Abstract
Methyltransferases that employ cobalamin cofactors, or their analogues the cobamides, as intermediates in catalysis of methyl transfer play vital roles in energy generation in anaerobic unicellular organisms. In a broader range of organisms they are involved in the conversion of homocysteine to methionine. Although the individual methyl transfer reactions catalyzed are simple SN2 displacements, the required change in coordination at the cobalt of the cobalamin or cobamide cofactors and the lability of the reduced Co+1 intermediates introduces the necessity for complex conformational changes during the catalytic cycle. Recent spectroscopic and structural studies on several of these methyltransferases have helped to reveal the strategies by which these conformational changes are facilitated and controlled.
Introduction
Although organic chemists have long been fascinated with the adenosylcobalamin-dependent enzymes that employ radical chemistry, it has become apparent over the last few decades that the majority of enzymes that employ cobalamin cofactors actually catalyze methyl transfer reactions and use methylcobalamin or its structural homologues the methylcobamides1 as intermediates in the methyl transfers. The discovery of the third domain of organisms, the Archaea, and the genome sequencing of many unicellular organisms, have contributed to our realization that methyl cobamides play essential roles in energy generation in many anaerobic organisms (reviewed in [1]). In contrast, in humans and other mammals, there is only one cobalamin-dependent methyltransferase, namely methionine synthase. Cobamides and cobalamin have not been found in fungi, or in higher plants, but many algae acquire cobalamins through a symbiotic relationship with bacteria, and they use cobalamin primarily as a cofactor for methionine synthase [2]. Table 1 lists some of the better-studied family members. However it is clear that there are many more still to be characterized. As an example, the recently determined genome sequence of Methanosarcina acetivorans contains 15 putative corrinoid protein sequences with unknown functions [3].
TABLE 1.
CORRINOID-DEPENDENT METHYLTRANSFERASES
| Enzyme complex | Protein designation | Organism | Reference |
|---|---|---|---|
| Methionine synthase | MetH | Escherichia coli Homo sapiens | [1] |
| Coenzyme M methyltransferases | |||
| monomethylamine:CoM | MtmBCA | Methanosarcina barkeri | [40,41] |
| dimethylamine:CoM | MtbB1CA | Methanosarcina barkeri | [42] |
| trimethylamine:CoM | MttB1CA | Methanosarcina barkeri | [43] |
| methanol:CoM | MtaBCA | Methanosarcina barkeri | [25] |
| dimethylsulfide:CoM | MtsAB | Methanosarcina barkeri | [44] |
| tetramethylammonium:CoM | MtqBCA | Methanococcoides sp. | [45] |
| Energy-conserving methyltetrahydromethanopterin: coenzyme M methyltransferase | MtrA-H | Methanobacterium thermoautotrophicum | [46] |
| Veratrol:H4folate O-demethylase | OdmABCD | Acetobacterium dehalogenans | [47,48] |
| Vanillate:H4folate O-methyltransferase | MtvABC | Moorella thermoaceticum | [49] |
| AcylCoA synthase/CO dehydrogenase | AcsABCDE | Moorella thermoaceticum | [20] |
| Methylchloride:H4folate methyltransferase | CmuAB | Methylobacterium sp. | [50] |
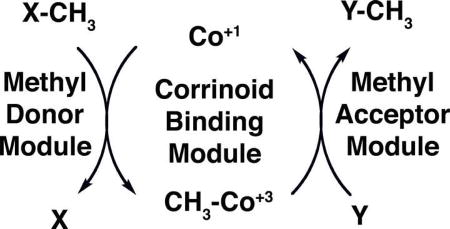
Methyl groups are transferred as carbocations from the cofactor to acceptor substrates, which are typically thiols such as homocysteine or coenzyme M (ethanethiol sulfonate), leaving the pair of electrons that formed the methyl-carbon bond of the cofactor so that the cobalt is now formally in the +1 oxidation state. The donor substrate then transfers a methyl group to the cofactor, reforming methylcobalamin. Substrate-binding components of these complexes may be either separate proteins, or modules of a protein that also contains the corrinoid-binding domain. Where structures of these substrate-binding components have been determined, they are TIM (α8β8) barrels (e.g. the homocysteine- and methyltetrahydrofolate-binding domains of methionine synthase [5]; the methyltetrahydrofolate-corrinoid-iron/sulfur protein methyltransferase that transfers methyl groups to the corrinoid-iron/sulfur protein [20]; the MtaB component of the methanol-coenzyme M methyltransferase from Methanosarcina barkeri [27••]; and the methylamine-binding domain (MtmB) of the monomethylamine:coenzyme M methyltransferase [28]).
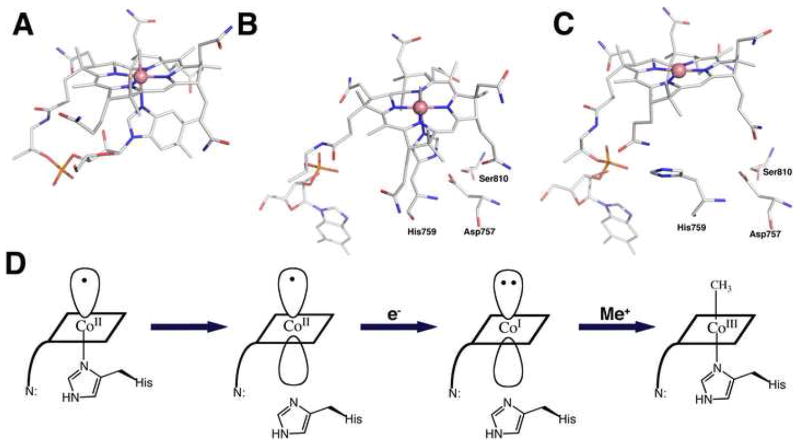
In methylcobalamin the dimethylbenzimidazole base is coordinated to the cobalt in the α-axial position as shown in Figure 1A. When methylcobalamin or methylcobamides are bound to methyltransferases, the nucleotide base is always dissociated. In most methyltransferases, a histidine ligand from the protein is coordinated in the α-axial position of the bound cofactor (Figure 1B), and a central issue currently being addressed in this field is the significance of this dramatic ligand substitution.
