Abstract
BCL11B is a transcriptional regulator with important role in T cell development and leukemogenesis. Recently we demonstrated that BCL11B controls expression from IL-2 promoter through direct binding to the US1 site. Here we provide evidence that BCL11B also participates in the activation of IL-2 gene expression by enhancing NF-kB activity in the context of TCR/CD28–triggered T cell activation. Enhanced NF-kB activation is not a consequence of BCL11B binding to the NF-kB response elements or association with the NF-kB-DNA complexes, but rather the result of higher translocation of NF-kB to the nucleus caused by enhanced degradation of the IkB. The enhanced IkB degradation in cells with increased levels of BCL11B was specific for T cells activated through TCR, but not through TNFα or UV, and was caused by higher activity of IkB kinase, as indicated by its higher phosphorylation. As BCL11B is a transcription factor we investigated whether expression of genes upstream of IkB kinase in the TCR/CD28 signaling pathway was affected by increased BCL11B expression, and found that Cot kinase mRNA levels were elevated. Cot kinase is known to promote enhanced IkB kinase activity which results in phosphorylation and degradation of the IkB inhibitors and activation of NF-kB. Implication of Cot kinase in BCL11B-mediated NF-kB activation in response TCR activation is supported by the fact that a Cot kinase dominant negative mutant or Cot kinase siRNA blocked BCL11B-mediated NF-kB activation. In support of our observations, we report that BCL11B enhances expression of several other NF-kB target genes, in addition to IL-2. In addition, we provide evidence that BCL11B associates with Cot kinase gene intron 2 to regulate its expression.
Keywords: BCL11B, CTIP2, NF-kB, Cot kinase, TCR signaling
INTRODUCTION
BCL11B, also known as CTIP2, is a C2H2 zinc finger transcription factor with critical role in T cell and brain development [1-3]. BCL11B was initially identified as a corepressor for COUP-TF nuclear receptors [4] and shown later to directly bind DNA and recruit the NuRD complex to repress expression from targeted promoters [5, 6]. In addition, we previously demonstrated that BCL11B participates in the transcriptional activation of IL-2 gene expression in response to TCR activation by direct binding to the US1 site in the IL-2 promoter [7]. IL-2 is the first cytokine whose expression is induced immediately after T cell activation through TCR/CD28 signaling [8]. Briefly, calcinerurin, a calcium/calmodulin-dependent serine/threonine phosphatase, and PKCθ are activated. The primary target for calcineurin is the nuclear factor of activated T cells (NF-AT) which is dephosphorylated and translocated to the nucleus, where it binds to the IL-2 promoter [9]. PKCθ is required for activation of the transcription factors NF-kB and AP-1 (Fos/Jun) [10, 11]. NF-kB activation requires a second costimulatory signal provided by CD28 receptor [12].
Here we demonstrate that BCL11B participates in the activation of IL-2 gene expression not only through binding to the US1 site, but also by enhancing the NF-kB activation in the context of TCR/CD28–triggered T cell activation. This process occurs without direct binding by BCL11B to the NF-kB response elements or association with NF-kB-DNA complexes, but rather indirectly through regulation of Cot kinase gene expression and consequent higher activation of IkB kinase. Cot/Tpl2/MAP3K8 is a mitogen-activated protein kinase kinase kinase which has been implicated in NF-kB activation and IL-2 gene expression by regulation of IKK complex in T lymphocytes, downstream of the CD28 pathway [13-15]. Our results demonstrate that a dominant negative mutant of Cot kinase [15] and Cot kinase siRNA inhibit BCL11B-mediated activation of NF-kB activity, supporting the idea that Cot kinase plays a role in BCL11B-mediated activation of NF-kB. Also we report that BCL11B activates additional NF-kB target genes in response to T cell activation.
MATERIALS AND METHODS
Plasmids
The pΔODLO 4xCD28RE-TRE-Luciferase, AP1-Luciferase and NFAT-Luciferase [16] and Cot kinase dominant negative (DN) (Cot S400A/S413A) [15] plasmids were kindly provided by Dr. Arthur Weiss. The NF-kB consensus (pNFkB-Luciferase) and Renilla (pRL-Luciferase) reporter vectors were purchased from Clontech. Flag-BCL11B was cloned into pRevTRE (Clontech) to generate pRevTRE-BCL11B plasmid. The MSCV-BCL11B plasmid was previously described [7].
