, but more than enough for the principal objective of preimplantation embryo screening: the detection of losses and gains affecting entire chromosomes.
The sizes of the probes are sufficiently large that each spot on the slide hybridizes many amplified DNA fragments derived from the region encompassed by the BAC. This is analogous to conventional CGH, where large numbers of test and reference DNA fragments anneal to all chromosomal regions. The annealing of multiple distinct DNA fragments, derived from the same region of the chromosome, reduces the influence of technical artifacts caused by preferential amplification and allele dropout (ADO). These problems frequently affect the amplification of individual DNA fragments in single cells and could produce false losses and gains of chromosomal material. However, in the case of BAC probes, each spot on the microarray provides an average result for the hundreds or thousands of amplified fragments annealed to it, diluting the effect of individual fragments with anomalous amplification.
The main drawbacks of BAC arrays are on the manufacturing side, where the nature of the probes and the way BAC arrays are fabricated can lead to batch-to-batch variation in performance and problems with reproducibility. For this reason, the manufacture of BAC arrays is best left to expert laboratories specializing in this technology. We have observed that some BAC clones do not perform reliably when used in conjunction with amplified material, highlighting the need for extensive validation experiments in order to identify and eliminate poorly performing probes. However, even after problematic BACs have been removed from the microarray, not all probes are expected to give accurate results in every experiment. Given the nature of WGA from a single cell, some random fluctuations in the test and reference fluorescence ratios are expected. For this reason, diagnoses cannot be based on the result from a single BAC, but must depend on an average fluorescence ratio obtained by pooling the data from several neighboring probes. The need to combine results limits the potential resolution of BAC arrays applied to single cells and increases the number of probes needed to obtain an accurate result from each chromosome.
Over the last few years, a variety of single cells, including polar bodies and blastomeres, have been successfully screened using BAC arrays (Wells et al., 2004; Stuerwald et al., 2007; Sanchez-Garcia et al., unpublished). It appears that most forms of WGA are compatible with this type of platform, although some caution should be exercised when utilizing DOP-PCR, as this method is also often used to enrich the DNA from the BAC clones as part of the microarray fabrication process. A commercially available array for screening human oocytes and embryos is now in the final stages of preclinical testing, with release expected toward the end of 2008 (Reprogenetics, unpublished data).
Microarray CGH using chromosome libraries
Another type of microarray, which has yielded aneuploidy data from single cells, utilizes chromosome-specific DNA libraries as probes. DNA samples derived from individual chromosomes can be obtained by microdissection (i.e. the removal of chromosomal material from a slide using micromanipulation techniques) or in some cases by flow sorting. The samples of chromosomal DNA are amplified using DOP-PCR and the repetitive sequences eliminated by negative subtraction hybridization with Cot-1 DNA and, for some chromosomes, centromere-specific repeat sequences. Following another round of amplification, DNA from each of the chromosome-specific libraries can be spotted onto the microarray (Hu et al., 2004).
This approach is promising for single cell analysis, as each spot on the microarray is composed of an extremely heterogeneous mixture of DNA fragments, derived from many sites along the length of each chromosome. As with BAC probes, the influence of DNA fragments affected by uneven amplification is likely to be averaged out and their negative effect on the data eliminated. The fact that each chromosome-specific probe is depleted of repetitive sequences should also improve the reliability of the diagnosis. Hybridization of repetitive DNA elements is a problem for both microarray-based and conventional CGH techniques, as some repeats are not entirely blocked by competitive in situ suppression with Cot-1 DNA. The number of repetitive elements on individual chromosomes can vary from patient to patient, producing spurious changes in the test:reference fluorescence ratio. This sometimes leads to difficulties enumerating specific chromosomes, particularly 19 and 22. With less repeat sequences in the probes, this microarray platform should be less susceptible to such problems.
Microarrays utilizing chromosome libraries are likely to be less expensive than alternative microarray platforms and may also turn out to be more diagnostically robust. However, there is one drawback, specifically a lack of resolution. While a chromosome library approach is sufficient for detecting aneuploidy affecting entire chromosomes, it will struggle to detect losses and gains involving smaller chromosome regions. De novo abnormalities of chromosome structure have been detected in ∼8% of human embryos (Fragouli et al., 2008). Although the clinical significance of these anomalies remains to be confirmed, they are generally considered to be a negative indicator of embryo viability and thus their detection is desirable. An inability to detect loss/gain of smaller chromosomal regions will also preclude application of this strategy to patients carrying reciprocal translocations (a structural chromosome rearrangement characterized by an exchange of material between two chromosome arms). Such patients produce a high proportion of embryos with aneuploidy affecting the chromosomal fragments involved and are frequently referred for PGD.