Figure 1. (A) The structure of methylcobalamin.
The cobalt in methylcobalamin, which is formally in the +3 oxidation state, is coordinated by four equatorial ligands contributed by the corrin ring and by two axial ligands. The β-axial ligand (the upper axial ligand in the orientation shown here) is the methyl anion in methylcobalalamin, while the lower axial ligand (the α ligand) in the free cofactor is a dimethylbenzimidazole nucleotide substituent of the D-ring of the corrin [32]. (B) The structure of methylcobalamin in the cobalamin-binding module of methionine synthase [6]. The dimethyl-benzimidazole nucleotide has dissociated, and cobalt is coordinated by His759 from the protein. His 759 is linked by a network of hydrogen bonds to Asp757 and Ser810. A signature Asp-X-His-X-X-Gly----Ser-Leu motif is diagnostic of the His-on state of cobalamin in many corrinoid proteins. While methylcobalamin is preferentially six-coordinate, cob(I)alamin is planar four-coordinate, due to the electron density of the lone pair of electrons that remain after transfer of the methyl group as a carbocation. As the cofactor cycles in catalysis between methylcobalamin and cob(I)alamin forms, the changes in preferred coordination geometry require that histidine dissociation be coupled with methyl transfer [33]. This histidine dissociation is accompanied by uptake of a proton, presumably due to protonation of the His-Asp-Ser ligand triad [34]. (C) The structure of cobalamin in the reactivation conformation of methionine synthase [10••]. In this structure Nε of His759 has rotated away from the cobalt of the cofactor, and is now 5.6 Å displaced from its position in (B). The structures in B and C are similarly oriented with respect to the cobalamin-binding domain while those in A and C are aligned based on the corrin rings. D. Cartoon of the change in cobalt coordination required for reduction of cob(II)alamin to cob(I)alamin and subsequent alkylation in His-on methyltransferases. Reactivation involves reduction of cob(II)alamin to cob(I)alamin and methylation of cob(I)alamin to regenerate methylcobalamin. In methionine synthase flavodoxin serves as the electron donor [35], and adenosylmethionine serves as the methyl donor for reactivation [7]. Cob(II)alamin is preferentially five-coordinate, and in methionine synthase His759 is coordinated in the α-axial position. Its reduction to cob(I)alamin is facilitated by dissociation of the histidine ligand. Binding of flavodoxin to the cob(II)alamin form of the enzyme results in dissociation of His759 from the cobalt, and uptake of a proton [36]. Following reduction to cob(I)alamin, methyl transfer from adenosylmethionine initially results in the formation of His-off methylcobalamin and the rate-limiting step in reactivation is the appearance of His-on methylcobalamin [37].
In this review, we will focus on a few family members that have been particularly well characterized both structurally and spectroscopically. These examples will serve to illustrate some of the features of this family and some of the issues that remain to be clarified.
Some well-studied examples of corrinoid-dependent methyltransferases
Cobalamin-dependent methionine synthase
Methionine synthase catalyzes the transfer of a methyl group from methyltetrahydrofolate to homocysteine, as illustrated in Figure 2A. The enzyme is a single polypeptide of 136 kDa, comprising four modules [4]. The first two modules are the homocysteine-binding acceptor domain and the methyltetrahydrofolate-binding donor domain, both of which are TIM (α8β8) barrels [5]. These domains retain the ability to catalyze methyl transfers to and from exogenous cobalamin: the homocysteine-binding domain catalyzes methyl group transfer from methylcobalamin to the bound acceptor to form methionine and cob(I)alamin, while the folate-binding domain catalyzes transfer from methyltetrahydrofolate to exogenous cob(I)alamin [4]. Thus the individual substrate-binding domains are methyltransferases in the same sense as the donor- and acceptor-binding polypeptides that react with separate corrinoid proteins in the enzyme complexes in Methanosarcina barkeri that are listed in Table 1.
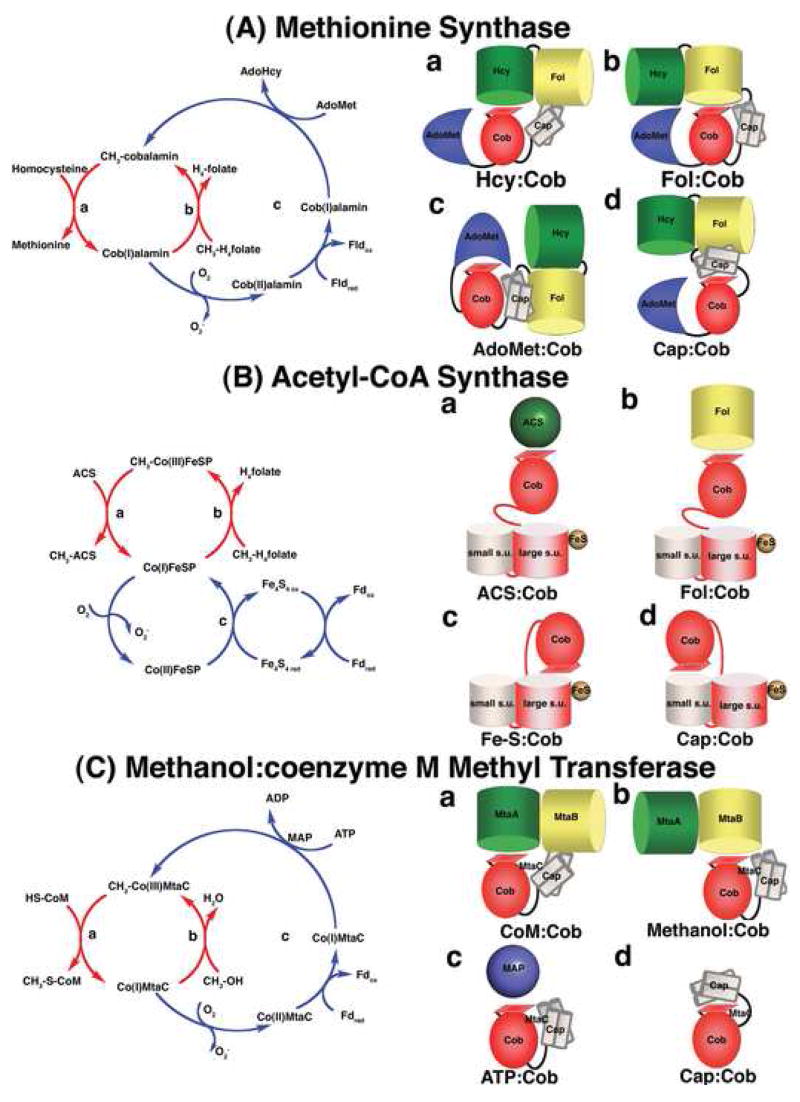
Figure 2. The catalytic cycles and proposed conformations of three well-studied methyltransferases.