Antibodies and biochemicals
Anti-BCL11B (B26−44) polyclonal antibodies were previously described [6]. Additional anti-BCL11B antibodies were purchased from Bethyl Laboratories. The mouse anti-actin antibodies were purchased from Sigma-Aldrich. The anti-human CD3 (OKT) and anti-human CD28 are from eBioscience. The antibodies against the following proteins are from SantaCruz Biotechnology: RelA (F-6), p105/p50 (H-119), IkBα (C-21), IkBβ (C-20), IKKα/β (H-470) and Cot. The phosphor-IkBα and phosphor-IKKα/β were purchased from Cell Signaling. Phorbol 12-myristate 13-acetate (PMA), ionomycin and MG132 were purchased from Sigma-Aldrich and used at the concentrations mentioned in the figure legends. Recombinant human TNFα was obtained from R&D Systems and used at a final concentration of 20ng/ml.
Cell lines
Jurkat and HeLa cells were obtained from ATCC. Jurkat cells were grown in RPMI 1640 media containing 2 mM L-glutamine, 10% heat-inactivated fetal bovine serum (HI-FBS), 50 U/ml penicillin and 50 μg/ml streptomycin. HeLa cells were grown in DMEM supplemented with 10% HI-FBS, 50 U/ml penicillin, 50 μg/ml streptomycin. The generation of the Jurkat MSCV and MSCV-BCL11B cell lines was described previously [7]. In a similar manner, HeLa TetON cells (Clontech) were transduced with pRevTRE-BCL11B retroviruses to generate the inducible cell line HeLa TetON-BCL11B.
Transient transfections and Luciferase Reporter Assays
Jurkat cells were transfected by electroporation as described previously [6, 7]. pRevTRE-BCL11B HeLa TetON cells were seeded on 6 cm plates (106 cells / plate) and transfected the next day with 2 μg of the reporter plasmids using Lipofectamine (Invitrogen). The cells were maintained in media supplemented with 50 ug/ml doxycyclin for 48 hours before harvesting. For reporter assays the cells were stimulated as indicated in figure legends and then harvested in Renilla Lysis Buffers (Promega). Luciferase activity was analyzed as previously described [6].
Gene knockdown by small interfering RNA (siRNA)
BCL11B and Cot specific and control nontargeting siRNAs were purchased from Santa Cruz Biotechnologies and Dharmacon. Jurkat cells (0.5 × 106 cells) were electroporated as describes previously [7]. The following day, cells were treated with PMA and ionomycin and then harvested for reporter assays or RNA preparation, as previously described [7].
Nuclear Fractionation, Reverse transcription and quantitative (q) PCR, and Chromatin immunoprecipitation were conducted as previously described [5-7].
RESULTS
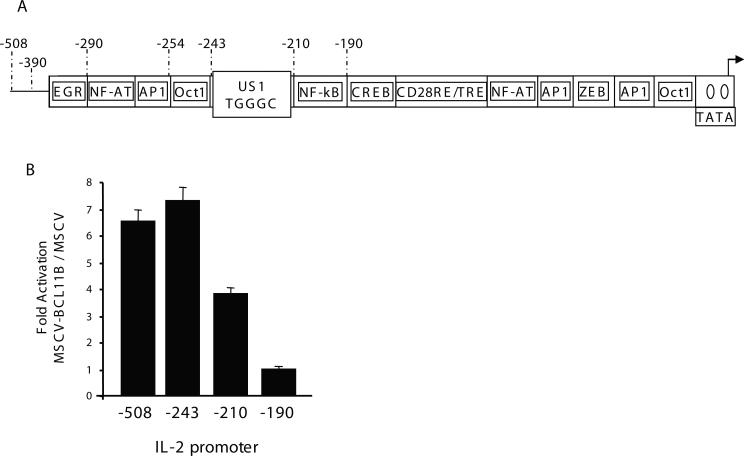
BCL11B regulates IL-2 promoter through site(s) located between −210 and −190, in addition to binding to the US1 site
Our previous results using populations of Jurkat cells stably transduced with retroviruses expressing GFP (MSCV) and Flag-BCL11B-IRES-GFP (MSCV-BCL11B) demonstrated that BCL11B regulates the IL-2 gene expression by binding directly to the US1 site located between −243 and −210 in the IL-2 promoter [7]. However, following deletion of the US1 site containing region, although the activity of the reporter decreased considerably, BCL11B continued to activate expression of the reporter, suggesting that site(s) downstream of −210 may be implicated in BCL11B-mediated activation of the IL-2 promoter (Fig. 1A and B). Deletion of the region between −210 and −190 resulted in approximately three-fold reduction of the BCL11B-mediated augmentation of the IL-2 promoter, bringing the levels of the reporter in MSCV-BCL11B and MSCV cells approximately equal (Fig. 1A and B). These results suggest that BCL11B regulates expression of IL-2 through site(s) located between −210 and −190, in addition to the US1 site.