Microarray CGH using oligonucleotide probes
A third variety of microarray platform utilizes oligonucleotides, which are synthesized in situ, directly on the surface of the solid support (i.e. the slide) that forms the base for the microarray. The manufacturing processes employed allow the production of high-density arrays with very consistent probe performance. Depending on the manufacturer, the probes typically vary from 25 to 85 nucleotides in length. One of the most promising oligonucleotide arrays is that offered by Agilent, utilizing probes 60 nucleotides in length. CGH analyses performed using Agilent arrays have successfully detected aneuploidy in single cells isolated from chromosomally abnormal cell lines, as well as in blastomeres derived from human embryos (Le Caignec et al., 2006). More recently, Agilent arrays have been used clinically, for the purpose of PGS, resulting in several pregnancies (Hellani et al., 2008). This represents the first successful clinical application of microarray technology to the detection of aneuploid embryos and confirms that array-CGH can permit rapid, comprehensive chromosome screening, without the need to cryopreserve embryos. The only drawback of using oligonucleotide arrays is that the small size of individual probes increases the risk that artifactual losses and gains will be seen, caused by errors introduced during WGA. It is possible to compensate for this problem by analyzing large numbers of probes, but this may increase the cost of the microarrays. Our data and that of others suggest that the Agilent perform is compatible with single cell array-CGH used in conjunction with several WGA techniques, including MDA and the Genomeplex method commercialized by Sigma (Le Caignec et al., 2006; Hellani et al., 2008; Alfarawati and Wells, unpublished).
Single nucleotide polymorphism microarrays
Another form of oligonucleotide array, capable of detecting aneuploidy, is based upon the analysis of single nucleotide polymorphisms (SNPs). SNPs are common polymorphic DNA sequences found throughout the genome. Most SNP-microarrays interrogate between 10 000 and 500 000 individual SNPs, located at numerous positions along the length of each chromosome. Although all of the microarray platforms discussed previously utilize an approach similar to conventional CGH, involving simultaneous hybridization of differentially labeled test and reference DNAs to the same microarray, SNP-microarrays employ a somewhat different strategy. Rather than being combined with the normal reference DNA, the amplified material from the test sample is hybridized separately, with reference DNA samples assessed in parallel. As test and reference DNAs hybridize to separate areas of the slide, both can be labelled with the same fluorochrome. Chromosomal copy number is calculated in two ways. Firstly, the alleles detected at each SNP locus are compared with those of the parents, revealing which parental chromosomes were inherited by the embryo. The inheritance of three distinct chromosomal haplotypes indicates the presence of a trisomic chromosome, while monosomies are revealed by homozygosity for all loci on the affected chromosome. Secondly, fluorescence intensities obtained for test and reference hybridizations are compared. If probes from a given chromosome display brighter signals for the test DNA than the reference DNA, an excess of chromosomal material (e.g. trisomy) is predicted. Conversely, reduced fluorescence for the test sample is associated with chromosome loss. Preliminary data suggests that aneuploidy screening using this strategy is accurate, although some questions remain concerning the reliability of trisomy detection. Not all trisomies are caused by the presence of three genetically distinct copies of the same chromosome (i.e. in some instances two of the three copies are identical). In such cases, detection by SNP-array is solely dependent on changes in fluorescence intensity. More data is required to determine the true accuracy of trisomy detection using SNP-arrays in a clinical scenario.
A significant advantage of SNP-microarrays is that the probes used provide genotype data in addition to chromosome copy number information. The simultaneous analysis of thousands of polymorphisms scattered throughout the genome produces a unique DNA fingerprint for each embryo tested. The DNA fingerprint allows parental origin to be confirmed, reducing the risk that a laboratory error could lead to embryos being transferred to the wrong patient. A further benefit of embryo fingerprinting is that any children born following the procedure can be tested and their fingerprints matched with those of the embryos transferred. This allows the embryo(s) that successfully produced children to be traced, providing an extremely powerful tool for research studies aimed at identifying factors that affect embryo implantation potential.