(A) Methionine synthase The cobalamin cofactor is alternately methylated by CH3-H4folate bound to the Fol domain (b, requiring the Fol:Cob conformation cartooned on the right) and demethylated by homocysteine bound to the Hcy domain (a, Hcy:Cob). Occasionally, the cob(I)alamin cofactor undergoes oxidation. Return of the inactive cob(II)alamin species to the catalytic cycle require a reductive reactivation, in which adenosylmethionine bound to the C-terminal domain (c, AdoMet:Cob) serves as the methyl donor [7]. In E. coli, flavodoxin serves as the source of electrons for reductive activation [35], while in humans the source of electrons is methionine synthase reductase, a protein with homology to both flavodoxin and NADPH-ferredoxin (flavodoxin) oxidoreductase [38,39]. The remaining conformation (d, Cap:Cob) has only been seen in crystals of the isolated cobalamin-binding module. While the cartoon shows all four domains, x-ray structures of the full length protein have never been obtained, and the cartoons are based on docking of the N-terminal and C-terminal halves of the protein in a plausible manner. (B) Acetyl-CoA synthase. The 5-hydroxymethylbenzimidazolyl cobamide cofactor of the corrinoid iron/sulfur protein (AcsCD) is alternately methylated by CH3-H4folate bound to the AcsE methyltransferase (b, Fol:Cob) and demethylated by transfer of the methyl group to Ni+1 in acetyl CoA synthase (a, ACS:Cob). When the cob(I)amide undergoes occasional oxidation to cob(II)amide, it is returned to the catalytic cycle by reduction by the Fe4-S4 cluster on the large subunit (AcsC) of the corrinoid iron/sulfur protein (c, Fe-S:Cob). This cluster is in turn re-reduced with electrons derived from ferredoxin or from enzymes coupled to ferredoxin. The remaining conformation (d, Cap:Cob) is the one seen in the isolated corrinoid iron-sulfur protein [18••]). (C) Methanol:coenzyme M methyltransferase. The corrinoid-binding protein MtaC forms a complex with MtaA (the coenzyme M-binding methyltransferase) and MtaB (the methanol-binding methyltransferase). During catalysis the complex cycles between CoM:Cob (a) and Methanol:Cob (b) conformations. The latter complex has been characterized crystallo-graphically in the absence of MtaA. Activation of the cob(II)amide cofactor requires reduction by ferredoxin and is coupled to ATP hydrolysis by the action of the MAP protein (c, ATP:Cob). The Cap:Cob conformation (d) is hypothetical.
The third module binds methylcobalamin, and consists of two domains: a Rossmann domain that interacts with the α-surface of the cofactor and the pendant dimethylbenzimidazole nucleotide, and a “cap” domain, a four helix-bundle that packs against the β-face of the cofactor [6]. The Cap:Cob conformation has only been seen in a proteolytic fragment containing residues 649–897, and it is not known if this conformation is assumed by the full-length enzyme. Access of substrates bound to the first two modules of the enzyme to the cobalamin cofactor clearly requires major conformational changes that would displace the cap and alternately position donor and acceptor modules above the cofactor [5]. These proposed conformers, Cap:Cob, Hcy:Cob and Fol:Cob, are illustrated as cartoons in Figure 2A.
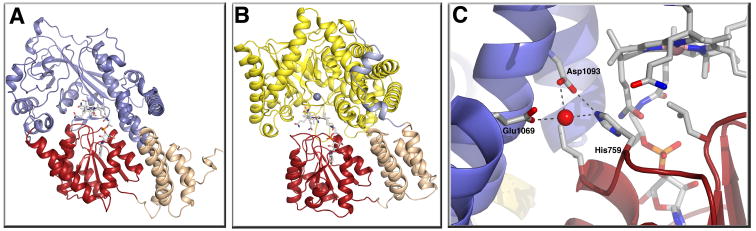
The final module in methionine synthase is required for reactivation of the enzyme when the cobalamin cofactor is oxidized to form cob(II)alamin [7]. X-ray structures of this module have been determined for both the E. coli [8] and human [9] enzymes. The structure of a C-terminal fragment of methionine synthase in the reactivation conformation (AdoMet:Cob, Figure 2A) is shown in Figure 3A [10••]. In contrast to the structure of the methylcobalamin form of methionine synthase in the His-on Cap:Cob conformation (Figure 1B), the cofactor in the reactivation conformation is now His-off (Figure 1C). Spectroscopic analysis of a full length His759Gly mutant indicated that in the AdoMet:Cob conformation the cob(II)alamin is five-coordinate, with a water in one of the axial positions [11]. Thus the model emerged of a change in protein conformation providing the energy needed to dissociate the histidine from cobalt when the enzyme enters the reactivation conformation. The structure of the fragment revealed another role for the histidine ligand. In this His-off form, the histidine now makes intermodular hydrogen-bonding interactions with residues in the activation module that are predicted to stabilize the activation conformation by 3–5 kcal/mol (Figure 3C). These intermodular interactions may explain the previously puzzling observation that while the methylcobalamin form of methionine synthase inter-converts between AdoMet:Cob and catalytic conformations, the cob(I)alamin form of the enzyme does not [12]. It will be extremely interesting to see whether similar intermodular interactions are made by histidine in the cob(I)alamin form of the Fol:Cob conformation. Such contacts could be the mechanism that prevents this catalytic intermediate from accessing adenosylmethionine and leading to futile cycling.
Figure 3. (A) Structure of the reactivation (AdoMet:Cob) conformation of methionine synthase[10••].
