Abstract
Preimplantation development shifts from a maternal to embryonic programme rapidly after fertilization. Although the majority of oogenetic products are lost during the maternal to embryonic transition (MET), several do survive this interval to contribute directly to supporting preimplantation development. Embryonic genome activation (EGA) is characterized by the transient expression of several genes that are necessary for MET, and while EGA represents the first major wave of gene expression, a second mid-preimplantation wave of transcription that supports development to the blastocyst stage has been discovered. The application of genomic approaches has greatly assisted in the discovery of stage specific gene expression patterns and the challenge now is to largely define gene function and regulation during preimplantation development. The basic mechanisms controlling compaction, lineage specification and blastocyst formation are defined. The requirement for embryo culture has revealed plasticity in the developmental programme that may exceed the adaptive capacity of the embryo and has fostered important research directions aimed at alleviating culture-induced changes in embryonic programming. New levels of regulation are emerging and greater insight into the roles played by RNA-binding proteins and miRNAs is required. All of this research is relevant due to the necessity to produce healthy preimplantation embryos for embryo transfer, to ensure that assisted reproductive technologies are applied in the most efficient and safest way possible.
Keywords: gene expression, embryo culture, in vitro fertilization, oocyte, blastocyst
Introduction
These are exciting times for mammalian preimplantation developmental biology. An unprecedented group of molecular tools are available for probing the molecular mechanisms that control preimplantation development. Studies have employed gene arrays and proteomics to define gene expression during preimplantation development leading to an astounding production of data that define the molecular programme controlling the first week of development. Our knowledge of the mechanisms controlling gene expression during this period has also increased exponentially with discoveries implicating chromatin remodelling, acetylation, methylation, transcription, deadenylation, RNA-binding proteins (RBPs), microRNAs, translation and protein phosphorylation as critical instruments that the early embryo employs to regulate its first week of development. Corresponding with these developments, is our increasing capacity to define specific gene function during preimplantation development by applying gene targeting, transgenics, down-regulation of gene expression by application of antisense and siRNA approaches and gain of function by injection of cRNAs. Collectively these advances have begun to unravel the secrets of preimplantation development, primarily in the mouse, but increasingly in other species such as the cow, sheep, pig and even the human.
These advances in our ability to define the mechanisms controlling preimplantation development are timely, as the necessity for understanding these mechanisms has never been greater, with our increasing application of assisted reproductive technologies to promote reproduction in humans and other important species such as livestock and also to endangered species to ensure their survival. In addition, there is not only a great need to characterize normal development, but to also understand the effects of exposure to abnormal culture environments on preimplantation development. This is necessary, since the production of embryos from all mammalian species requires the removal of either gametes and/or embryos from the reproductive tract. This requirement has fostered a research enterprise that has led to the understanding that the gene expression patterns of cultured embryos can vary quite significantly from that of in vivo derived embryos. We are concerned about these differences because research has correlated these differences with abnormalities that range from early embryo loss to placental defects, fetal growth abnormalities and even increased susceptibility to disease in later life. Thus, it is imperative that we understand the mechanisms controlling normal development and also to assess the adaptive capacity of cultured embryos so that we can increasingly be assured that the environments within which we place gametes and embryos will not exceed these adaptive capacities and result in the dire consequences presented above.
The purpose of this review is to summarize our current understanding of the gene expression patterns that arise during preimplantation development and review our current understanding of the mechanisms that control mRNA expression and stability during early development. We will also introduce emerging areas of research interest including microRNAs and RBP control of mRNA stability and translation. We will briefly consider the effects of culture environment on gene expression patterns and mechanisms controlling gene expression. The field has expanded so rapidly that we do not claim to be comprehensive in this single review. The reader, if interested, should also be directed to several other excellent recent reviews on this topic that understandably emphasize other aspects of this topic to greater or lesser extents than the present review covers (Gandolfi and Gandolfi, 2001; Schultz, 2005; Bettegowda et al., 2008).
Genomic profiling of mRNA pools during preimplantation development
Preimplantation development includes the free-living period of mammalian development extending from the 1-cell zygote to blastocyst attachment or implantation into the uterine wall (Fig. 1). It generally encompasses the first week of development and includes critical events such as the transition from maternal to embryonic genome activation (EGA), compaction, and cavitation or blastocyst formation. The main accomplishment of preimplantation development is to produce an implantation-competent embryo for the initiation of pregnancy. To accomplish this goal, the very first epithelium, the trophectoderm (TE), must form, as this epithelium becomes the outer layer of the blastocyst, and interacts with the endometrium to initiate implantation. The TE is also the progenitor of the embryonic component of the placenta. A second cell type that remains pluripotent, the inner cell mass (ICM), is the progenitor of the embryo proper. These cells remain enclosed in the blastocyst surrounded by both mural and polar TE. Thus, preimplantation development is not primarily concerned with embryogenesis but rather TE differentiation, for placental mammals must first conquer this requirement for implantation and placental development before embryogenesis can proceed.
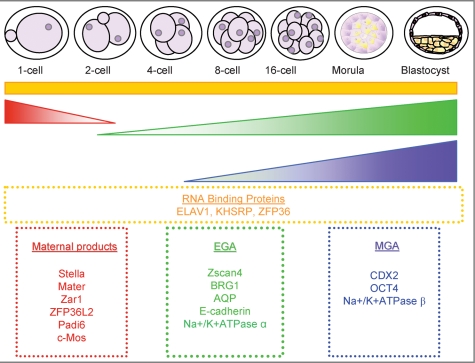
Figure 1:
Characterization of global gene expression patterns during preimplantation development.
Three groups published stage specific gene array analysis of gene expression in mouse preimplantation embryos in 2004 (Hamatani et al., 2004; Wang et al., 2004; Zeng et al., 2004). Two main waves of gene expression were observed corresponding with EGA and a second MGA that contributed to events associated with TE differentiation and blastocyst formation (Hamatani et al., 2004). Hamatani et al. (2004), Wang et al. (2004) and Zeng et al. (2004) are landmark studies in the characterization of gene expression patterns during preimplantation development as they have produced extensive catalogues of global stage specific gene expression patterns. General gene expression patterns are indicated; loss of maternal mRNAs (red); activation of embryonic genome (green); MGA (purple) and continuous expression (orange). Examples of genes expressed in each category are indicated in boxes.
Recent studies have completely transformed our approach to studying gene expression during early development by bringing the global perspective that gene arrays can bring to gene expression studies (Dobson et al., 2004; Hamatani et al., 2004; Tanaka and Ko, 2004; Wang et al., 2004; Misirlioglu et al., 2006; Rinaudo et al., 2006; Giritharan et al., 2007; Vallee et al., 2008). Three groups published stage specific gene array analysis of gene expression in mouse preimplantation embryos in 2004 (Hamatani et al., 2004; Wang et al., 2004; Zeng et al., 2004; Fig. 1). Hamatani et al. (2004) employed an NIA 60-mer oligo microarray, enriched in stem cells and preimplantation stage embryo transcripts, while Wang et al. (2004) and Zeng et al. (2004) used Affymetrix 25-mer DNA microarray systems. Even more recent studies use the new mouse Affymetrix 430 2.0 array that is believed to cover the complete mouse genome (Giritharan et al., 2007). Hamatani et al. (2004) reported that 12 179/21 939 genes displayed a significant change in expression during mouse preimplantation development. Wang et al. (2004) found that over 4000 genes changed in expression over 5-fold during development and evidence that the expression of a large number of genes was regulated at the transcription, adenylation or mRNA half-life levels. Two main waves of gene expression were observed corresponding with EGA and a second mid-preimplantation gene activation (MGA) that contributed to events associated with TE differentiation and blastocyst formation (Hamatani et al., 2004; Fig. 1). Thus, Hamatani et al. (2004), Wang et al. (2004) and Zeng et al. (2004) are landmark studies in the characterization of gene expression patterns during preimplantation development as they have produced extensive catalogues of global stage specific gene expression patterns. The challenge in the field now has moved from asking whether a particular gene is expressed and when during preimplantation development; to understanding what factors affect its expression and what function does it contribute to preimplantation development. Global gene expression analysis has also been applied to contrast gene expression between cultured and in vivo derived mouse embryos (Rinaudo et al., 2006; Giritharan et al., 2007); to examine the effects of oxygen atmosphere on mouse preimplantation embryo gene expression patterns (Rinaudo et al., 2006); and has been extended to contrast global gene expression profiles in other species such as the cow (Sirard et al., 2005; Misirlioglu et al., 2006), pig (Whitworth et al., 2005) and the human preimplantation embryo (Dobson et al., 2004). Future applications of this technology will include contrasting gene expression between wild-type and gene deleted or knocked down ovaries or embryos (Murchison et al., 2005; Shin et al., 2005; Joshi et al., 2007) and in antagonist and agonist treatment experiments to determine the specific influences of individual regulatory genes and signalling pathways in controlling gene expression during preimplantation development. We must however increase our bioinformatic capacity to efficiently process all the information generated by these types of experiments and to a certain degree we must begin to think more along the lines of ‘systems biology’ where we consider the implications that result from affecting gene pools rather than outcomes based upon single gene analysis. This represents a large challenge for future developments in this field.