One of the most widely used SNP-microarrays is produced by Affymetrix. This platform employs short probes, 25 nucleotides in length, which are synthesized on the surface of the array using photolithographic techniques. A software package (CNAT, Affymetrix) calculates chromosomal copy number based on fluorescence intensity of probes mapping to each chromosome. An alternative platform is that offered by Illumina. Rather than directly affixing SNP-specific probes to a slide, or synthesizing oligonucletides in situ, the Illumina strategy involves coating 3 µm silica beads with hundreds of thousands of copies of an oligonucleotide probe. The beads are held in microwells on either of two substrates: fiber optic bundles or silica slides. As with Affymetrix microarrays, aneuploidy can be detected by analyzing the signal intensity for probes derived from each chromosome. Additionally, both platforms generate DNA fingerprints, allowing embryo identification.
At the time of writing, no clinical cases of PGD or PGS utilizing SNP-microarrays have been reported in the literature. However, encouraging data from preclinical studies has begun to emerge. The Affymetrix 250K platform, an array comprising 250 000 different SNPs dispersed across all chromosomes, has been employed for the analysis of single blastomeres, successfully detecting several aneuploid chromosomal configurations in non-transferred embryos (Treff et al., 2007). Additionally, Scott et al. have presented interesting data from a prospective ‘non-selection’ trial using the same platform (Scott et al., 2008). In that study, cells were biopsied from preimplantation embryos and assessed using SNP microarrays, providing a reliable DNA fingerprint and revealing the presence of aneuploidy. Unfortunately, the chromosome screening results were not available until after transfer to the uterus had taken place and could not therefore be used to assist embryo selection. Upon analysis of the microarray data, it was later discovered that some of the embryos transferred had been aneuploid. Fetal DNA was obtained from any pregnancies that resulted and a DNA fingerprint produced, allowing the embryos that formed pregnancies to be identified. Remarkably, the negative predictive value of aneuploidy screening was found to be 100% (0/31 embryos with aneuploidy detected in the biopsied cell formed an ongoing pregnancy, 26 failing to implant and 5 miscarrying). The positive predictive value was also encouraging, 42.9% of embryos diagnosed normal formed an ongoing pregnancy. The fact that positive predictive value fell short of 100% emphasizes that, while extremely important, aneuploidy is not the only problem impacting embryo viability and pregnancy.
As well as the promising data obtained using Affymetrix microarrays, Illumina SNP-microarrays have also yielded encouraging results. Analysis of ∼370 000 SNPs scattered throughout the genome has permitted accurate detection of chromosomal losses and gains in single cells derived from cytogenetically characterized aneuploid cell lines and blastomeres derived from human embryos (Kearns et al., 2008; Rabinowitz et al., 2008). Furthermore, analysis of DNA samples from the mother and father, in addition to DNA from the embryo, has made it possible to follow the inheritance of each parental chromosome. Not only does this provide interesting scientific information, such as revealing the parental origin of extra chromosomes in trisomic embryos, but it also yields data on the inheritance of specific genetic loci, including those associated with disease (Kearns et al., 2008).
In general, disease causing mutations are not detected directly using SNP-microarrays (although this is technically possible in some cases). Rather they are detected indirectly via a linkage approach. This involves the genotyping of SNPs located in close proximity to the mutation site, on the same chromosome (i.e. linked polymorphisms). It is usually possible to identify specific alleles, which do not cause the disease, but are always inherited along with it and can therefore be used to infer a diagnosis. Proof of principle studies has shown that accurate data concerning the inheritance of disease-associated polymorphisms can be obtained from single cells using SNP-microarrays (Handyside et al., 2008). This potentially opens up the possibility of using SNP-microarray platforms for the concomitant detection of chromosome anomalies and single gene disorders. Traditionally, the simultaneous diagnosis of aneuploidy and gene mutations has been problematic for PGD, as the FISH techniques used for chromosome screening and the PCR methods employed for single gene testing are incompatible. A limited chromosome screen is possible using PCR, but such protocols fail to screen most of the chromosomes and have a limited ability to detect errors arising in meiosis II or after fertilization.