Figure 1. BCL11B regulates expression of IL-2 through site(s) located between −210 and −190 in addition to the US1 site.
(A) Schematic representation of the IL-2 promoter. (B) Ratios of Luciferase reporter assays between MSCV-BCL11B and MSCV Jurkat cells transfected with the IL-2 promoter deletion mutants. Reporter assays were conducted after treatment of the cells with PMA/ionomycin for 8 hours. The quantification represents the means of three independent experiments ± SD.
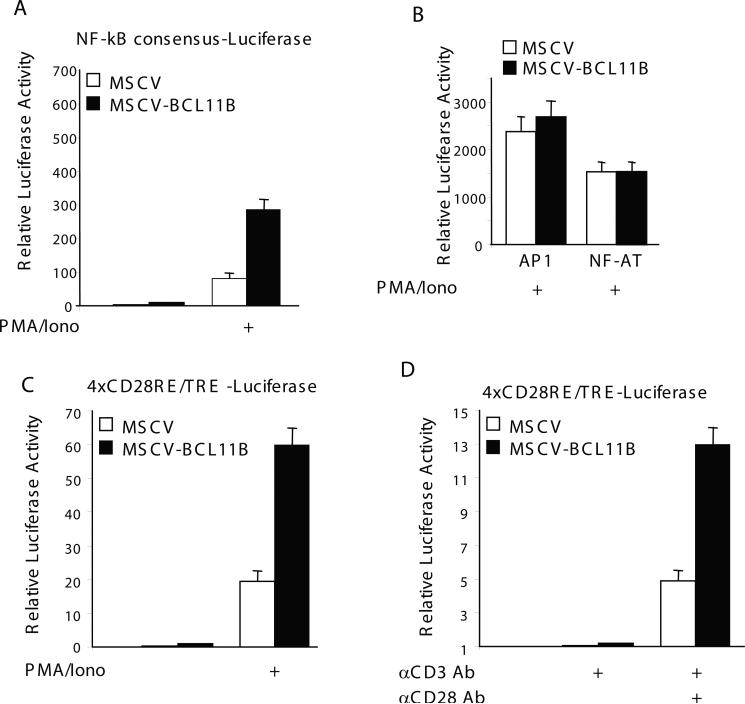
BCL11B activates transcription from the NF-KB sites in the context of TCR activation
The −210 through −190 IL-2 promoter does not contain any US1 or consensus BCL11B sites, however it contains an NF-kB site, located upstream of −190 and designated distal NF-kB site (Fig. 1A) [11, 15]. To investigate whether BCL11B may regulate the −210−190 through the NF-kB site, we conducted reporter assays with a construct containing the consensus NF-kB site. The results showed enhanced expression of the NF-kB reporter in MSCV-BCL11B Jurkat cells compared to MSCV, suggesting that BCL11B activates the expression driven by NF-kB site (Fig. 2A). Conversely, reporter constructs controlled by NFAT or AP1 showed similar levels of activation in MSCV-BCL11B and control cells (Fig. 2B), indicating that the BCL11B-mediated activation is specific for NF-kB.