Maternal to embryonic transition and EGA
We will now focus attention to the mechanisms controlling mRNA transcript abundance during early development. Our discussion begins in the oocyte as oocyte cytoplasmic maturation is required to complete nuclear maturation, fertilization and preimplantation development and thus provides a foundation for implantation, initiation of pregnancy and fetal development (Gandolfi and Gandolfi, 2001; Sirard et al., 2006). Oocyte cytoplasmic maturation includes the storage of mRNAs, proteins, substrates and nutrients that result in the attainment of oocyte developmental competence that ensures embryonic developmental competence (Gandolfi and Gandolfi, 2001; Krisher, 2004; Sirard et al., 2006). Transcription and storage of maternal mRNA occurs during follicular growth but ceases as the germinal vesicle undergoes breakdown during meiosis (Brevini-Gandolfi et al., 1999; Gandolfi and Gandolfi, 2001; Fair et al., 2004). The embryo is dependent on stored maternal mRNA until at least the maternal–zygotic transition, when transcription of embryonic genes begins with a major burst occurring at the 2-cell stage in mice (Flach et al., 1982; Brevini-Gandolfi et al., 1999; Gosden, 2002; Schultz, 2002). One of the greatest mysteries of the mammalian developmental programme is why is it necessary to store and accumulate mRNAs and proteins during oogenesis to simply then activate a programme that results largely in their demise shortly after fertilization within the first few cleavage divisions? Degradation of maternal mRNA stores and mRNA deadenylation shortly after fertilization results in a dramatic turnover of maternal mRNA during early cleavage stages with up to a 40% decline in bulk maternal RNA and up to a 70% decrease in maternal poly A+ mRNA (Bachvarova and De Leon, 1980; Piko and Clegg, 1982; Fig. 1). Of that decrease in poly A+ mRNA, some is degraded and some is deadenylated (Bachvarova, 1985; Paynton et al., 1988). Many of the maternal messages degraded immediately after fertilization are likely detrimental to further development (Alizadeh et al., 2005). For example, c-mos is an important factor for oocyte maturation and is present from the oocyte to fertilized oocyte (Paules et al., 1989). However, when c-mos mRNA was reintroduced into Xenopus 2-cell embryo, developmental arrest resulted (Sagata et al., 1989). Most of the maternal mRNAs that decline early after fertilization contain a cytoplasmic polyadenylation element (CPE) close to the nuclear poly A+ signal in the 3′UTR (Oh et al., 2000; Alizadeh et al., 2005).
It is challenging to determine whether disappearance of mRNAs is due to degradation or deadenylation. Many protocols use oligo(dT) for reverse transcription; however, mRNAs without a long enough poly A+ tail are inefficiently transcribed (Schultz et al., 1999; Lequarre et al., 2004; Schultz, 2005), thus polyadenylation of a message may increase its detection level (Wang et al., 2004, Zeng et al., 2004). In addition, some procedures only capture poly A+ mRNA (Wrenzycki et al., 1999; Gutierrez-Adan et al., 2004; Tanaka and Ko, 2004; Shin et al., 2005) and deadenylated mRNAs will be underrepresented in the sample (Lequarre et al., 2004; Thelie et al., 2007). Underrepresentation of deadenylated messages could also be a problem when using oligo(dT) amplification for generating microarray data (Su et al., 2007; Thelie et al., 2007). Therefore, absence or reduction in particular maternal mRNAs must be confirmed to occur by degradation by northern blotting, or reverse transcription with gene-specific (Bachvarova, 1985; Oh et al., 2000; Alizadeh et al., 2005), random primers (Su et al., 2007; Thelie et al., 2007). It is critical that these methodological approaches be applied in tandem to decipher the contributions of adenlyation status to overall transcript stability (Paynton et al., 1988; Paynton and Bachvarova 1994; Oh et al., 2000).
Despite the low rates of transcription at the 1-cell stage, recruitment and polyadenylation of maternal mRNAs likely results in the higher detection of 1500 transcripts in 1-cell embryos than other stages (Oh et al., 2000). On the other hand, most of the highly up-regulated genes at the 2-cell stage are newly transcribed (Zeng and Schultz, 2005; Fig. 1). Furthermore, polyadenylation is closely linked in time with translation (Huarte et al., 1987; Stutz et al., 1998; Oh et al., 2000). Timely translation of maternal mRNAs provides the coordination of protein synthesis required for the maternal to embryonic transition (MET) (Oh et al., 2000). In general, polyadenylation is associated with translational activation, while deadenylation is associated with translational silencing (Oh et al., 2000; Piccioni et al., 2005). Polyadenylation and translation are controlled mostly by regions in the 5′ and 3′UTRs (reviewed by (Piccioni et al., 2005)). Silencing or masking factors bind to mRNAs and keep them from being translated (Stutz et al., 1998; Gosden, 2002). These factors are likely to be post-translationally modified to allow dissociation from mRNAs to allow their later translation (Schultz, 1993; Stutz and Rosbash, 1998). Maternal mRNAs with long poly A+ tails but lacking a U-rich CPE are deadenylated by default during oocyte maturation. Some transcripts with shorter poly A+ tails are stable. While others, such as hypoxanthine-quanidine phosphoribosyl transferase, with a CPE and a short poly A+ tail are polyadenylated during maturation. Studies in this field have extended to also investigate the influences of varying culture environments on mRNA deadenylation and mRNA turnover/stability (Brevini-Gandolfi et al., 1999; Gandolfi and Gandolfi, 2001; Vigneault et al., 2007).
It would appear that the mammalian preimplantation developmental programme has evolved to ensure that the EGA occurs very early in development, and thus from the first cleavage divisions onward, the embryo assumes a great control over its own developmental fate by the expression of gene products from its own genome (Fig. 1). Despite this being the general pattern, it would be inaccurate to suggest that maternal transcripts serve little role in ensuring development proceeds properly in mammalian embryos, as not all maternal gene products are degraded during the MET, some are degraded later at the mouse 4- and 8-cell stages and are probably necessary for the EGA (Hamatani et al., 2004). Up to 10% of labelled maternal mRNAs persist until the blastocyst stage of development (Bachvarova and De Leon, 1980), and thus could have a critical role that extends beyond ensuring that EGA proceeds in an optimal fashion. Several important recent studies have defined several maternal gene products that are required to support early development such as Stella, (also known as developmental pluripotency associated 3, Dppa3 (Payer et al., 2003)), MATER (maternal product that embryos require, also known as Nalp5 (Tong et al., 2000a,b)), Zar1 (Zygote arrest 1 (Wu et al., 2003a,b)) and ZFP36L2 (a tristetraprolin-like RBP family member (Ramos et al., 2004; Fig. 1)). These are all dispensable for growth up to the mature follicle stage, and embryo development is affected in gene knock-outs.
Embryonic genome activation
Early studies suggested that EGA represented a global and promiscuous activation of genes whose regulated repression was then necessary to establish the preimplantation developmental programme (Schultz, 1993; Ma et al., 2001). More recent transcriptomic analyses have demonstrated that EGA is in fact a highly regulated gene expression programme (Hamatani et al., 2004; Wang et al., 2004; Zeng et al., 2004). Genes involved in transcription and RNA metabolism are overrepresented at the EGA (Zeng et al., 2004). Evidence suggests that most of the newly synthesized EGA transcripts are quickly translated (Flach et al., 1982). Epigenetic chromatin remodelling is now proposed as the main mechanism via which the EGA occurs, where epigenetic marks involve post-translational modifications (methylation, acetylation, phosphorylation and ubiquitination) of nucleosomal histones, DNA methylation and non-histone proteins that bind to chromatin. In general, transcriptionally inactive heterochromatin is characterized by deacetylated histones, methylation of histone H3 lysine 9 and DNA methylation, whereas acetylation of H3 and H4 histones, methylation of histone H3 lysine 4 and low level of DNA methylation are associated with active gene expression. These modifications alter the chromatin structure to provide access for the regulatory transcriptional factors that control gene expression. Interestingly, demethylation and remethylation events are not uniform across all species, and differences are seen between pig, cow, sheep, mice and even humans. For an excellent review on chromatin remodelling in the early embryo, see Fulka et al. (2008).