The only drawbacks of analysis using SNP-microarrays are a lack of diagnostic accuracy at individual SNP loci and the high expense of the microarrays and labeling techniques. The WGA methods, required to generate sufficient DNA from a single cell for subsequent microarray analysis, exacerbate problems such as ADO and preferential amplification. This causes many of the SNP loci to be incorrectly genotyped. Fortunately, the huge number of sites tested means that accurate chromosomal haplotypes can still be constructed, and the inheritance of chromosomal regions containing mutations deduced, despite the presence of many errors. However, statistical processing of the data is necessary in order to identify correctly genotyped SNPs from those giving unreliable results and extract meaningful data (Handyside et al., 2008; Rabinowitz et al., 2008). Inevitably, the elimination of large numbers of incorrectly genotyped loci from the data set has the effect of reducing the resolution of the microarray and prevents reliable diagnosis based upon direct analysis of individual mutations.
Currently, SNP arrays represent an expensive option for oocyte/embryo screening. With commercial subsidy, it is possible to offer this method of screening to a limited population of patients. However, it is not yet clear whether this platform can be adapted to create an inexpensive, widely applicable test. Additionally, many of the existing software interfaces will need modification prior to extensive clinical application, in order to make analysis less laborious and time-consuming.
Summary
Accurate methods for the simultaneous analysis of all 24 types of chromosome (22 autosomes, X and Y) look set to usher in a new era of embryo evaluation. Recent data clearly indicate that comprehensive chromosomal screening assists the identification of viable embryos for transfer to the uterus, leading to improved IVF outcomes (Schoolcraft et al., 2008a). The dramatic improvement in implantation rate is particularly noteworthy and will be an extremely useful tool for maintaining high pregnancy rates while reducing the number of embryos transferred each cycle. The introduction of highly reliable vitrification techniques for embryo cryopreservation has removed the last remaining obstacle to the widespread application of conventional CGH and the number of clinical cases utilizing this screening method is expanding rapidly. Microarray methods of chromosome screening also continue to show technical improvement. Platforms utilizing BACs, chromosome-specific libraries, oligonucleotides and SNPs have all succeeded in detecting aneuploidies in single blastomeres and are now being clinically applied. It remains to be seen which microarray approach will ultimately provide the optimal combination of accuracy, speed and cost, but given the rapid evolution of these technologies a cost-effective and reliable test seems close at hand. For the time being, however, conventional CGH remains the test of choice for comprehensive aneuploidy screening.
Funding
D.W. is funded by NIHR Biomedical Research Centre Programme.
References
- Angell RR, Aitken RJ, van Look PF, Lumsden MA, Templeton AA. Chromosome abnormalities in human embryos after in vitro fertilization. Nature. 1983;303:336–338. doi: 10.1038/303336a0. [DOI] [PubMed] [Google Scholar]
- Bergh T, Ericson A, Hillensjo T, Nygren KG, Wennerholm UB. Deliveries and children born after in-vitro fertilisation in Sweden 1982–1995: a retrospective cohort study. Lancet. 1999;354:1579–1585. doi: 10.1016/S0140-6736(99)04345-7. [DOI] [PubMed] [Google Scholar]
- Colls P, Escudero T, Cekleniak N, Sadowy S, Cohen J, Munné S. Increased efficiency of preimplantation genetic diagnosis for infertility using ‘no result rescue. Fertil Steril. 2007;88:53–61. doi: 10.1016/j.fertnstert.2006.11.099. [DOI] [PubMed] [Google Scholar]