Figure 2. BCL11B activates transcription from NFKB sites, but not from AP1 or NFAT.
MSCV (open bars) and MSCV-BCL11B (solid bars) Jurkat cells were transfected with pNFkB-Luc(A), AP1-Luciferase or NFAT-luciferase (B), or 4xRE/AP1-Luciferase (C and D) and Renilla Luciferase plasmids. 24 hrs posttransfection, the cells were stimulated with 50 ng/ml PMA plus 1 μM Ionomycin for 6 hrs (A, B and C), or crosslinked anti-CD3 antibody (OKT3) (10 μg/ml) or anti-CD3 antibody plus soluble anti-CD28 antibody (2 μg/ml) for 10 hrs (D). Renilla Luciferase activity was used for normalization.
NF-kB also binds the CD28RE/TRE response element in the IL-2 promoter and participates in the costimulatory activation of the IL-2 gene expression [17]. The CD28RE/TRE site is located downstream of −190 (Fig. 1A) and plays a critical role in CD28-mediated full activation of IL-2 promoter [17]. Its participation in the costimulatory activation of IL-2 promoter can only be evaluated when the −210 IL-2 promoter is intact, as it requires the distal NF-kB site [11, 15]. However, the activation of multimers of CD28RE/TRE can be evaluated in the context of a TATA-like promoter in T lymphocytes, in response to activation through TCR/CD28 or PMA and ionomycin [16]. We therefore tested the activity of a reporter controlled by the CD28RE/TRE composite element in the context of a TATA-like promoter and found that in response to stimulation with PMA/ionomycin the activity of this composite site was higher in MSCV-BCL11B cells compared to MSCV control (Fig. 2C). We further investigated whether enhanced activation of NF-kB mediated by BCL11B occurs also in conditions considered closer to a physiological activation, through treatment with anti-CD3 and anti–CD28 antibodies. Previous observations demonstrated that activation of NF-kB requires costimulatory signals generated through CD28 [16]. When TCR was stimulated alone through anti-CD3 treatment, the level of expression of the reporter was minimal, as expected (Fig. 2D). When cells were activated by both anti-CD3 and anti-CD28 antibodies, the Luciferase activity was higher in MSCV-BCL11B Jurkat cells compared to control (Fig. 2D). These data suggest that enhanced activation of NF-kB mediated by BCL11B requires CD28 costimulatory pathways.
These results taken together demonstrate that BCL11B augments transcription from NF-kB sites in the context of activation of T cells through TCR/CD28.
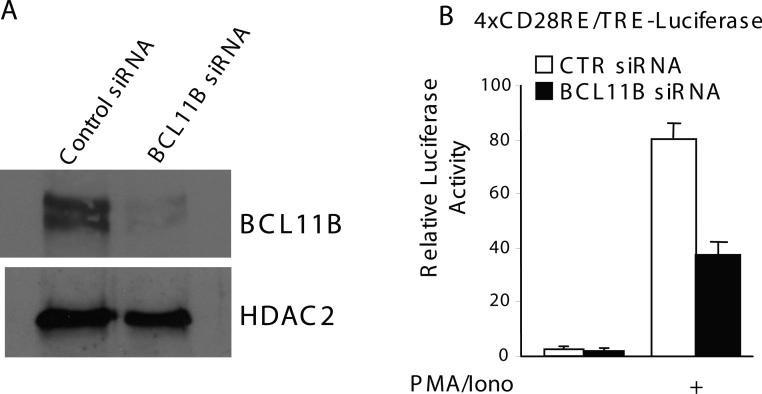
BCL11B knockdown reduces the transcriptional activation controlled by NF-kB response elements
Previously we found that knockdown of BCL11B causes a significant decrease of IL-2 promoter activation [7]. To demonstrate that endogenous BCL11B plays a role in the activation of transcription from NFKB sites, we used siRNA to decrease the level of endogenous BCL11B (Fig. 3A). Knockdown of BCL11B resulted in a reduction of CD28RE/TRE composite element activity, demonstrating that endogenous BCL11B is involved in the regulation of NF-kB activation (Fig. 3B).