This structure was obtained with a fragment of the enzyme containing only the cobalamin-binding and adenosylmethionine-binding modules. The AdoMet-binding module is shown in blue, the cobalamin-binding domain in red, and the cap in tan. To reduce the conformational flexibility yet at the same time keeping the active site intact, a strategy of disulfide cross-linking between the two modules was used. Two cysteine mutations, Ile690Cys (in the cap domain) and Gly743Cys (in the cobalamin-binding domain) were introduced. The resulting disulfide cross-link sufficiently favored the conversion from His-on to His-off such that the protein crystallized in the AdoMet:Cob conformation. Nε of His-759 has moved 5.6 Å away from the cobalt towards the AdoMet binding module and is involved in an intermodular hydrogen bonding contact. (B) Structure of the MtaBC complex [27••] of methanol:coenzyme M methyltranferase, shown in the same orientation as methionine synthase in (A). MtaB forms a decorated TIM barrel (yellow), with the zinc atom (gray sphere) located at the C-terminal opening of the barrel in close proximity to the corrinoid of MtaC. The zinc is ligated by two cysteines and a glutamate, while the identity of the fourth ligand remains unclear. The sequences containing these three ligands bear no resemblance to sequences associated with zinc binding in other family members, despite the fact that the closest structural relative of MtaB is the homocysteine-binding domain of methionine synthase. Methanol is not present in the MtaBC complex, but if coordinated to the zinc by its hydroxyl oxygen, could be positioned appropriately for methyl transfer to the cobalt of the 5-hydroxybenzimidazolyl cobamide of MtaC, MtaC consists of two domains, a corrinoid-binding domain (red) and a four helix bundle or cap (tan) with an N-terminal extension (light blue). (C) Details of the intermodular interaction of His-off methionine synthase. His759 forms a hydrogen bond to Glu1069 and a water mediated hydrogen bond with Asp1093. This contact is expected to stabilize the His-off form in the AdoMet:Cob conformation.
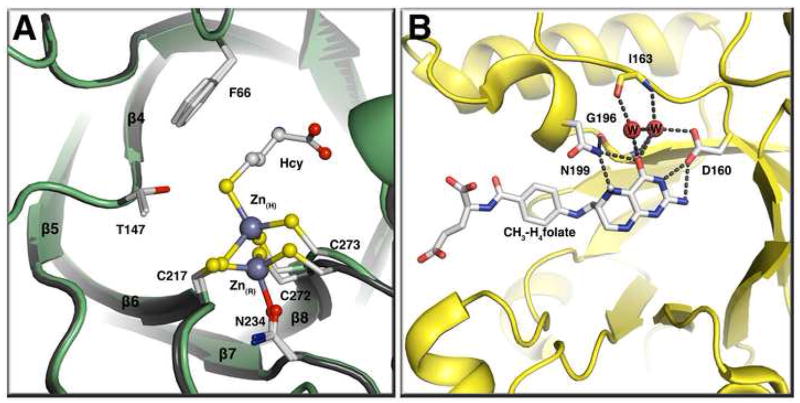
Methionine synthase, like many other corrinoid-dependent methyltransferases, transfers its methyl groups to a sulfur atom in the acceptor substrate homocysteine. At physiological pH values, homocysteine is present as the thiol rather than the more nucleophilic thiolate. Activation of the thiol substrates has been shown to involve a catalytically essential zinc ion. Zinc was found in the homocysteine-binding domain of cobalamin-dependent methionine synthase, where it was shown to be coordinated by three cysteines from the protein [13]. Addition of homocysteine resulted in a change in coordination from three thiol residues and one oxygen or nitrogen ligand to four thiol residues, and the displaced ligand was initially assumed to be a water molecule. However, very recently, it was shown that the fourth ligand in the unliganded enzyme was actually an Asn234, and that addition of homocysteine results in the displacement of this ligand with inversion of geometry around the zinc site and a considerable movement of the zinc away from Asn234 [14**]. Zinc is presumed to serve as a Lewis acid to stabilize the thiolate of homocysteine. However the inversion of geometry at the zinc site (Figure 4A) suggests that it may play a role not only in nucleophilic activation of the thiolate for methyl transfer but also in facilitating product dissociation as homocysteine is converted to the weaker thiol ether ligand methionine by methylation.
Figure 4. (A) The homocysteine binding domain of methionine synthase in the presence or absence of homocysteine[14**].
This panel shows a tructure superposition of the MetH resting state (gray) and the Hcy bound state (green) with Zn(R) representing the zinc atom at the resting state (no substrates bound) and Zn(H) the zinc atom at the Hcy bound state. Binding of homocysteine leads to an inversion of geometry at the active-site zinc and the displacement of Asn234 from the zinc coordination sphere. The zinc moves 2 Å on binding of homocysteine. (B) The CH3-H4folate-binding domain of the corrinoid iron-sulfur protein methyltransferase AcsE [41•]. CH3-H4folate is unprotonated in this structure and N5 accepts a hydrogen bond from Asn199.
Corrinoid iron-sulfur protein—a component of the acylCoA synthase complex
The corrinoid iron-sulfur protein (AcsCD) serves as an intermediate in the transfer of a methyl group between methyltetrahydrofolate and a nickel center of acylCoA synthase (Figure 2B). This methyl group is ultimately carbonylated and incorporated into acetylCoA. Methyltetrahydrofolate is bound to a donor methyltransferase, AcsE, which catalyzes methyl transfer to the cobamide (corrinoid) bound to AcsCD. Uniquely, in this system the acceptor substrate is a metal center on acylCoA synthase. The enzyme consists of two subunits, the larger of which contains the 4Fe-4S center.
Initial spectroscopic studies established that the cobamide cofactor of AcsCD exhibited a “base-off” spectrum under all conditions [15]. Optical and electron paramagnetic spectroscopy can be used to distinguish cob(II)alamin with an axial nitrogen ligand from “base-off” cob(II)alamin, but can not distinguish between four coordinate cob(II)alamin and five-coordinate cob(II)alamin with an oxygen ligand such as water coordinated ito one of the axial positions. Magnetic circular dichroism and computational analysis can be used to distinguish these two alternatives [16] and these methods were used to show that the corrinoid iron-sulfur protein in the Co(II) form is actually five-coordinate with a water axial ligand [17•]. Magnetic circular dichroism is also useful for distinguishing six-coordinate methylcobalamin with an axial water ligand from five-coordinate methylcobalamin, and the “base-off” methylated corrinoid iron-sulfur protein was shown to be six coordinate [17•].