- Cummins JM, Breen TM, Harrison KL, Shaw JM, Wilson LM, Hennessey JF. A formula for scoring human embryo growth rates in in vitro fertilization: its value in predicting pregnancy and in comparison with visual estimates of embryo quality. J In Vitro Fert Embryo Transf. 1986;3:284–295. doi: 10.1007/BF01133388. [DOI] [PubMed] [Google Scholar]
- Delhanty JD, Griffin DK, Handyside AH, Harper J, Atkinson GH, Pieters MH, Winston RM. Detection of aneuploidy and chromosomal mosaicism in human embryos during preimplantation sex determination by fluorescent in situ hybridisation, (FISH) Hum Mol Genet. 1993;2:1183–1185. doi: 10.1093/hmg/2.8.1183. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Bianchi V, Delhanty J, Patrizio P, Wells D. Gene expression analysis of human oocytes: towards a non-invasive diagnosis of meiotic aneuploidy. Hum Reprod. 2007;22:i32. doi: 10.1093/molehr/gaq033. O-078. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod. 2008;23:2596–2608. doi: 10.1093/humrep/den287. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Wells D, Thornhill A, Serhal P, Faed MJ, Harper JC, Delhanty JD. Comparative genomic hybridization analysis of human oocytes and polar bodies. Hum Reprod. 2006;21:2319–28. doi: 10.1093/humrep/del157. [DOI] [PubMed] [Google Scholar]
- Gerris J. The near elimination of triplets in IVF. Reprod Biomed Online. 2007;15(Suppl 3):40–44. doi: 10.1016/s1472-6483(10)62250-x. [DOI] [PubMed] [Google Scholar]
- Gianaroli L, Magli MC, Ferraretti AP, Fiorentino A, Garrisi J, Munné S. Preimplantation genetic diagnosis increases the implantation rate in human in vitro fertilization by avoiding the transfer of chromosomally abnormal embryos. Fertil Steril. 1997;68:1128–1131. doi: 10.1016/s0015-0282(97)00412-3. [DOI] [PubMed] [Google Scholar]
- Gianaroli L, Magli MC, Ferraretti AP, Munné S. Preimplantation diagnosis for aneuploidies in patients undergoing in vitro fertilization with a poor prognosis: identification of the categories for which it should be proposed. Fertil Steril. 1999;72:837–44. doi: 10.1016/s0015-0282(99)00377-5. [DOI] [PubMed] [Google Scholar]
- Griffin DK, Wilton LJ, Handyside AH, Winston RM, Delhanty JD. Dual fluorescent in situ hybridisation for simultaneous detection of X and Y chromosome-specific probes for the sexing of human preimplantation embryonic nuclei. Hum Genet. 1992;89:18–22. doi: 10.1007/BF00207035. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Mateo C, Benet J, Wells D, Colls P, Bermúdez MG, Sánchez-García JF, Egozcue J, Navarro J, Munné S. Aneuploidy study of human oocytes first polar body comparative genomic hybridization and metaphase II fluorescence in situ hybridization analysis. Hum Reprod. 2004;19:2859–2868. doi: 10.1093/humrep/deh515. [DOI] [PubMed] [Google Scholar]
- Handyside AH, Thornhill AR, Affara NA, Harton GL, Mariani BD, Griffin DK. Recombination mapping: a universal molecular karyotyping method for preimplantation genetic diagnosis of inherited disease. Fertil Steril. 2008;90(Suppl 1):S24. [Google Scholar]
- Hassold TJ, Jacobs PA. Annu Rev Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet. 2007;16:R203–R208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- Hellani A, Abu-Amero K, Azouri J, El-Akoum S. Successful pregnancies after application of array-CGH in PGS aneuploidy screening. Reprod Biomed Online. 2008 doi: 10.1016/s1472-6483(10)60413-0. [DOI] [PubMed] [Google Scholar]
- Hu DG, Webb G, Hussey N. Aneuploidy detection in single cells using DNA array-based comparative genomic hybridization. Mol Hum Reprod. 2004;10:283–289. doi: 10.1093/humrep/gah038. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Kamiguchi Y, Rosenbusch B, Sterzik K, Mikamo K. Chromosomal analysis of unfertilized human oocytes prepared by a gradual fixation-air drying method. Hum Genet. 1993;90:533–541. doi: 10.1007/BF00217454. [DOI] [PubMed] [Google Scholar]
- Katz-Jaffe M, Stevens J, Kearns W, Gardner D, Schoolcraft W. Relationship between embryonic secretome and chromosomal abnormalities in human IVF. Fertil Steril. 2006;86:S57. [Google Scholar]