Figure 3. Endogenous BCL11B is required for full activation of transcription from the NF-kB site.
Jurkat cells were transfected with 4xRE/AP1-Luciferase plasmid and the nontargeting siRNA (open bars) or BCL11B specific (solid bars) siRNAs. (A) Western blot analysis of BCL11B after transfection of Jurkat cells with BCL11B specific or nontargeting siRNAs. (B) Luciferase activity was measured in cells treated or not with 50 ng/ml PMA and 1 μM Ionomycin for 7 hrs.
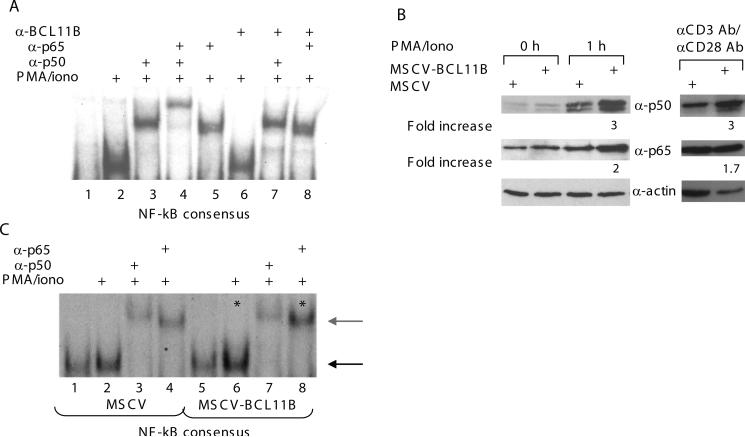
Augmentation of transcription from NF-kB sites by BCL11B is a consequence of increased nuclear levels of p50 and p65 in T lymphocytes activated through TCR
BCL11B can regulate the NF-kB sites in the IL-2 promoter by direct binding to the sites, through association with NF-kB or by increasing the NF-kB activation. We first evaluated by EMSA whether BCL11B directly binds the NF-kB sites and found that BCL11B did not bind either the consensus, distal NF-kB, nor the RE/AP1 sites (data not shown). We then used EMSA to determine whether BCL11B is present in the NF-kB-DNA complexes and cooperatively binds the NF-kB sites together with NF-kB. The results show that p65 (RelA)/p50-DNA complex does not contain BCL11B, as it was not shifted by anti-BCL11B antibodies (Fig. 4A). Of note that the anti-BCL11B antibodies are appropriate for EMSAs, as demonstrated by our previous studies [7]. These results show that BCL11B neither binds NF-kB sites, nor interacts with the NF-kB (p50/p65)–DNA complexes.
Figure 4. BCL11B enhances nuclear levels of p50 and p65, as well as binding of NF-kB consensus sites following T cell activation.
(A) EMSA using a labeled consensus NF-kB oligonucleotides and nuclear extracts prepared from Jurkat cells stimulated (lanes 2 to 8) or not (lane 1) with 50 ng/ml PMA and 1 μM Ionomycin for 1 hr. For the supershift experiments the nuclear extracts were incubated with the indicated antibodies for 15 min prior to incubation with the labeled oligonucleotide probe. (B) MSCV and MSCV-BCL11B Jurkat cells were stimulated or not with 50 ng/ml PMA and 1 μM Ionomycin for 1 hr (left panel), or with anti-CD3 and anti-CD28 (right panel). The nuclear extracts were analyzed by immunoblotting for p65 and p50 proteins. Actin was used as a loading control. Numbers indicate fold increase in the levels of p50 and p65 after normalization to actin, evaluated through densitometry. (C) EMSA using labeled consensus NF-kB oligonucleotides and nuclear extracts prepared from MSCV (lanes 1 through 4) and MSCV-BCL11B (lanes 5 through 8) Jurkat cells stimulated (lanes 2 to 4 and 6 to 8) or not (lanes 1 and 5) with 50 ng/ml PMA and 1 μM Ionomycin for 1 hr. For the supershift experiments the nuclear extracts were incubated with the indicated antibodies for 15 min prior to incubation with the labeled probe. NFkB-DNA complexes are indicated by the black arrow and the supershifted complexes by the gray arrow. Enhancement of binding as a result of BCL11B overexpression is indicated by asterisks.