The x-ray structure of a closely related corrinoid iron-sulfur protein from Carboxydothermus hydrogenoformans revealed that the cobamide cofactor (presumably in the Co+2 oxidation state) is five coordinate as expected from the spectroscopy, with a water in the β-axial position [18••]. The small subunit is a TIM barrel, while the larger subunit consists of three domains: an N-terminal domain that binds the iron-sulfur center, a central domain that is a TIM barrel, and a C-terminal domain with a Rossmann fold. The cobamide is positioned between the small subunit and the C-terminal domain of the large subunit. In this position, access to the cofactor by either the methyltransferase or acylCoA synthase would be blocked, so a conformational change would be needed to expose the cobalt (Figure 2B). Thus this conformation may be analogous to the Cap:Cob conformation seen in the cobalamin-binding module of methionine synthase. The iron-sulfur center is about 52 Å away from the cobalt in this structure. Since earlier studies had indicated that this center is responsible for the reduction of Co+2 to Co+1 during reductive activation of the corrinoid iron-sulfur protein [19], a conformational change would also be required to bring the corrinoid cofactor and the iron-sulfur center into proximity for electron transfer.
Like methionine synthase, the corrinoid iron-sulfur protein accepts a methyl group from methyltetrahydrofolate. The structure of the methyltetrahydrofolate-binding methyltransferase AcsE has recently been determined [20,21••]. There is no obvious proton donor for protonation of the tetrahydrofolate-leaving group, and instead, the nitrogen of Asn199 forms hydrogen bonds to both N5 and O4 of methyltetrahydrofolate (Figure 4B). Mutation of Asn199 results in a 25,000-fold decrease in kcat/Km, with only a minor effect on the binding of methyltetrahydrofolate. The authors propose that protonation of methyltetrahydrofolate would lead to the formation of a new hydrogen bond to the oxygen of this asparagyl residue in which N5-H would serve as the donor, a proposal that is very similar to the mechanism proposed for stabilization of the transition state accompanying protonation of hypoxanthine in purine nucleotide phosphorylase [22]. The interactions of methyltetrahydrofolate with the folate-binding module of methionine synthase are also very similar [5], although in this case, mutations of the homologous active site asparagyl residue have not been characterized.
Methanol-coenzyme M methyltransferase
The enzyme complex from Methanosarcina barkeri consists of three subunits: MtaC contains the 5-hydrobenzimidazolylcobalamide cofactor that interacts with the methanol-binding donor methyltransferase MtaB and the coenzyme M-binding acceptor methyltransferase MtaA. Both MtaB [23] and MtaA [24] require zinc for activity, and in both cases the zinc is thought to mediate electrophilic activation of the bound substrate. The MtaC subunit shows 35% homology with the cobalamin-binding domain of MetH, and the α-position of the corrinoid is ligated by His136 from this subunit [25]. A structure of a homologue of MtaC, specified by orf1948 from Moorella thermoacetica, has been determined [26], but this structure was obtained from crystals of an N-terminally truncated fragment lacking the “cap” domain. In M. barkeri, MtaB and MtaC form a tight complex, the structure of which was recently determined [27••]. This represents the first structure in which a substrate-binding module is docked with the corrinoid-binding module, and is illustrated in Figure 3B. In this structure, the “cap” of MtaC is displaced in a fashion similar to that seen in the structure of the AdoMet:Cob conformation of methionine synthase (Figure 3A), allowing MtaB to occupy the β-face of the cofactor.
Methanol, with a pKa of ~16, is an extremely challenging substrate to activate for methyl transfer. More generally, donor substrates, in contrast to acceptor substrates, often provide very poor leaving groups for methyl transfer and require substantial activation. These donor substrates include aromatic O-methyl ethers, methanol and various methylamines. A major remaining issue in the field concerns the mechanisms for activation of these challenging methyl donors. While zinc is essential for the activation of methanol, it is lacking in the donor substrate-binding sites of other methyltransferases. In monomethylamine methyltransferase a unique active site residue, pyrrolysine, is thought to be involved in covalent catalysis to activate the amine leaving group and facilitate methyl transfer [28]. And in methyltetrahydrofolate-binding domains, the strategy described above for the corrinoid iron-sulfur protein of forming a new hydrogen bond between protonated methyltetrahydrofolate and an active site asparagine appears to be operative. These diverse strategies for activation are reflected in the lack of sequence homologies in the various donor substrate binding domains.
Role of His as the lower axial ligand to cobalamin
Why is the dimethylbenzimidazole nucleotide always dissociated when cobamides bind to methyltransferases and why in some, but not all of these enzymes, is a histidine from the protein coordinated? We know from model studies that the affinity of the coordinated base for the cobalt is highly sensitive to the electron-donating properties of the upper axial ligand in cob(III)alamins and also to the oxidation state of the cobalt [29,30]. Thus the conversion of base-on to base-off cob(III)alamin is highly unfavorable when a weakly electron donating ligand like water is present in the β-position, and much less unfavorable with strongly electron donating methyl or propyl ligands [30]. The equilibrium between base-on and base-off forms of cob(II)alamin is intermediate between those measured for methylcobalamin and propylcobalamin [29]. Measurements of these equilibria with methionine synthase establish that the trends are remarkably similar, even though in this case the His-on/His-off transition is accompanied by major conformational changes [31•]. The equilibrium between the His-on and His-off forms of enzyme in the methylcobalamin state favors the His-on form by about 2.5 kcal/mol. In contrast, when the enzyme is in the cob(II)alamin form, the equilibrium shifts to favor the His-on form by only 0.5 kcal/mol. Hence, the histidine ligand serves as a sensor of the alkylation and/or oxidation state of the cofactor, and allows access to the activation conformation only when cob(II)alamin forms. This striking finding requires that the free energies needed for transition from one protein conformation to another be relatively small, so that the affinity of the histidine for the cobalt in different alkylation and oxidation states exerts a dominant effect. These studies also showed that flavodoxin binding further biases the equilibrium towards the reactivation conformation by preferentially binding to this protein conformation.