- Kearns WG, Pen R, Benner A, Kittai A, Widra E, Leach R. SNP microarray genetic analyses to determine 23-chromosome ploidy, structural chromosome aberrations and genome-wide scans to identify disease risks from a single embryonic cell. Fertil Steril. 2008;90(Suppl 1):S23. [Google Scholar]
- Kuliev A, Cieslak J, Ilkevitch Y, Verlinsky Y. Chromosomal abnormalities in a series of 6,733 human oocytes in preimplantation diagnosis for age-related aneuploidies. Reprod Biomed Online. 2003;6:54–59. doi: 10.1016/s1472-6483(10)62055-x. [DOI] [PubMed] [Google Scholar]
- Kuliev A, Verlinsky Y. Meiotic and mitotic nondisjunction: lessons from preimplantation genetic diagnosis. Hum Reprod Update. 2005;10:401–407. doi: 10.1093/humupd/dmh036. [DOI] [PubMed] [Google Scholar]
- Le Caignec C, Spits C, Sermon K, De Rycke M, Thienpont B, Debrock S, Staessen C, Moreau Y, Fryns JP, Van Steirteghem A, et al. Single-cell chromosomal imbalances detection by array CGH. Nucleic Acids Res. 2006;34:e68. doi: 10.1093/nar/gkl336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastenbroek S, Twisk M, van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, Vogel NE, Arts EG, de Vries JW, Bossuyt PM, et al. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:9–17. doi: 10.1056/NEJMoa067744. [DOI] [PubMed] [Google Scholar]
- McArthur SJ, Leigh D, Marshall JT, de Boer KA, Jansen RP. Pregnancies and live births after trophectoderm biopsy and preimplantation genetic testing of human blastocysts. Fertil Steril. 2005;84:1628–1636. doi: 10.1016/j.fertnstert.2005.05.063. [DOI] [PubMed] [Google Scholar]
- McArthur SJ, Leigh D, Marshall JT, Gee AJ, De Boer KA, Jansen RP. Blastocyst trophectoderm biopsy and preimplantation genetic diagnosis for familial monogenic disorders and chromosomal translocations. Prenat Diagn. 2008;28:434–442. doi: 10.1002/pd.1924. [DOI] [PubMed] [Google Scholar]
- Menasha J, Levy B, Hirschhorn K, Kardon NB. Incidence and spectrum of chromosome abnormalities in spontaneous abortions: new insights from a 12-year study. Genet Med. 2005;7:251–263. doi: 10.1097/01.gim.0000160075.96707.04. [DOI] [PubMed] [Google Scholar]
- Munné S, Lee A, Rosenwaks Z, Grifo J, Cohen J. Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum Reprod. 1993;8:2185–2191. doi: 10.1093/oxfordjournals.humrep.a138001. [DOI] [PubMed] [Google Scholar]
- Munné S, Magli C, Adler A, Wright G, de Boer K, Mortimer D, Tucker M, Cohen J, Gianaroli L. Treatment-related chromosome abnormalities in human embryos. Hum Reprod. 1997;12:780–784. doi: 10.1093/humrep/12.4.780. [DOI] [PubMed] [Google Scholar]
- Munné S, Magli C, Cohen J, Morton P, Sadowy S, Gianaroli L, Tucker M, Márquez C, Sable D, Ferraretti AP, et al. Positive outcome after preimplantation diagnosis of aneuploidy in human embryos. Hum Reprod. 1999;14:2191–2199. doi: 10.1093/humrep/14.9.2191. [DOI] [PubMed] [Google Scholar]
- Munné S, Sandalinas M, Escudero T, Velilla E, Walmsley R, Sadowy S, Cohen J, Sable D. Improved implantation after preimplantation genetic diagnosis of aneuploidy. Reprod Biomed Online. 2003;7:91–97. doi: 10.1016/s1472-6483(10)61735-x. [DOI] [PubMed] [Google Scholar]
- Munné S, Chen S, Fischer J, Colls P, Zheng X, Stevens J, Escudero T, Oter M, Schoolcraft B, Simpson JL, et al. Preimplantation genetic diagnosis reduces pregnancy loss in women aged 35 years and older with a history of recurrent miscarriages. Fertil Steril. 2005;84:331–335. doi: 10.1016/j.fertnstert.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Munné S, Cohen J, Simpson JL. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;a 357:1769–1770. doi: 10.1056/NEJMc076314. [DOI] [PubMed] [Google Scholar]
- Munné S, Gianaroli L, Tur-Kaspa I, Magli C, Sandalinas M, Grifo J, Cram D, Kahraman S, Verlinsky Y, Simpson JL. Substandard application of preimplantation genetic screening may interfere with its clinical success. Fertil Steril. 2007;b 88:781–784. doi: 10.1016/j.fertnstert.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Obradors A, Fernández E, Oliver-Bonet M, Rius M, de la Fuente A, Wells D, Benet J, Navarro J. Birth of a healthy boy after a double factor PGD in a couple carrying a genetic disease and at risk for aneuploidy: case report. Hum Reprod. 2008;23:1949–1956. doi: 10.1093/humrep/den201. [DOI] [PubMed] [Google Scholar]