It has been demonstrated that NF-kB is translocated to the nucleus, as a result of its activation [18]. We therefore evaluated the nuclear levels of NF-kB in response to T cell activation by PMA/ionomycin or anti-CD3 and anti–CD28. Because at early time points of TCR activation, p65(RelA)-p50 complex is the major NF-kB complex present in the nucleus [19], we evaluated the nuclear levels of p50 and p65 (RelA) and found that they were increased in MSCV-BCL11B Jurkat cells compared to control (Fig. 4B). In addition, we observed that DNA-NF-kB complex formation was enhanced in MSCV-BCL11B cells compared to the control (Fig. 4C). These results show that the regulation of NF-kB activity by BCL11B is a consequence of higher levels of p50 and p65 in the nucleus of activated cells, and occurs without association of BCL11B with NF-kB binding sites or with NF-kB -DNA complexes.
BCL11B enhances IkB degradation in response to TCR, but not UV-mediated activation
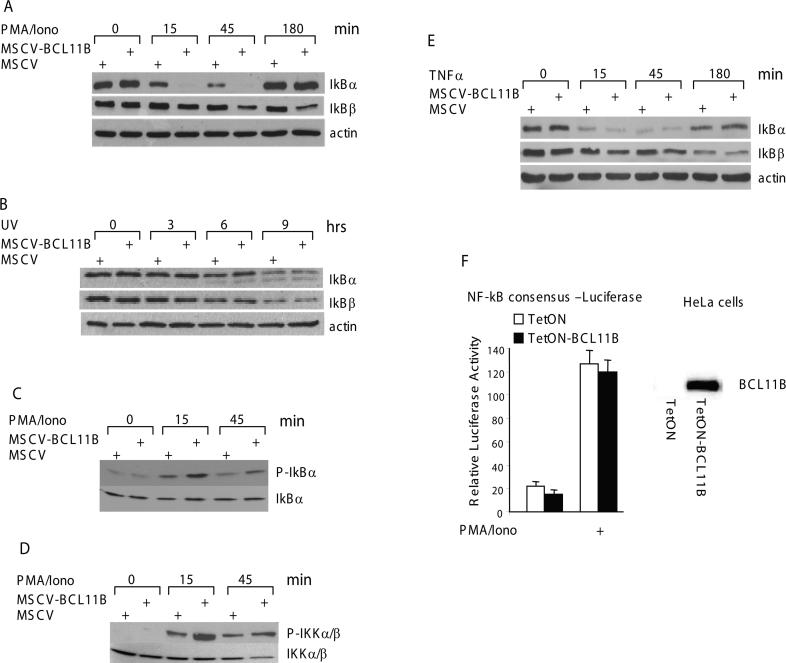
We next wanted to determine the cause of enhanced NF-kB nuclear translocation in the presence of higher levels of BCL11B. Nuclear translocation of NF-kB is a direct consequence of IkB degradation [20]. As enhanced expression of BCL11B leads to increased nuclear translocation of p65 and p50, we investigated whether IkB degradation was elevated in MSCV-BCL11B cells compared to control. In resting conditions, the levels of IkBα and IkBβ were similar in both MSCV-BCL11B and MSCV cells (Fig. 5A, upper blot, 0 hr). However, in response to activation with PMA/ionomycin, IkBα degradation occurred significantly faster in MSCV-BCL11B cells (Fig. 5A, upper blot, 15 and 45 min). Although the IkBβ degradation occurred with slower kinetics, BCL11B enhanced its degradation as well (Fig. 5A, middle panel). Therefore these results suggest that the augmentation of NF-kB activity by BCL11B in cells activated by PMA/ionomycin occurs through enhanced degradation of IkB. Degradation of IkBα/β triggered by TCR/CD28 or PMA/ionomycin treatments is a three step process. First, the IkB is phosphorylated by the IkB kinase (IKK). Second, the phospho-IkB is ubiquitinated, and third, the ubiquitinated IkB is degraded by the 26S proteasome [20]. Since the level of IkB was reduced in MSCV-BCL11B compared to control cells, we wanted to investigate in which of these steps BCL11B is involved. Previously it was demonstrated that UV-C radiation triggers ubiquitin-dependent degradation of IkB, through a process independent of IKK activation [21, 22]. If BCL11B is implicated in ubiquitination and/or degradation of IKBα/β, then its sustained expression would result in enhanced degradation of IkB, regardless of how the cells are activated, i.e., through TCR/CD28 or UV treatment. Our results indicate that IkBα/β degradation in response to UV radiation occurred with the same rate in MSCV-BCL11B and control MSCV Jurkat cells (Fig. 5B), suggesting therefore that BCL11B does not enhance the ubiquitination and/or proteosome-mediated degradation of IkBα/β proteins.