According to this view, a major function of the histidine ligand is to control access to the reactivation (AdoMet:Cob) conformation and thus prevent futile cycling. We know that the degree of futile cycling is actually very low in methionine synthase [12]. Why is there no requirement for histidine in the corrinoid iron-sulfur protein? As shown in Figure 2B, this enzyme, uniquely, can be reactivated by simple reduction of the cob(II)amide to a cob(I)amide, without the necessity to couple reduction to an exergonic reaction like methylation by adenosylmethionine or ATP hydrolysis. Thus futile cycling is not a problem for the corrinoid iron/sulfur protein.
A remaining challenge will be to obtain structures of a complete methyltransferase complex in each of its major conformational states. Because it has proved extremely difficult to crystallize mobile multi-domain or multi-subunit proteins, immobilization strategies (e.g. disulfide cross-linking) may be useful. We are also challenged to determine whether histidine ligation/displacement actually facilitates catalytic methyl transfers. Comparisons of the reactivity of cobamides and cobalamin with their parent methyltransferase modules in trans may be misleading, because the methylcobamides are then 6-coordinate, with an axial water ligand, whereas the appropriate comparison for methyl transfer in the physiological catalytic cycle may be with a transiently 5-coordinate MeCbl. It is tempting to believe that histidine association would provide considerable stabilization to the transition state during methyl transfer to cob(I)alamin.
Conclusions
A salient property of the cobalamin- and cobamide-dependent methyltransferases is the displacement of the dimethylbenzimidazole nucleotide of the cofactor on binding to the apoprotein and in most cases, its replacement by a histidyl ligand from the protein. The recent structural and spectroscopic studies described in this review have helped to clarify the role of the histidine ligand. It acts as a sensor of the oxidation and alkylation state of the cobalt of the cofactor, controlling access to the appropriate protein conformations at various stages of the catalytic cycle.
Acknowledgments
This work was supported by National Institutes of Health grants GM24908 (RGM) and GM048533 (Janet Smith, P. I.). We acknowledge a great debt to the late Professor Martha Ludwig, who played a central role in the elucidation of the structural features of methionine synthase and in understanding what the structures revealed in terms of function.
Footnotes
Cobamides differ from cobalamins in the nature of the base in the nucleotide that is pendant from the corrin ring: in cobalamins the base is 5,6-dimethylbenzimidazole, while in other cobamides alternative bases are present, e.g. 5-hydroxybenzimidazole, 5-methoxybenzimidazole, or adenosine. Both cobalamins and cobamides are corrinoids, a more general term for compounds with a cyclic tetrapyrrole based on the corrin skeleton, and proteins containing these cofactors are often referred to as corrinoid proteins.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended readings
- 1.Matthews RG. Corrinoid- and cobalamin-dependent methyltransferases. In: Sigal A, Sigal H, Sigal RKO, editors. Metal-carbon bonds in enzymes and cofactors. Royal Society of Chemistry; 2009:in press. vol 6 of “Metal Ions in Life Sciences”. [Google Scholar]
- 2.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 3.Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002;12:532–542. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goulding CW, Postigo D, Matthews RG. Cobalamin-dependent methionine synthase is a modular protein with distinct regions for binding homocysteine, methyltetrahydrofolate, cobalamin, and adenosylmethionine. Biochemistry. 1997;36:8082–8091. doi: 10.1021/bi9705164. [DOI] [PubMed] [Google Scholar]
- 5.Evans JC, Huddler DP, Hilgers MT, Romanchuk G, Matthews RG, Ludwig ML. Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase. Proc Natl Acad Sci USA. 2004;101:3729–3736. doi: 10.1073/pnas.0308082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig M. How a protein binds B12: A 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 7.Drummond JT, Huang S, Blumenthal RM, Matthews RG. Assignment of enzymic function to specific protein regions of cobalamin-dependent methionine synthase from Escherichia coli. Biochemistry. 1993;32:9290–9295. doi: 10.1021/bi00087a005. [DOI] [PubMed] [Google Scholar]
- 8.Dixon MM, Huang S, Matthews RG, Ludwig M. The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12. Structure. 1996;4:1263–1275. doi: 10.1016/s0969-2126(96)00135-9. [DOI] [PubMed] [Google Scholar]
- 9.Wolthers KR, Toogood HS, Jowitt TA, Marshall KR, Leys D, Scrutton NS. Crystal structure and solution characterization of the activation domain of human methionine synthase. FEBS J. 2007;274:738–750. doi: 10.1111/j.1742-4658.2006.05618.x. [DOI] [PubMed] [Google Scholar]
- 10••.Datta S, Koutmos M, Pattridge KA, Ludwig ML, Matthews RG. A disulfide-stabilized conformer of methionine synthase reveals an unexpected role for the histidine ligand of the cobalamin cofactor. Proc Natl Acad Sci U S A. 2008;105:4115–4120. doi: 10.1073/pnas.0800329105. A disulfide cross-link was engineered to stabilize the AdoMet:Cob conformation and enable the crystallization of the C-terminal half of methionine synthase. The structure provides the first view of His759 in the His-off AdoMet:Cob conformation. In this conformation, His759 is involved in intermodular hydrogen bonding interactions with the AdoMet-binding domain that are proposed to play a role in stabilization of the activation conformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liptak MD, Brunold TC. Spectroscopic and computational studies of Co1+ cobalamin: spectral and electronic properties of the “superreduced” B12 cofactor. J Am Chem Soc. 2006;128:9144–9156. doi: 10.1021/ja061433q. [DOI] [PubMed] [Google Scholar]