- Pellestor F, Andréo B, Arnal F, Humeau C, Demaille J. Maternal aging and chromosomal abnormalities: new data drawn from in vitro unfertilized human oocytes. Hum Genet. 2003;112:195–203. doi: 10.1007/s00439-002-0852-x. [DOI] [PubMed] [Google Scholar]
- Pinborg A, Loft A, Schmidt L, Andersen AN. Morbidity in a Danish national cohort of 472 IVF/ICSI twins, 1132 non-IVF/ICSI twins and 634 IVF/ICSI singletons: health-related and social implications for the children and their families. Hum Reprod. 2003;18:1234–1243. doi: 10.1093/humrep/deg257. [DOI] [PubMed] [Google Scholar]
- Puissant F, Van Rysselberge M, Barlow P, Deweze J, Leroy F. Embryo scoring as a prognostic tool in IVF treatment. Hum Reprod. 1987;2:705–708. doi: 10.1093/oxfordjournals.humrep.a136618. [DOI] [PubMed] [Google Scholar]
- Rabinowitz M, Johnson DS, Salzman J, Banjevic M, Cinnioglu C, Behr B. Reliable concurrent calling of multiple genetic alleles and 24-chromosome ploidy without embryo freezing using Parental Support™ technology (PS) Fertil Steril. 2008;90(Suppl 1):S23. [Google Scholar]
- Sakkas D, Gardner DK. Noninvasive methods to assess embryo quality. Curr Opin Obstet Gynecol. 2005;17:283–288. doi: 10.1097/01.gco.0000169106.69881.3e. [DOI] [PubMed] [Google Scholar]
- Sandalinas M, Márquez C, Munné S. Spectral karyotyping of fresh, non-inseminated oocytes. Mol Hum Reprod. 2002;8:580–585. doi: 10.1093/molehr/8.6.580. [DOI] [PubMed] [Google Scholar]
- Schoolcraft WB, Katz-Jaffe MG, Stevens J, Rawlins M, Munne S. Preimplantation aneuploidy testing for infertile patients of advanced maternal age: a randomized prospective trial. Fertil Steril. 2008;a 8 doi: 10.1016/j.fertnstert.2008.05.029. [DOI] [PubMed] [Google Scholar]
- Schoolcraft WB, Fragouli E, Stevens J, Munne S, Katz-Jaffe M, Wells D. Dramatically increased embryo implantation and high pregnancy rates achieved after comprehensive chromosomal screening of in vitro fertilized embryos. 2008;b submitted for publication. [Google Scholar]
- Scott RT, Miller KA, Olivares R, Su J, Fratterelli JL, Treff NR. Microarray based 24 chromosome preimplantation genetic diagnosis (mPGD) is highly predictive of the reproductive potential of human embryos: a prospective blinded non-selection trial. Fertil Steril. 2008;90(Suppl 1):S22–S23. [Google Scholar]
- Sher G, Keskintepe L, Keskintepe M, Ginsburg M, Maassarani G, Yakut T, Baltaci V, Kotze D, Unsal E. Oocyte karyotyping by comparative genomic hybridization provides a highly reliable method for selecting ‘competent’ embryos, markedly improving in vitro fertilization outcome: a multiphase study. Fertil Steril. 2007;87:1033–1040. doi: 10.1016/j.fertnstert.2006.08.108. [DOI] [PubMed] [Google Scholar]
- Staessen C, Platteau P, Van Assche E, Michiels A, Tournaye H, Camus M, Devroey P, Liebaers I, Van Steirteghem A. Comparison of blastocyst transfer with or without preimplantation genetic diagnosis for aneuploidy screening in couples with advanced maternal age: a prospective randomized controlled trial. Hum Reprod. 2004;19:2849–2858. doi: 10.1093/humrep/deh536. [DOI] [PubMed] [Google Scholar]
- Stromberg B, Dahlquist G, Ericson A, Finnstrom O, Koster M, Stjernqvist K. Neurological sequelae in children born after in-vitro fertilisation: a population-based study. Lancet. 2002;359:461–465. doi: 10.1016/S0140-6736(02)07674-2. [DOI] [PubMed] [Google Scholar]
- Stuerwald N, Wells D, Cohen J, Munne S. Comprehensive aneuploidy screening in single cells using microarray comparative genomic hybridization methods: implications for preimplantation genetic diagnosis. Fertil Steril. 2007;88:S86–S87. [Google Scholar]