In the mouse, the pattern that has been defined is one where at fertilization, the maternal genome is packaged with histones already displaying various modifications (acetylation or methylation) in different regions of the genome and a high level of DNA methylation. In the paternal pronucleus, protamines are first replaced with histones that are more acetylated than those inherited by the maternal genome but evidence of early histone methylation appears soon after this incorporation. An active demethylation of the paternal DNA then occurs before DNA replication and only some specific regions of heterochromatin around centromeres, inhibitor of apoptosis (IAP) retrotransposons and paternally methylated imprinted genes escape it (Morgan et al., 2005). Both parental genomes are thus epigenetically unique, which may underlie the early transcriptional activation of the paternal genome. Following fertilization, the entire embryonic genome undergoes a passive DNA demethylation resulting in a largely demethylated genome by the morula stage (Morgan et al., 2005). Methylation of the genome reasserts itself at the blastocyst stage.
In recent years, several genes have been identified as having a role in EGA. These include genes that regulate early transcription of the zygotic genome, RNA stabilization up to the 2-cell stage as well as genes involved in RNA translation at the 2-cell stage (Fig. 1). For example, Zscan4 is exclusively expressed in late 2-cell embryos as well as embryonic stem (ES) cells (Falco et al., 2007). Zscan4 mRNA is highly expressed at the 2-cell stage but not expressed at all at the 3-cell stage indicating that it has a very short half-life and is only transiently expressed. Functional analysis of Zscan4 using RNAi knock-down revealed that Zscan4 is indeed important for the 2-cell stage progression to the 4-cell stage, timely progression to the blastocyst stage, proper blastocyst hatching and implantation (Falco et al., 2007). Furthermore, Zscan4 knock-down embryos showed reduced ability to hatch from the zona pellucida, and when reintroduced into the uterus of pseudo-pregnant females, they were unable to implant compared with control-injected embryos (Falco et al., 2007).
One of the first chromatin remodelling genes to be identified during early development is a maternally derived transcript called BRG1, also known as Smarca4 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4) (Bultman et al., 2006). BRG1 is the enzymatic subunit of the SWI/SNF complex and BRG1 null embryos display preimplantation embryo lethality and die at the blastocyst stage (Bultman et al., 2000; Bultman et al., 2006). However, heterozygous females are able to accumulate maternal transcripts of BRG1 which may support development and survival to the blastocyst stage. BRG1 conditional knock-outs produce oocytes that develop normally, and could be fertilized, which indicates that BRG1 did not affect oocyte maturation or fertility. However, 78% of the maternally depleted BRG1 embryos arrest at the 2-cell stage compared with only 3% of control embryos (Bultman et al., 2006). Transcription was also reduced in BRG1 maternally depleted embryos by up to 30% (Bultman et al., 2006). Genes involved in cell cycle, transcription and mRNA processing were particularly affected including the transcription-requiring complex, an accepted marker of EGA (Schultz, 2002). The SWI/SNF complex is involved in chromatin remodelling via histone modification, therefore changes in dimethyl H3K4 were investigated due to its correlation with transcriptional activity (Bultman et al., 2006). Maternally depleted embryos of BRG1 showed a 61% reduction in dimethylated H3K4 compared with controls (Bultman et al., 2006).
For successful MET, we must also consider the importance of RNA translation, not just transcription. Recently, peptidyl arginine deiminase 6 (PADI6) was found to associated with oocyte cytoplasmic lattices (CPLs) (Yurttas et al., 2008). PADI6 plays a role in establishing or maintaining the CPL, which in turn affects ribosome localization at the 2-cell stage (Yurttas et al., 2008). Padi6 −/− females are infertile, and although the oocytes can be fertilized, they block at the 2-cell stage. Furthermore, CPLs do not form in the oocyte, which indicates that they may be playing a role in EGA. Padi6 −/− embryos expressed lower levels of total rRNA compared with control embryos, and as a result, proper genome activation was disrupted (Yurttas et al., 2008). These results underscore the importance of lattice structures in the developing embryo and, in particular, have identified a gene, PADI6, directly involved in correct EGA. These are but a few examples of the exciting discoveries that are defining the global and specific mechanisms controlling genome activation in the early embryo. Following EGA, the embryos proceed with the next major event that establishes cell polarity in the outer embryonic blastomeres and triggers the programme that supports the differentiation of these cells into the epithelial TE.
Lineage specification during preimplantation development
Embryo compaction represents the first morphogenetic indication of cell lineage specification in the early embryo. Despite the intriguing possibility that even 2-cell blastomeres may display asymmetries and some level of cell lineage bias (Piotrowska et al., 2001; Plusa et al., 2005), compaction is still coincident with the loss of blastomere totipotency and the initiation of the TE developmental programme. Compaction is defined by an increase in cell-to-cell contact between embryonic blastomeres. It is promoted by the establishment of adherens junctions consisting of E-cadherin and catenin complexes (Fleming et al., 2000; Johnson and McConnell, 2004). Compaction is initiated at the 8-cell stage in the mouse but the timing varies across species (Telford et al., 1990). In all cases, compaction results in the formation of a morula, which creates a topology that forms outer cells and inner cells that are completely surrounded by the outer cells, which are the progenitors of the TE and ICM, respectively (Fleming et al., 2000; Johnson and McConnell, 2004). The main model that defines this event is called ‘the cell polarity model’ (Tarkowski and Wroblewska, 1967; Johnson and Ziomek, 1981). The cell polarity model predicts that cell fate is established at the 8-cell stage in the mouse and propagated by symmetrical or asymmetrical cell divisions that either generate two polar cells by dividing a radial polarity axis or an outer polar and an inner apolar cell (Johnson and McConnell, 2004; Yamanaka et al., 2006). Adherens junction components, the PAR complex (PAR 1, 3 and 6), atypical protein kinase C and tight junction-associated proteins are key mediators of these events, but what factors actually regulate lineage specification in the early embryo? Recent research applied to the mouse has established that TE and ICM differentially express several lineage-specific transcription factors (Fig. 2). Cdx2 becomes restricted to the TE and is required for TE formation (Yamanaka et al., 2006). In contrast, Oct4 and Nanog become restricted to and influence ICM fate (Yamanaka et al., 2006; Fig. 2). This research has led to the formation of a model that proposes mutual antagonism between Oct4 and Cdx2 in supporting the formation of TE and ICM fates in the blastocyst (Yamanaka et al., 2006). The focus of research in this area is now directed at understanding the target genes that these transcription factors control and how these transcription factors regulate the cell fate of ES cell lines.
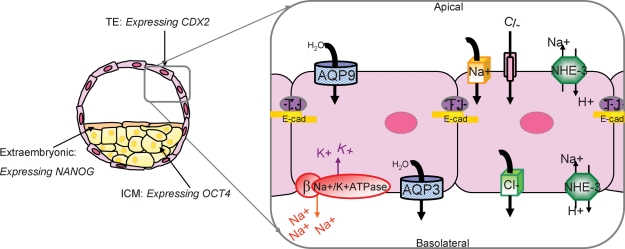
Figure 2:
Preimplantation embryo cell lineage specification and blastocyst formation model.
Recent research applied to the mouse has established that TE and ICM differentially express several lineage-specific transcription factors. Cdx2 becomes restricted to the TE and is required for TE formation (Yamanaka et al., 2006). In contrast, Oct4 and Nanog become restricted to and influence ICM fate (Yamanaka et al., 2006). This research supports a model that proposes mutual antagonism between Oct4 and Cdx2 in supporting the formation of TE and ICM fates in the blastocyst (Yamanaka et al., 2006). Our research has been directed at understanding the mechanisms that control blastocyst formation, which is dependent upon TE differentiation, as it is the ion and water transport functions of the TE that mediate the fluid dynamics that control cavitation (Watson et al., 1992; Watson and Barcroft, 2001). Na/K-ATPase, AQP (water channels) and tight junction (TJ) proteins have established roles in coordinating blastocyst formation (Watson et al., 1992; Watson and Barcroft, 2001). Our model predicts that blastocyst formation is dependent upon the polarized distribution of the Na/K-ATPα1β1ase confined to the basolateral membrane domains of the TE. This establishes a trans-TE ion gradient that facilitates movement of water across the epithelium facilitated by the presence of both apical and basolateral AQPs. These events combined with the establishment of a TE tight junctional seal to prevent the loss of fluid out of the embryo through paracellular routes result in the expansion of the embryo and the formation of the blastocyst (Watson and Barcroft, 2001) E-cad, E-cadherin; NHE-3, sodium–hydrogen exchanger 3.