- 12.Jarrett JT, Huang S, Matthews RG. Methionine synthase exists in two distinct conformations that differ in reactivity toward methyltetrahydrofolate, adenosylmethionine, and flavodoxin. Biochemistry. 1998;37:5372–5382. doi: 10.1021/bi9730893. [DOI] [PubMed] [Google Scholar]
- 13.Goulding CW, Matthews RG. Cobalamin-dependent methionine synthase from Escherichia coli: involvement of zinc in homocysteine activation. Biochemistry. 1997;36:15749–15757. doi: 10.1021/bi971988l. [DOI] [PubMed] [Google Scholar]
- 14••.Koutmos M, Pejchal R, Bomer TM, Matthews RG, Smith JL, Ludwig ML. Metal active site elasticity linked to activation of homocysteine in methionine synthases. Proc Natl Acad Sci USA. 2008;105:3286–3291. doi: 10.1073/pnas.0709960105. The zinc atom in MetH is shown to be ligated by three cysteines and a asparagyl residue in the resting enzyme and to expel the asparagyl ligand with an inversion in its geometry as homocysteine is bound. Associated with inversion, the zinc moves more than 2Å upon homocysteine binding. This unexpected flexiblity in the metal center is proposed to facilitate methyltransfer and product release. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ragsdale SW, Lindahl PA, Munck E. Mossbauer, EPR, and optical studies of the corrinoid/iron-sulfur protein involved in the synthesis of acetyl coenzyme A by Clostridium thermoaceticum. J Biol Chem. 1987;262:14289–14297. [PubMed] [Google Scholar]
- 16.Stich TA, Buan NR, Brunold TC. Spectroscopic and computational studies of Co2+corrinoids: Spectral and electronic properties of the biologically relevant base-on and base-off forms of Co2+Cobalamin. J Am Chem Soc. 2004;126:9735–9749. doi: 10.1021/ja0481631. [DOI] [PubMed] [Google Scholar]
- 17•.Stich TA, Seravalli J, Venkateshrao S, Spiro TG, Ragsdale SW, Brunold TC. Spectroscopic studies of the corrinoid/iron-sulfur protein from Moorella thermoacetica. J Am Chem Soc. 2006;128:5010–5020. doi: 10.1021/ja054690o. Elegant spectroscopic/computational studies show that in the cobalt(III) and cobalt(II) states the enzyme-bound corrinoid is base-off with water as the axial ligand. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Svetlitchnaia T, Svetlitchnyi V, Meyer O, Dobbek H. Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl- CoA synthesis. Proc Natl Acad Sci USA. 2006;103:14331–14336. doi: 10.1073/pnas.0601420103. The crystal structure reveals the relative position of the smaller subunit above the bound base-off cobalamin moiety with a water molecule occupying the β-axial position. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menon S, Ragsdale SW. Role of the [4Fe-4S] cluster in reductive activation of the cobalt center of the corrinoid iron-sulfur protein from Clostridium thermoaceticum during acetate biosynthesis. Biochemistry. 1998;37:5689–5698. doi: 10.1021/bi9727996. [DOI] [PubMed] [Google Scholar]
- 20.Doukov T, Seravelli J, Stezowski JJ, Ragsdale SW. Crystal structure of a methyltetrahydrofolate- and corrinoid-dependent methyltransferase. Structure. 2000;8:817–830. doi: 10.1016/s0969-2126(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 21••.Doukov TI, Hemmi H, Drennan CL, Ragsdale SW. Structural and kinetic evidence for an extended hydrogen-bonding network in catalysis of methyl group transfer: Role of an active site asparagine residue in activation of methyl transfer by methyltransferases. Biochemistry. 2007;282:6609–6618. doi: 10.1074/jbc.M609828200. This paper reveals the network required for protonation of methyltetrahydrofolate in the corrinoid iron-sulfur protein methyltransferase AcsE. The hydrogen bonding of the enzyme-methyltetrahydrofolate complex is shown. Asn199 is proposed serve as a hydrogen bond acceptor to N5 of protonated methyltetrahydrofolate, by analogy with the role of Asn in purine nucleotide phosphorylase. Mutation of Asn199 is shown to decrease the activity by ~20,000-fold, but to have only a minor effect on the binding of methyltetrahydrofolate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Federov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JD, Gainsford GJ, Larese JA, Schramm VL, et al. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- 23.Sauer K, Thauer RK. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri: zinc dependence and thermodynamics of the methanol:cob(I)alamin methyltransferase reaction. Eur J Biochem. 1997;249:280–285. doi: 10.1111/j.1432-1033.1997.t01-1-00280.x. [DOI] [PubMed] [Google Scholar]
- 24.Sauer K, Thauer RK. Methyl-coenzyme M formation in methanogenic archaea. Involvement of zinc in coenzyme M activation. Eur J Biochem. 2000;267:2498–2504. doi: 10.1046/j.1432-1327.2000.01245.x. [DOI] [PubMed] [Google Scholar]
- 25.Sauer K, Thauer TK. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri: Identification of the active-site histidine in the corrinoid-harboring subunit MtaC by site-directed mutagenesis. Eur J Biochem. 1998;253:698–705. doi: 10.1046/j.1432-1327.1998.2530698.x. [DOI] [PubMed] [Google Scholar]
- 26.Das A, Fu Z-Q, Tempel W, Liu Z-J, Chang J, Chen L, Lee D, Zhou W, Xu H, Shaw N, et al. Characterization of a corrinoid protein involved in the C1 metabolism of strict anaerobic bacterium Moorella thermoacetica. Proteins. 2007;67:167–176. doi: 10.1002/prot.21094. [DOI] [PubMed] [Google Scholar]
- 27••.Hagemeier CH, Kruer M, Thauer RK, Warkentin E, Ermler U. Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex. Proc Natl Acad Sci USA. 2006;103:18917–18922. doi: 10.1073/pnas.0603650103. In this paper, the structure for the cobalamin-containing MtaC protein docked with its partner MtaB, which contains a binding site for coenzyme M has been determined. This is thus far the only structure available for a corrinoid-dependent methyltransferase docked with a substrate-binding domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 29.Brown KL, Peck-Siler S. Heteronuclear NMR studies of cobalamins. 9 Temperature-dependent NMR of organocobalt corrins enriched in 13C in the organic ligand and the thermodynamics of the base-on/base-off reaction. Inorg Chem. 1988;27:3548–3555. [Google Scholar]