- Sunde A. Significant reduction of twins with single embryo transfer in IVF. Reprod Biomed Online. 2007;15(Suppl 3):28–34. doi: 10.1016/s1472-6483(10)62248-1. [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjöld M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- Treff NR, Su J, Mavrianos J, Bergh PA, Miller KA, Scott RT. Accurate 23 chromosome aneuploidy screening in human blastomeres using single nucleotide polymorphism (SNP) microarrays. Fertil Steril. 2007;88:S1. [Google Scholar]
- Van Royen E, Mangelschots K, De Neubourg D, Valkenburg M, Van de Meerssche M, Ryckaert G, Eestermans W, Gerris J. Characterization of a top quality embryo, a step towards single-embryo transfer. Hum Reprod. 1999;14:2345–2349. doi: 10.1093/humrep/14.9.2345. [DOI] [PubMed] [Google Scholar]
- Veiga A, Calderón G, Santaló J, Barri PN, Egozcue J. Chromosome studies in oocytes and zygotes from an IVF programme. Hum Reprod. 1987;2:425–430. doi: 10.1093/oxfordjournals.humrep.a136562. [DOI] [PubMed] [Google Scholar]
- Voullaire L, Slater H, Williamson R, Wilton L. Chromosome analysis of blastomeres from human embryos by using comparative genomic hybridization. Hum Genet. 2000;106:210–217. doi: 10.1007/s004390051030. [DOI] [PubMed] [Google Scholar]
- Voullaire L, Wilton L, McBain J, Callaghan T, Williamson R. Chromosome abnormalities identified by comparative genomic hybridization in embryos from women with repeated implantation failure. Mol Hum Reprod. 2002;8:1035–1041. doi: 10.1093/molehr/8.11.1035. [DOI] [PubMed] [Google Scholar]
- Walker MC, Murphy KE, Pan S, Yang Q, Wen SW. Adverse maternal outcomes in multifetal pregnancies. Br J Obstet Gynaecol. 2004;111:1294–1296. doi: 10.1111/j.1471-0528.2004.00345.x. [DOI] [PubMed] [Google Scholar]
- Wells D, Delhanty J. Evaluating comparative genomic hybridisation (CGH) as a strategy for preimplantation diagnosis of unbalanced chromosome complements. Eur J Hum Genet. 1996;4(Suppl 1):125. [Google Scholar]
- Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod. 2000;6:1055–1062. doi: 10.1093/molehr/6.11.1055. [DOI] [PubMed] [Google Scholar]
- Wells D, Sherlock JK, Handyside AH, Delhanty JD. Detailed chromosomal and molecular genetic analysis of single cells by whole genome amplification and comparative genomic hybridisation. Nucleic Acids Res. 1999;27:1214–1218. doi: 10.1093/nar/27.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D, Escudero T, Levy B, Hirschhorn K, Delhanty JD, Munné S. First clinical application of comparative genomic hybridization and polar body testing for preimplantation genetic diagnosis of aneuploidy. Fertil Steril. 2002;78:543–549. doi: 10.1016/s0015-0282(02)03271-5. [DOI] [PubMed] [Google Scholar]
- Wells D, Bermudez MG, Steuerwald N, Chu L, Weier U, Cohen J, Munne S. Microarrays for analysis and diagnosis of human embryos. In: Papp Z, Rodeck C, editors. Recent Advances in Prenatal Genetic Diagnosis. Medimond; 2004. pp. 9–17. [Google Scholar]
- Wells D, Fragouli E, Bianchi V, Obradors A, Delhanty J, Patrizio P. Comprehensive characterization of gene expression in human oocytes and identification of perturbations associated with meiotic aneuploidy, maturation and infertile pathology. Fertil Steril. 2006;86:S40–S41. [Google Scholar]
- Wells D, Fragouli E, Stevens J, Munne S, Schoolcraft W, Katz-Jaffe M. High pregnancy rate after comprehensive chromosomal screening of blastocysts. Fertil Steril. 2008;90:S80. doi: 10.1016/j.fertnstert.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Wilton L, Williamson R, McBain J, Edgar D, Voullaire L. Birth of a healthy infant after preimplantation confirmation of euploidy by comparative genomic hybridization. N Engl J Med. 2001;345:1537–1541. doi: 10.1056/NEJMoa011052. [DOI] [PubMed] [Google Scholar]
- Wilton L, Voullaire L, Sargeant P, Williamson R, McBain J. Preimplantation aneuploidy screening using comparative genomic hybridization or fluorescence in situ hybridization of embryos from patients with recurrent implantation failure. Fertil Steril. 2003;80:860–868. doi: 10.1016/s0015-0282(03)01162-2. [DOI] [PubMed] [Google Scholar]