Our research has been directed at understanding the mechanisms that control blastocyst formation, which is dependent upon TE differentiation, as it is the ion and water transport functions of the TE that mediate the fluid dynamics that control cavitation (Watson et al., 1992; Watson and Barcroft, 2001; Fig. 2). Na/K-ATPase, aquaporins (AQPs; water channels) and tight junction proteins have established roles in coordinating blastocyst formation (Watson et al., 1992; Watson and Barcroft, 2001). Our model predicts that blastocyst formation is dependent upon the polarized distribution of the Na/K-ATPα1β1ase confined to the basolateral membrane domains of the TE (Fig. 2). This establishes a trans-TE ion gradient that facilitates movement of water across the epithelium facilitated by the presence of both apical and basolateral AQPs (Fig. 2). These events combined with the establishment of a TE tight junctional seal to prevent the loss of fluid out of the embryo through paracellular routes result in the expansion of the embryo and the formation of the blastocyst (Kidder and McLachlin, 1985; Watson and Barcroft, 2001).
Na/K-ATPase enzyme activity increases just prior to blastocyst formation (Watson et al., 1992; Watson and Barcroft, 2001). The expression of all principal Na/K-ATPase isozymes has been determined, as well as the functions of Na/K-ATPase α1 and β1 subunits in supporting blastocyst formation in the mouse (Watson et al., 1990; MacPhee et al., 2000; Barcroft et al., 2004; Madan et al., 2007). In addition, AQPs and their role in facilitating blastocyst formation in the mouse have been investigated (Barcroft et al., 2003; Offenberg and Thomsen, 2005). Interestingly, ATP1β1 and Aqp8 mRNAs were two of the genes up-regulated during the 4- to 8-cell stages (Hamatani et al., 2004), many hours prior to the morphological changes of compaction. Aqp8 mRNA is also down-regulated in cultured mouse blastocysts compared with in vivo derived blastocysts in several microarray studies (Rinaudo and Schultz, 2004; Rinaudo et al., 2006; Giritharan et al., 2007) as were Aqp3 and 9 (Giritharan et al., 2007).
In addition to these critical gene products, recent studies have uncovered additional waves of gene expression, one of which is now called the MGA, which includes 3329 genes whose expression first increase at the 4- and 8-cell stages (Hamatani et al., 2004). A further characterization of TS versus ES cells indicated that genes such as Gata3, IRX3, EndoA, Bmp8b, Sin 3b and Gata6 are all more highly expressed in TS cells than in ES cells, indicating that they may play specific roles in TE cell differentiation (Tanaka et al., 2002; Hamatani et al., 2004). Among the other genes up-regulated during the MGA were several tight junction, gap junction, Na/K-ATPase and AQP gene family members (Hamatani et al., 2004). Genes up-regulated from the morula to blastocyst stages included skeletal development/morphogenesis, lipid binding, glucose metabolism/energy pathway, actin binding/cytoskeletal and steroid metabolism genes (Hamatani et al., 2004; Tanaka and Ko, 2004; Zeng et al., 2004). Thus, the array studies have been invaluable in confirming the physiological and molecular data implicating ion transporters, cell junctions and water channels in mediating blastocyst formation and have also identified many other important candidates that must now be investigated to define their functional contributions to preimplantation development, TE differentiation and blastocyst formation.
Thus, the principal challenge that remains is to place all of these stage specific gene expression patterns into a functional context during preimplantation development. Fortunately, there have been great advances made in recent years to apply RNA silencing methods and injection of cRNAs into early embryos to explore both loss and gain of function to identify gene function in the early embryo. In addition, the development and characterization of specific and potent pharmacological agents that target specific gene function and signalling pathways have been invaluable in defining gene function during preimplantation development. We have applied both strategies for investigating the role of Na/K-ATPase subunits and also mitogen-activated protein kinase pathway (MAPK) function during preimplantation development. Our studies have determined that siRNA knock-down of the Na/K-ATPase β1 subunit results in a blockade of development at the morula stage, as only 3% of β1 subunit down-regulated embryos undergo cavitation (Madan et al., 2007). The application of pharmacological approaches using cytokine suppressive anti-inflammatory drugs (CSAIDs) has resulted in our identification of the p38 MAPK pathway as having both an essential role in supporting preimplantation development and also a regulatory role to allow the embryo to respond to exposure to stressful culture environments (Natale et al., 2004; Paliga et al., 2005; Fong et al., 2007; Hickson et al., 2007). Treatment of 2- or 8-cell embryos with active CSAID results in a reversible blockade of development at the 8- to 16-cell stage (Natale et al., 2004; Paliga et al., 2005). This blockade is associated with a loss of filamentous actin in each blastomere, which reforms following removal of the CSAID from the culture medium allowing the resumption of normal development to the blastocyst stage (Paliga et al., 2005). In addition, increasing the osmolarity of culture medium increased p38 MAPK pathway activation (Fong et al., 2007). We have also used a similar approach to discover that p38 MAPK increases AQP 3 and 9 expression (mRNA and protein) in response to hyperosmotic stress, but only when the medium is made hyperosmotic with sucrose and not with glycerol (Bell and Watson, unpublished results). These types of studies highlight approaches that are required to explore and define gene function during preimplantation development. Methods for defining gene function during preimplantation development are now readily available and future studies must focus on taking the information acquired from genomic studies of gene expression patterns and translating them into a functional understanding of the precise roles played by these genes during preimplantation development. This information is required to fully understand and develop ways of assessing embryonic developmental potential prior to embryo transfer. This ability must be developed, as it represents the main breakthrough for human IVF practitioners to fully implement strategies such as single embryo transfer to promote healthy singleton pregnancies following the application of assisted reproductive technologies to treat infertility.
Influence of culture on embryonic transcripts and preimplantation programming
The application of assisted reproductive technologies to all mammalian species requires the exposure of gametes and early embryos to culture environments. There has been a concerted effort applied to constant improvement of culture environments since embryo culture was first developed in the early 1960s; however, even with all of the advances and improvements, culture conditions have not been fully optimized for any mammalian species including the mouse to date. While it is possible to achieve impressive frequencies of development to the blastocyst stage for a variety of mammalian species in vitro, blastocyst development does not represent the ideal end-point for assessing culture influences on development. One of most profound new understandings that has emerged from studies conducted over the past 15 years is that cultured embryos display significant changes in their gene expression patterns and these changes affect not only survival during the preimplantation period but also affect the ability of the embryo to implant, proceed normally through fetal development and also post-partum health and susceptibility to disease in later life. It is imperative that the mechanisms underlying these effects of culture on development be defined and the knowledge obtained from such an understanding be applied directly to improving embryo culture further. This is especially relevant considering the need to apply single embryo blastocyst transfer in human assisted reproductive procedures to reduce the incidence of multiple pregnancies that often are associated with low birthweight infants and premature delivery. The literature is replete with studies that have now documented significant changes in mRNA transcript levels in cultured versus in vivo derived embryos. These effects of culture have been studied and documented in a variety of mammalian species including the human, and have also been extended to oocyte maturation in addition to culture influences on in vivo matured and fertilized, cultured zygotes (Watson et al., 2000; Lonergan et al., 2003; Russell et al., 2006).
While the documentation of these differences in transcript levels between cultured and in vivo derived embryos is sound, the interpretation and understanding of the functional significance of these changes is not well understood. It is necessary to correlate these changes in gene expression with pregnancy outcomes. While this is understandably very difficult to do, El-Sayed et al. (2006) conducted an experiment where the relationship between the transcriptional profiles of embryos measured in cell biopsies was correlated with pregnancy outcome. The results indicated that embryos that successfully generated pregnancies and successful fetuses displayed up-regulated expression of genes necessary for implantation (COX2, CDX2), carbohydrate metabolism (ALOX15), growth factor (BMP15), signal transduction (PLAU) and placental development (PLAC8), while embryos that failed to generate a pregnancy overexpressed inflammatory cytokines (TNF), protein amino acid binding (EEF1A1), transcription factors (MSX1, PTTG1) and glucose metabolism genes (El-Sayed et al., 2006). We have highlighted this study as it indicates the need for extending experimental end-points to fully assess and understand the impact of exposure to culture environments on embryonic and fetal programming.
Our efforts in this regard have begun to focus on understanding the mechanisms and signalling pathways that preimplantation embryos activate in response to exposure to culture environments (Fig. 3). There is an assumption in the literature that the majority of variations in gene expression patterns observed between cultured and in vivo derived embryos are a consequence of changes in gene transcription. However, it is equally likely that these changes stem from variations in the mechanisms that cells employ to regulate mRNA stability and half-life (Fig. 3). Thus, our research efforts are now being directed towards understanding the role of RBPs during preimplantation development, so that we may test the hypothesis that the culture-induced changes in mRNA abundance are regulated by RBP-mediated changes in target mRNA stability (Fig. 3). These studies will not only address culture-induced mechanisms controlling gene expression in the early embryo, but also will increase our understanding of mRNA transcript regulation in general during preimplantation development.