- 30.Brown KL, Hakimi JM, Nuss DM, Montejano YD, Jacobsen DW. Acid-base properties of a-ribazole and the thermodynamics of dimethylbenzimidazole association in alkylcobalamins. Inorg Chem. 1984;27:3548–3555. [Google Scholar]
- 31•.Fleischhacker AS, Matthews RG. Ligand trans influence governs conformation in cobalamin-dependent methionine synthase. Biochemistry. 2007;46:12382–12392. doi: 10.1021/bi701367c. Access to the AdoMet:Cob conformation of methionine synthase is governed by the trans effect conveyed by the upper axial ligand to the cobalt of cobalamin, as well as by the net charge on the cobalt. Increasing electron density on the cobalt, whether induced by the trans axial ligand or by reduction of the cobalt, favors entrance into the AdoMet:Cob conformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randaccio L, Furlan M, Geremia S, Slouf M, Srnova I, Toffoli D. Similarities and differences between cobalamins and cobalaoximes. Accurate structural determination of methylcobalamin and of LiCl- and KCl-containing cyanocobalamins by synchrotron radiation. Inorg Chem. 2000;39:3403–3413. doi: 10.1021/ic0001199. CCDC 148899. [DOI] [PubMed] [Google Scholar]
- 33.Bandarian V, Pattridge KA, Lennon BW, Huddler DR, Matthews RG, Ludwig ML. Domain alternation switches B12-dependent methionine synthase to the activation conformation. Nat Struct Biol. 2002;9:53–56. doi: 10.1038/nsb738. [DOI] [PubMed] [Google Scholar]
- 34.Jarrett JT, Choi CY, Matthews RG. Changes in protonation associated with substrate binding and cob(I)alamin formation in cobalamin-dependent methionine synthase. Biochemistry. 1997;36:15739–15748. doi: 10.1021/bi971987t. [DOI] [PubMed] [Google Scholar]
- 35.Hall DA, Jordan-Starck TC, Loo RO, Ludwig ML, Matthews RG. Interaction of flavodoxin with cobalamin-dependent methionine synthase. Biochemistry. 2000;39:10711–10719. doi: 10.1021/bi001096c. [DOI] [PubMed] [Google Scholar]
- 36.Hoover DM, Jarrett JT, Sands RH, Dunham WR, Ludwig ML, Matthews RG. Interaction of Escherichia coli cobalamin-dependent methionine synthase and its physiological partner flavodoxin: Binding of flavodoxin leads to axial ligand dissociation from the cobalamin cofactor. Biochemistry. 1997;36:127–138. doi: 10.1021/bi961693s. [DOI] [PubMed] [Google Scholar]
- 37.Jarrett JT, Hoover DM, Ludwig ML, Matthews RG. The mechanism of adenosylmethionine-dependent activation of methionine synthase: A rapid kinetic analysis of intermediates in reductive methylation of cob(II)alamin enzyme. Biochemistry. 1998;37:12649–12658. doi: 10.1021/bi9808565. [DOI] [PubMed] [Google Scholar]
- 38.LeClerc D, Wilson A, Dumas R, Gafuik C, Song D, Watkins D, Heng HHQ, Rommens JM, Scherer SW, Rosenblatt DS, et al. Cloning and mapping of a cDNA for methionine synthase reductase, a flavoprotein defective in patients with homocystinuria. Proc Natl Acad Sci, U S A. 1998;95:3059–3064. doi: 10.1073/pnas.95.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olteanu H, Banerjee R. Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation. J Biol Chem. 2001;276:35558–35563. doi: 10.1074/jbc.M103707200. [DOI] [PubMed] [Google Scholar]
- 40.Burke SA, Krzycki JA. Reconstitution of monomethylamine:coenzyme M methyl transfer with a corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J Biol Chem. 1997;272:16570–16577. doi: 10.1074/jbc.272.26.16570. [DOI] [PubMed] [Google Scholar]
- 41.Burke SA, Lo SL, Krzycki JA. Clustered genes encoding the methyltransferases of methanogenesis from monomethylamine. J Bacteriol. 1998;180:3432–3440. doi: 10.1128/jb.180.13.3432-3440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson DJ, Jr, Gorlatova N, Grahame DA, Krzycki JA. Reconstitution of dimethylamine:coenzyme M methyl transfer with a discrete corrinoid protein and two methyltranserases purified from Methanosarcina barkeri. J Biol Chem. 2000;275:29053–29060. doi: 10.1074/jbc.M910218199. [DOI] [PubMed] [Google Scholar]
- 43.Paul L, Ferguson DJ, Kryzycki JA. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J Bacteriol. 2000;182:2520–2529. doi: 10.1128/jb.182.9.2520-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tallant TC, Paul L, Krzycki JA. The MtsA subunit of the methylthiol:coenzyme M methyltransferase of Methanosarcina barkeri catalyses both half-reactions of corrinoid dependent dimethylsulfide:coenzyme M methyl transfer. J Biol Chem. 2001;276:4485–4493. doi: 10.1074/jbc.M007514200. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka K. Anaerobic degradation of tetramethylammonium by a newly isolated marine methanogen. J Ferment Bioeng. 1994;78:386–388. [Google Scholar]
- 46.Gottschalk G, Thauer RK. The Na+-translocating methyltransferase complex from methanogenic archaea. Biochim Biophys Acta. 2001;1505:28–36. doi: 10.1016/s0005-2728(00)00274-7. [DOI] [PubMed] [Google Scholar]
- 47.Kaufmann F, Wohlfarth G, Diekert G. O-Demethylase from Acetobacterium dehalogenans: Cloning, sequencing, and active expression of the gene encoding the corrinoid protein. Eur J Biochem. 1998;257:5125–5521. doi: 10.1046/j.1432-1327.1998.2570515.x. [DOI] [PubMed] [Google Scholar]
- 48.Kaufmann F, Wohlfarth G, Diekert G. O-Demethylase from Acetobacterium dehalogenans. Substrate specificity and function of the participating proteins. Eur J Biochem. 1998;253:706–711. doi: 10.1046/j.1432-1327.1998.2530706.x. [DOI] [PubMed] [Google Scholar]
- 49.Naidu D, Ragsdale SW. Characterization of a three-component vanillate O-demethylase from Moorella thermoacetica. J Bacteriol. 2001;183:3272–3281. doi: 10.1128/JB.183.11.3276-3281.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanelli T, Messmer M, Studer A, Vuilleumier S, Leisinger T. A corrinoid-dependent catabolic pathway for growth of a Methylobacterium strain with chloromethane. Proc Nat Acad Sci. 1999;96:4615–4620. doi: 10.1073/pnas.96.8.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]