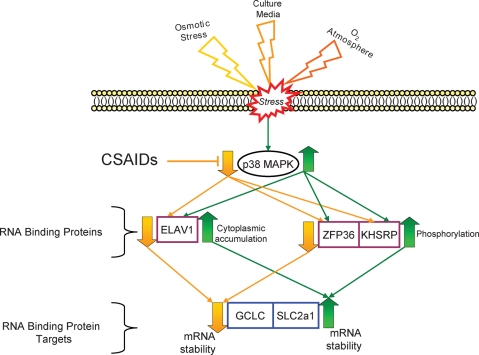
Figure 3:
MAPK regulation of RBPs and control of mRNA stability.
We are investigating the hypothesis that exposure to stressful environments activates p38MAPK activity in preimplantation embryos which in turn regulates RBP (Elavl1, ZFP36 and KHSRP) activity and the stability of their target mRNAs (GCLC and SLC2a1). This mechanism may be among the principal routes that preimplantation embryos utilize to adapt to exposure to changing environments and may contribute to the variation in mRNA levels that are reported to occur due to placement of embryos into suboptimal culture environments.
Regulation of mRNA transcripts during preimplantation development—RBPs
RBPs influence RNA editing, localization, stability and translation (Saunders and Barber, 2003). Moreover, regulated mRNA stability occurs in mammalian cells in response to nutrient levels, hormones and environmental stresses such as hypoxia and fluctuations in temperature and misregulation may occur in cancer or disease (reviewed by Guhaniyogi and Brewer, 2001). Up to 380 RBPs have been identified in the mouse genome (McKee et al., 2005). Except for MSY2 and DAZL, there has been little research on the role of mammalian RBPs in mammalian oocytes or early embryos (Ruggiu et al., 1997; Yu et al., 2003; Yang et al., 2005). MSY2 compromises ∼2% of total protein in the mouse oocyte. Both MSY2 mRNA and protein are degraded by the mouse 2-cell stage (Yu et al., 2001). In female MSY2 knock-out mice, there is early loss of follicles and oocytes, a lack of ovulation and infertility (Yang et al., 2005). MSY2 is likely to be a major control of maternal mRNA stability and translation in mice (Yu et al., 2001). DAZL is a member of the DAZ gene family (deleted in azoospermia), first identified in infertile men (Reijo et al., 1995), and has a role in regulating translation in germ cells (Collier et al., 2005). Female mouse knock-outs of DAZL have no follicles or oocytes and are infertile, while homozygous knock-out males have an absence of germ cells and even heterozygous males have reduced sperm counts and sperm abnormalities (Ruggiu et al., 1997). This suggests that the mammalian RBP DAZL is important for germ cell survival.
We have determined that several RBPs are expressed during both mouse and cow early embryonic development including Staufens 1 and 2 (STAU1 and STAU2), embryonic lethal abnormal vision-like 1 and 2 (ELAVL1 and ELAVL2, previously known as HuR, HuB), spermatid perinuclear RNA-binding protein (STRBP), deleted in azoospermia-like (DAZL), CUG triplet repeat RNA-binding protein 1 (CUGBP1, previously known as CUG-BP, EDEN-BP), smaug (SAMD4), zing finger protein 36 (ZFP36, previously known as tristetraprolin or TTP) and KH-type splicing regulatory protein (KHSRP) (Calder et al., unpublished results; Calder et al., 2008).
These results are important because studies have determined that ELAVL1, ZFP36 and KHSRP RBPs are downstream targets of p38 MAPK α/β signalling and that they bind to AU-rich elements (AREs) in the 3′UTR of target mRNAs, a process that results in either stabilization or turnover of target mRNAs (Lal et al., 2004; Raineri et al., 2004; Briata et al., 2005; Fechir et al., 2005; Linker et al., 2005; Song et al., 2005; Chen et al., 2006; Cherry et al., 2006; Chou et al., 2006; Gantt et al., 2006; Marderosian et al., 2006; Fig. 3). Regulation of mRNA turnover is a critical component determining abundance of mRNA transcripts and thus regulation of gene expression in general (Piccioni et al., 2005). The best characterized cis-elements controlling mRNA stability are the AREs (Lal et al., 2004; Raineri et al., 2004; Chen et al., 2006). AREs orchestrate exosomal degradation of mRNAs. mRNAs containing AREs are usually rapidly turned over in resting cells but are readily stabilized through interactions with 3′UTR-binding RBPs (Lal et al., 2004; Raineri et al., 2004; Linker et al., 2005; Chen et al., 2006). Several RBPs that bind to AREs and regulate mRNA turnover have been identified. ELAV RBPs bind to AREs and stabilize mRNA transcripts (Lal et al., 2004; Song et al., 2005; Chen et al., 2006), while ZFP36 and KHSRP RBPs also bind to AREs but instead promote the rapid decay of mRNA transcripts (Briata et al., 2005; Fechir et al., 2005; Linker et al., 2005; Chen et al., 2006; Chou et al., 2006; Marderosian et al., 2006). Increased cytoplasmic localization of ELAVL1 protein is associated with increased oxidative stress and enhanced MAPKAPK2/3 phosphorylation (Lal et al., 2004; Raineri et al., 2004; Cherry et al., 2006).
Studies have indicated that ZFP36 phosphorylation is regulated by p38 MAPK signalling and that phosphorylated ZFP36 displays reduced RNA-binding activity associated with reduced target mRNA degradation (Fechir et al., 2005; Song et al., 2005; Marderosian et al., 2006). The ZFP36 gene has been knocked out in mice; however, the main phenotype is arthritis and there is no reported reproductive phenotype (Taylor et al., 1996). Our results indicate an increase in expression at the mouse 4-cell stage (Calder et al., unpublished results). However, in the knockout of a ZFP36 family member, ZFP36L2, nulls are infertile because embryos block at the 2-cell stage (Ramos et al., 2004). Because of these interesting features, ZFP36 family members should be more thoroughly investigated in the early mouse embryo, their roles are likely to be in degradation of maternal mRNAs.
KHSRP also regulates mRNA turnover by binding to AREs, and interactions with the exosome and deadenylases (Linker et al., 2005; Chou et al., 2006). Studies have determined that stability of target mRNAs (iNOS) is increased when KHSRP is immunodepleted and that KHSRP co-immunoprecipitates with ZFP36 on target mRNAs and also competes directly with ELAVL1 for ARE-binding sites on target mRNAs (Linker et al., 2005; Chou et al., 2006). In addition, KHSRP is phosphorylated by p38 MAPK and its phosphorylation is accompanied by reduced binding to target mRNAs and increased target mRNA stability (Linker et al., 2005; Chou et al., 2006).
We are testing a model that predicts that ZFP36 and KHSRP may therefore oppose ELAVL1 binding and that p38 MAPK α/β signalling may increase mRNA stability by both reducing ZFP36 and KHSRP binding and increasing ELAVL1 cytoplasmic distribution (Fechir et al., 2005; Song et al., 2005; Marderosian et al., 2006; Fig. 3). Our results demonstrate that gene products encoding these RBPs are present in mouse oocytes and early embryos. Furthermore, two microarray studies have shown that abundance of several RBP mRNAs and mRNA processing and metabolism are affected by culture (Whitworth et al., 2005; Giritharan et al., 2007). The proposed studies will establish a foundation for investigating the roles of RBPs during early development and, more importantly, their collective roles in regulating mRNA transcripts during early embryogenesis. We predict that this route is one of the primary mechanisms being affected in embryonic responses to culture environment and alteration of mRNA half-life likely represents a fundamental mechanism controlling undesired changes in preimplantation embryo and fetal programming.
MicroRNAs
In addition to transcription, adenylation, deadenylation, methylation, acetylation and RBPs, the discovery of hundreds of microRNAs (miRNAs—20–22 nucleotides) as regulators of post-transcriptional events has defined yet another level of mRNA regulation that could have a major impact on defining the programme controlling preimplantation development (Blakaj and Lin, 2008; Kedde and Agami, 2008). These short mRNAs have the ability to target and regulate the stability and translation of individual and pools of target mRNAs, thus are likely to have profound effects on embryonic gene expression patterns. Interest in this area began following investigation of the mouse phenotype resulting from the deletion of Dicer 1, an enzyme required for miRNA production in cells. One study which generated Dicer nulls by homologous recombination showed that embryos failed to progress to the 2-cell stage (Murchison et al., 2005; Tang et al., 2007), and the lack of miRNAs resulted in persistence of maternal mRNAs. However, knock-down of Dicer by siRNA did not result in early embryonic lethality, and embryos developed to the blastocyst stage but with reduced expression of OCT3/4, Nanog and SOX2 transcription factors (Cui et al., 2007) that may have contributed to later mortality and loss of stem cells seen in an earlier study (Bernstein et al., 2003). In zebrafish, miRNA-430 is one of the most abundant miRNAs, with hundreds of possible targets, and accumulates at the MET (Giraldez et al., 2006). When Dicer was knocked out, miRNA-430 is not present and maternal transcripts were not deadenylated and degraded in a timely fashion (Giraldez et al., 2006). We believe that the investigation of the functions of specific miRNAs represents an important new avenue for understanding the mechanisms controlling early development. The possibility becomes even more intriguing when one considers that recent studies have demonstrated that RBPs and miRNAs may co-operate with or counteract each other to control transcript stability and translation (Kedde and Agami, 2008; Yu and Hecht, 2008). These truly are exciting times for this type of research.
Summary
The preimplantation developmental programme shifts from a maternal to embryonic programme rapidly after fertilization. Although the majority of oogenetic products are lost during the MET, several do survive this interval to contribute directly to supporting preimplantation development. EGA is characterized by the transient expression of a few genes that are likely necessary for MET, and while EGA represents the first major wave of gene expression, there are several more, including the MGA, that supports development to the blastocyst stage. These events are regulated by changes in chromatin remodelling that include variations in DNA methylation and histone acetylation. The application of genomic approaches have greatly assisted in the discovery of stage specific gene expression patterns and the challenge now is to largely define gene function and regulation during preimplantation development. The basic mechanisms controlling compaction, lineage specification and blastocyst formation are defined; however, much more knowledge is required to understand how the specific gene products controlling these events are regulated and the function of most genes is not understood to date. The requirement for embryo culture has revealed plasticity in the developmental programme that may exceed the adaptive capacity of the embryo and has fostered important research directions aimed at alleviating culture-induced changes in embryonic programming. New levels of regulation are emerging and greater insight into the roles played by RBPs and miRNAs is required to fully understand the regulation of transcript stability and translation during preimplantation development. All of this exciting research is relevant due to the necessity to produce healthy and competent preimplantation embryos for embryo transfer, to ensure that assisted reproductive technologies are applied in the most efficient and safest way possible.
Funding
Unpublished studies conducted by the authors and referred to in this review were supported by research operating funds obtained from the Natural Sciences and Engineering Research Council (NSERC) of Canada and the Canadian Institutes of Health Research (CIHR) awarded to AJW.
References
- Alizadeh Z, Kageyama S, Aoki F. Degradation of maternal mRNA in mouse embryos: selective degradation of specific mRNAs after fertilization. Mol Reprod Dev. 2005;72:281–290. doi: 10.1002/mrd.20340. [DOI] [PubMed] [Google Scholar]
- Bachvarova R. Gene expression during oogenesis and oocyte development in mammals. Dev Biol (N Y 1985) 1985;1:453–524. doi: 10.1007/978-1-4615-6814-8_11. [DOI] [PubMed] [Google Scholar]
- Bachvarova R, De Leon V. Polyadenylated RNA of mouse ova and loss of maternal RNA in early development. Dev Biol. 1980;74:1–8. doi: 10.1016/0012-1606(80)90048-2. [DOI] [PubMed] [Google Scholar]
- Barcroft LC, Offenberg H, Thomsen P, Watson AJ. Aquaporin proteins in murine trophectoderm mediate transepithelial water movements during cavitation. Dev Biol. 2003;256:342–354. doi: 10.1016/s0012-1606(02)00127-6. [DOI] [PubMed] [Google Scholar]
- Barcroft LC, Moseley AE, Lingrel JB, Watson AJ. Deletion of the Na/K-ATPase alpha1-subunit gene (Atp1a1) does not prevent cavitation of the preimplantation mouse embryo. Mech Dev. 2004;121:417–426. doi: 10.1016/j.mod.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bettegowda A, Lee KB, Smith GW. Cytoplasmic and nuclear determinants of the maternal-to-embryonic transition. Reprod Fertil Dev. 2008;20:45–53. doi: 10.1071/rd07156. [DOI] [PubMed] [Google Scholar]
- Blakaj A, Lin H. Piecing together the mosaic of early mammalian development through microRNAs. J Biol Chem. 2008;283:9505–9508. doi: 10.1074/jbc.R800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevini-Gandolfi TA, Favetta LA, Mauri L, Luciano AM, Cillo F, Gandolfi F. Changes in poly(A) tail length of maternal transcripts during in vitro maturation of bovine oocytes and their relation with developmental competence. Mol Reprod Dev. 1999;52:427–433. doi: 10.1002/(SICI)1098-2795(199904)52:4<427::AID-MRD12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, Puri PL, Gherzi R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol Cell. 2005;20:891–903. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder MD, Madan P, Watson AJ. Bovine oocytes and early embryos express Staufen and ELAVL RNA-binding proteins. Zygote. 2008;16:161–168. doi: 10.1017/S096719940700456X. [DOI] [PubMed] [Google Scholar]
- Chen YL, Huang YL, Lin NY, Chen HC, Chiu WC, Chang CJ. Differential regulation of ARE-mediated TNFalpha and IL-1beta mRNA stability by lipopolysaccharide in RAW264.7 cells. Biochem Biophys Res Commun. 2006;346:160–168. doi: 10.1016/j.bbrc.2006.05.093. [DOI] [PubMed] [Google Scholar]
- Cherry J, Karschner V, Jones H, Pekala PH. HuR, an RNA-binding protein, involved in the control of cellular differentiation. In Vivo. 2006;20:17–23. [PubMed] [Google Scholar]
- Chou CF, Mulky A, Maitra S, Lin WJ, Gherzi R, Kappes J, Chen CY. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol Cell Biol. 2006;26:3695–3706. doi: 10.1128/MCB.26.10.3695-3706.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XS, Shen XH, Kim NH. Dicer1 expression in preimplantation mouse embryos: involvement of Oct3/4 transcription at the blastocyst stage. Biochem Biophys Res Commun. 2007;352:231–236. doi: 10.1016/j.bbrc.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C, Pera RA. The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet. 2004;13:1461–1470. doi: 10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- El-Sayed A, Hoelker M, Rings F, Salilew D, Jennen D, Tholen E, Sirard MA, Schellander K, Tesfaye D. Large-scale transcriptional analysis of bovine embryo biopsies in relation to pregnancy success after transfer to recipients. Physiol Genomics. 2006;28:84–96. doi: 10.1152/physiolgenomics.00111.2006. [DOI] [PubMed] [Google Scholar]
- Fair T, Murphy M, Rizos D, Moss C, Martin F, Boland MP, Lonergan P. Analysis of differential maternal mRNA expression in developmentally competent and incompetent bovine two-cell embryos. Mol Reprod Dev. 2004;67:136–144. doi: 10.1002/mrd.10385. [DOI] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechir M, Linker K, Pautz A, Hubrich T, Forstermann U, Rodriguez-Pascual F, Kleinert H. Tristetraprolin regulates the expression of the human inducible nitric-oxide synthase gene. Mol Pharmacol. 2005;67:2148–2161. doi: 10.1124/mol.104.008763. [DOI] [PubMed] [Google Scholar]
- Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1982;1:681–686. doi: 10.1002/j.1460-2075.1982.tb01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TP, Ghassemifar MR, Sheth B. Junctional complexes in the early mammalian embryo. Semin Reprod Med. 2000;18:185–193. doi: 10.1055/s-2000-12557. [DOI] [PubMed] [Google Scholar]
- Fong B, Watson PH, Watson AJ. Mouse preimplantation embryo responses to culture medium osmolarity include increased expression of CCM2 and p38 MAPK activation. BMC Dev Biol. 2007;7:2–17. doi: 10.1186/1471-213X-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulka H, St John JC, Fulka J, Hozak P. Chromatin in early mammalian embryos: achieving the pluripotent state. Differentiation. 2008;76:3–14. doi: 10.1111/j.1432-0436.2007.00247.x. [DOI] [PubMed] [Google Scholar]
- Gandolfi TA, Gandolfi F. The maternal legacy to the embryo: cytoplasmic components and their effects on early development. Theriogenology. 2001;55:1255–1276. doi: 10.1016/s0093-691x(01)00481-2. [DOI] [PubMed] [Google Scholar]
- Gantt KR, Cherry J, Richardson M, Karschner V, Atasoy U, Pekala PH. The regulation of glucose transporter (GLUT1) expression by the RNA binding protein HuR. J Cell Biochem. 2006;99:565–574. doi: 10.1002/jcb.20950. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish miRNA-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Giritharan G, Talbi S, Donjacour A, Di Sebastiano F, Dobson AT, Rinaudo PF. Effect of in vitro fertilization on gene expression and development of mouse preimplantation embryos. Reproduction. 2007;134:63–72. doi: 10.1530/REP-06-0247. [DOI] [PubMed] [Google Scholar]
- Gosden RG. Oogenesis as a foundation for embryogenesis. Mol Cell Endocrinol. 2002;186:149–153. doi: 10.1016/s0303-7207(01)00683-9. [DOI] [PubMed] [Google Scholar]
- Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Adan A, Rizos D, Fair T, Moreira PN, Pintado B, de la Fuente J, Boland MP, Lonergan P. Effect of speed of development on mRNA expression pattern in early bovine embryos cultured in vivo or in vitro. Mol Reprod Dev. 2004;68:441–448. doi: 10.1002/mrd.20113. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hickson JA, Fong B, Watson PH, Watson AJ. PP2Cdelta (Ppm1d, WIP1), an endogenous inhibitor of p38 MAPK, is regulated along with Trp53 and Cdkn2a following p38 MAPK inhibition during mouse preimplantation development. Mol Reprod Dev. 2007;74:821–834. doi: 10.1002/mrd.20688. [DOI] [PubMed] [Google Scholar]
- Huarte J, Belin D, Vassalli A, Strickland S, Vassalli JD. Meiotic maturation of mouse oocytes triggers the translation and polyadenylation of dormant tissue-type plasminogen activator mRNA. Genes Dev. 1987;1:1201–1211. doi: 10.1101/gad.1.10.1201. [DOI] [PubMed] [Google Scholar]
- Johnson MH, McConnell JM. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol. 2004;15:583–597. doi: 10.1016/j.semcdb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Johnson MH, Ziomek CA. Induction of polarity in mouse 8-cell blastomeres: specificity, geometry, and stability. J Cell Biol. 1981;91:303–308. doi: 10.1083/jcb.91.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Davies H, Sims LP, Levy SE, Dean J. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev Biol. 2007;7:67–79. doi: 10.1186/1471-213X-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Agami R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle. 2008;7:899–903. doi: 10.4161/cc.7.7.5644. [DOI] [PubMed] [Google Scholar]
- Kidder GM, McLachlin JR. Timing of transcription and protein synthesis underlying morphogenesis in preimplantation mouse embryos. Dev Biol. 1985;112:265–275. doi: 10.1016/0012-1606(85)90397-5. [DOI] [PubMed] [Google Scholar]
- Krisher RL. The effect of oocyte quality on development. J Anim Sci. 2004;82(E-Suppl):E14–E23. doi: 10.2527/2004.8213_supplE14x. [DOI] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequarre AS, Traverso JM, Marchandise J, Donnay I. Poly(A) RNA is reduced by half during bovine oocyte maturation but increases when meiotic arrest is maintained with CDK inhibitors. Biol Reprod. 2004;71:425–431. doi: 10.1095/biolreprod.103.026724. [DOI] [PubMed] [Google Scholar]
- Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan P, Rizos D, Gutierrez-Adan A, Moreira PM, Pintado B, de la Fuente J, Boland MP. Temporal divergence in the pattern of messenger RNA expression in bovine embryos cultured from the zygote to blastocyst stage in vitro or in vivo. Biol Reprod. 2003;69:1424–1431. doi: 10.1095/biolreprod.103.018168. [DOI] [PubMed] [Google Scholar]
- Ma J, Svoboda P, Schultz RM, Stein P. Regulation of zygotic gene activation in the preimplantation mouse embryo: global activation and repression of gene expression. Biol Reprod. 2001;64:1713–1721. doi: 10.1095/biolreprod64.6.1713. [DOI] [PubMed] [Google Scholar]
- MacPhee DJ, Jones DH, Barr KJ, Betts DH, Watson AJ, Kidder GM. Differential involvement of Na(+), K(+)-ATPase isozymes in preimplantation development of the mouse. Dev Biol. 2000;222:486–498. doi: 10.1006/dbio.2000.9708. [DOI] [PubMed] [Google Scholar]
- Madan P, Rose K, Watson AJ. Na/K-ATPase beta1 subunit expression is required for blastocyst formation and normal assembly of trophectoderm tight junction-associated proteins. J Biol Chem. 2007;282:12127–12134. doi: 10.1074/jbc.M700696200. [DOI] [PubMed] [Google Scholar]
- Marderosian M, Sharma A, Funk AP, Vartanian R, Masri J, Jo OD, Gera JF. Tristetraprolin regulates Cyclin D1 and c-Myc mRNA stability in response to rapamycin in an Akt-dependent manner via p38 MAPK signaling. Oncogene. 2006;25:6277–6290. doi: 10.1038/sj.onc.1209645. [DOI] [PubMed] [Google Scholar]
- McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14–22. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misirlioglu M, Page GP, Sagirkaya H, Kaya A, Parrish JJ, First NL, Memili E. Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc Natl Acad Sci USA. 2006;103:18905–18910. doi: 10.1073/pnas.0608247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale DR, Paliga AJ, Beier F, D'souza SJ, Watson AJ. p38 MAPK signaling during murine preimplantation development. Dev Biol. 2004;268:76–88. doi: 10.1016/j.ydbio.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Offenberg H, Thomsen PD. Functional challenge affects aquaporin mRNA abundance in mouse blastocysts. Mol Reprod Dev. 2005;71:422–430. doi: 10.1002/mrd.20306. [DOI] [PubMed] [Google Scholar]
- Oh B, Hwang S, McLaughlin J, Solter D, Knowles BB. Timely translation during the mouse oocyte-to-embryo transition. Development. 2000;127:3795–3803. doi: 10.1242/dev.127.17.3795. [DOI] [PubMed] [Google Scholar]
- Paliga AJ, Natale DR, Watson AJ. p38 mitogen-activated protein kinase (MAPK) first regulates filamentous actin at the 8–16-cell stage during preimplantation development. Biol Cell. 2005;97:629–640. doi: 10.1042/BC20040146. [DOI] [PubMed] [Google Scholar]
- Paules RS, Buccione R, Moschel RC, Vande Woude GF, Eppig JJ. Mouse Mos protooncogene product is present and functions during oogenesis. Proc Natl Acad Sci USA. 1989;86:5395–5399. doi: 10.1073/pnas.86.14.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, Thresher R, Dixon JP, Zahn D, Colledge WH, Carlton MB, Nakano T, Surani MA. Stella is a maternal effect gene required for normal early development in mice. Curr Biol. 2003;13:2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Paynton BV, Bachvarova R. Polyadenylation and deadenylation of maternal mRNAs during oocyte growth and maturation in the mouse. Mol Reprod Dev. 1994;37:172–180. doi: 10.1002/mrd.1080370208. [DOI] [PubMed] [Google Scholar]
- Paynton BV, Rempel R, Bachvarova R. Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev Biol. 1988;129:304–314. doi: 10.1016/0012-1606(88)90377-6. [DOI] [PubMed] [Google Scholar]
- Piccioni F, Zappavigna V, Verrotti AC. Translational regulation during oogenesis and early development: the cap-poly(A) tail relationship. C R Biol. 2005;328:863–881. doi: 10.1016/j.crvi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Piko L, Clegg KB. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev Biol. 1982;89:362–378. doi: 10.1016/0012-1606(82)90325-6. [DOI] [PubMed] [Google Scholar]
- Piotrowska K, Wianny F, Pedersen RA, Zernicka-Goetz M. Blastomeres arising from the first cleavage division have distinguishable fates in normal mouse development. Development. 2001;128:3739–3748. doi: 10.1242/dev.128.19.3739. [DOI] [PubMed] [Google Scholar]
- Plusa B, Hadjantonakis AK, Gray D, Piotrowska-Nitsche K, Jedrusik A, Papaioannou VE, Glover DM, Zernicka-Goetz M. The first cleavage of the mouse zygote predicts the blastocyst axis. Nature. 2005;434:391–395. doi: 10.1038/nature03388. [DOI] [PubMed] [Google Scholar]
- Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos SB, Stumpo DJ, Kennington EA, Phillips RS, Bock CB, Ribeiro-Neto F, Blackshear PJ. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131:4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- Rinaudo P, Schultz RM. Effects of embryo culture on global pattern of gene expression in preimplantation mouse embryos. Reproduction. 2004;128:301–311. doi: 10.1530/rep.1.00297. [DOI] [PubMed] [Google Scholar]
- Rinaudo PF, Giritharan G, Talbi S, Dobson AT, Schultz RM. Effects of oxygen tension on gene expression in preimplantation mouse embryos. Fertil Steril. 2006;86:1252–1265. doi: 10.1016/j.fertnstert.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, Dorin J, Cooke HJ. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- Russell DF, Baqir S, Bordignon J, Betts DH. The impact of oocyte maturation media on early bovine embryonic development. Mol Reprod Dev. 2006;73:1255–1270. doi: 10.1002/mrd.20553. [DOI] [PubMed] [Google Scholar]
- Sagata N, Watanabe N, Vande Woude GF, Ikawa Y. The c-mos proto-oncogene product is a cytostatic factor responsible for meiotic arrest in vertebrate eggs. Nature. 1989;342:512–518. doi: 10.1038/342512a0. [DOI] [PubMed] [Google Scholar]
- Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- Schultz RM. Regulation of zygotic gene activation in the mouse. Bioessays. 1993;15:531–538. doi: 10.1002/bies.950150806. [DOI] [PubMed] [Google Scholar]
- Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- Schultz RM. From egg to embryo: a peripatetic journey. Reproduction. 2005;130:825–828. doi: 10.1530/rep.1.00902. [DOI] [PubMed] [Google Scholar]
- Schultz RM, Davis W, Jr, Stein P, Svoboda P. Reprogramming of gene expression during preimplantation development. J Exp Zool. 1999;285:276–282. doi: 10.1002/(sici)1097-010x(19991015)285:3<276::aid-jez11>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Shin SY, Lee HJ, Ko DS, Lee HC, Park WI. The regulators of VEGF expression in mouse ovaries. Yonsei Med J. 2005;46:679–686. doi: 10.3349/ymj.2005.46.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard MA, Dufort I, Vallee M, Massicotte L, Gravel C, Reghenas H, Watson AJ, King WA, Robert C. Potential and limitations of bovine-specific arrays for the analysis of mRNA levels in early development: preliminary analysis using a bovine embryonic array. Reprod Fertil Dev. 2005;17:47–57. doi: 10.1071/rd04113. [DOI] [PubMed] [Google Scholar]
- Sirard MA, Richard F, Blondin P, Robert C. Contribution of the oocyte to embryo quality. Theriogenology. 2006;65:126–136. doi: 10.1016/j.theriogenology.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Song IS, Tatebe S, Dai W, Kuo MT. Delayed mechanism for induction of gamma-glutamylcysteine synthetase heavy subunit mRNA stability by oxidative stress involving p38 mitogen-activated protein kinase signaling. J Biol Chem. 2005;280:28230–28240. doi: 10.1074/jbc.M413103200. [DOI] [PubMed] [Google Scholar]
- Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- Stutz A, Conne B, Huarte J, Gubler P, Volkel V, Flandin P, Vassalli JD. Masking, unmasking, and regulated polyadenylation cooperate in the translational control of a dormant mRNA in mouse oocytes. Genes Dev. 1998;12:2535–2548. doi: 10.1101/gad.12.16.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y-Q, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, Eppig JJ. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302:104–117. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TS, Ko MS. A global view of gene expression in the preimplantation mouse embryo: morula versus blastocyst. Eur J Obstet Gynecol Reprod Biol. 2004;115(Suppl 1):S85–S91. doi: 10.1016/j.ejogrb.2004.01.026. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Kunath T, Kimber WL, Jaradat SA, Stagg CA, Usuda M, Yokota T, Niwa H, Rossant J, Ko MSH. Gene profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 2002;12:1921–1928. doi: 10.1101/gr.670002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowski AK, Wroblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol. 1967;18:155–180. [PubMed] [Google Scholar]
- Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev. 1990;26:90–100. doi: 10.1002/mrd.1080260113. [DOI] [PubMed] [Google Scholar]
- Thelie A, Papillier P, Pennetier S, Perreau C, Traverso JM, Uzbekova S, Mermillod P, Joly C, Humblot P, Dalbies-Tran R. Differential regulation of abundance and deadenylation of maternal transcripts during bovine oocyte maturation in vitro and in vivo. BMC Dev Biol. 2007;7:125–137. doi: 10.1186/1471-213X-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000;a 26:267–268. doi: 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- Tong ZB, Nelson LM, Dean J. Mater encodes a maternal protein in mice with a leucine-rich repeat domain homologous to porcine ribonuclease inhibitor. Mamm Genome. 2000;b 11:281–287. doi: 10.1007/s003350010053. [DOI] [PubMed] [Google Scholar]
- Vallee M, Aiba K, Piao Y, Palin MF, Ko MS, Sirard MA. Comparative analysis of oocyte transcript profiles reveals a high degree of conservation among species. Reproduction. 2008;135:439–448. doi: 10.1530/REP-07-0342. [DOI] [PubMed] [Google Scholar]
- Vigneault C, Gilbert I, Sirard MA, Robert C. Using the histone H2a transcript as an endogenous standard to study relative transcript abundance during bovine early development. Mol Reprod Dev. 2007;74:703–715. doi: 10.1002/mrd.20665. [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Watson AJ, Barcroft LC. Regulation of blastocyst formation. Front Biosci. 2001;6:D708–D730. doi: 10.2741/watson. [DOI] [PubMed] [Google Scholar]
- Watson AJ, Damsky CH, Kidder GM. Differentiation of an epithelium: factors affecting the polarized distribution of Na+,K(+)-ATPase in mouse trophectoderm. Dev Biol. 1990;141:104–114. doi: 10.1016/0012-1606(90)90105-r. [DOI] [PubMed] [Google Scholar]
- Watson AJ, Kidder GM, Schultz GA. How to make a blastocyst. Biochem Cell Biol. 1992;70:849–855. doi: 10.1139/o92-133. [DOI] [PubMed] [Google Scholar]
- Watson AJ, De Sousa P, Caveney A, Barcroft LC, Natale D, Urquhart J, Westhusin ME. Impact of bovine oocyte maturation media on oocyte transcript levels, blastocyst development, cell number, and apoptosis. Biol Reprod. 2000;62:355–364. doi: 10.1095/biolreprod62.2.355. [DOI] [PubMed] [Google Scholar]
- Whitworth KM, Agca C, Kim JG, Patel RV, Springer GK, Bivens NJ, Forrester LJ, Mathialagan N, Green JA, Prather RS. Transcriptional profiling of pig embryogenesis by using a 15-K member unigene set specific for pig reproductive tissues and embryos. Biol Reprod. 2005;72:1437–1451. doi: 10.1095/biolreprod.104.037952. [DOI] [PubMed] [Google Scholar]
- Wrenzycki C, Herrmann D, Carnwath JW, Niemann H. Alterations in the relative abundance of gene transcripts in preimplantation bovine embryos cultured in medium supplemented with either serum or PVA. Mol Reprod Dev. 1999;53:8–18. doi: 10.1002/(SICI)1098-2795(199905)53:1<8::AID-MRD2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 2003;a 33:187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
- Wu X, Wang P, Brown CA, Zilinski CA, Matzuk MM. Zygote arrest 1 (Zar1) is an evolutionarily conserved gene expressed in vertebrate ovaries. Biol Reprod. 2003;b 69:861–867. doi: 10.1095/biolreprod.103.016022. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Ralston A, Stephenson RO, Rossant J. Cell and molecular regulation of the mouse blastocyst. Dev Dyn. 2006;235:2301–2314. doi: 10.1002/dvdy.20844. [DOI] [PubMed] [Google Scholar]
- Yang J, Medvedev S, Yu J, Tang LC, Agno JE, Matzuk MM, Schultz RM, Hecht NB. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc Natl Acad Sci USA. 2005;102:5755–5760. doi: 10.1073/pnas.0408718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Hecht NB. The DNA/RNA-binding protein, translin, binds microRNA122a and increases its in vivo stability. J Androl. 2008;29:572–579. doi: 10.2164/jandrol.108.005090. [DOI] [PubMed] [Google Scholar]
- Yu J, Hecht NB, Schultz RM. Expression of MSY2 in mouse oocytes and preimplantation embryos. Biol Reprod. 2001;65:1260–1270. doi: 10.1095/biolreprod65.4.1260. [DOI] [PubMed] [Google Scholar]
- Yu J, Hecht NB, Schultz RM. Requirement for RNA-binding activity of MSY2 for cytoplasmic localization and retention in mouse oocytes. Dev Biol. 2003;255:249–262. doi: 10.1016/s0012-1606(02)00094-5. [DOI] [PubMed] [Google Scholar]
- Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, Coonrod SA. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol. 2005;283:40–57. doi: 10.1016/j.ydbio.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]