Figure 5. BCL11B enhances IkB phosphorylation and degradation, and IkB kinase phosphorylation in a TCR-dependent manner.
(A) MSCV and MSCV-BCL11B Jurkat cells were treated with 50 ng/ml PMA and 1 μM ionomycin for the indicated time points. IkB degradation was analyzed by Western blot analysis of cytoplasmic fractions with specific antibodies. Actin was used as loading control. (B) Same as in (a) except that the cells were irradiated with 80 J/m2. (C) MSCV and MSCV-BCL11B Jurkat cells were pretreated with 2.5 μM MG132 for 16 hrs followed by stimulation with 50 ng/ml PMA and 1 μM ionomycin for the indicated time points and P-IkBα was recognized with specific antibodies. The same membrane was stripped and total amount of IkBα was detected by reprobing with an anti-IkBα antibody. (D) MSCV and MSCV-BCL11B Jurkat cells were treated with 50 ng/ml PMA and 1 μM Ionomycin for the indicated time points. The P-IKKα/β were detected with specific antibodies. The total amount of IKKα/β proteins is shown on the same membrane. (E) MSCV and MSCV-BCL11B Jurkat cells were treated with 20 ng/ml TNFα for the indicated time points. IkB degradation was analyzed by Western blot analysis of cytoplasmic fractions with specific antibodies. Actin was used as loading control. (F) (Left panel) HeLa cells ectopically expressing BCL11B (solid bars) or not (open bars) were transfected with pNF-kB-Luc and Renilla Luciferase plasmids. 24 hrs posttransfection the cells were stimulated with 50 ng/ml PMA and 1 μM ionomycin for 6 hrs and the luciferase activity was evaluated. (Right panel) Western blot showing ectopic expression of BCL11B in Hela cells.
BCL11B enhances IkB and IkB kinase phosphorylation in T lymphocytes activated though TCR
We then tested whether the phosphorylation of IkB is enhanced by BCL11B overexpression in conditions of TCR activation. In these experiments the degradation of IkB was blocked by pretreatment of the cells with the proteasome inhibitor MG132, and the phosphorylated IkBα was analyzed in MSCV-BCL11B and control cells after stimulation with PMA/ionomycin. The levels of phosphorylated IkBα were significantly higher in MSCV-BCL11B cells, suggesting that IKK may be more active in these cells (Fig. 5C). To directly demonstrate that the enhanced phosphorylation of IkB is a consequence of increased activation of IKK, we tested the levels of phosphorylated IKK, known to be activated through phosphorylation following T cell activation through TCR/CD28 or PMA/ionomycin treatments. The levels of phosphorylated IKKα/β were higher in MSCV-BCL11B cells compared to control (Fig. 5D). These results taken together demonstrate that enhanced degradation of IkB in MSCV-BCL11B cells is a consequence of enhanced phosphorylation, and consequently activation of IKK.
BCL11B does not participate in the TNFα-mediated activation of NF-kB in T lymphocytes
In addition to TCR/CD28 stimulation, IKK can be activated by TNFα, but the upstream components of the two pathways are different [23]. We therefore investigated whether BCL11B enhances NF-kB activation by a mechanism specific for TCR/CD28 or PMA/ionomycin activated-pathway, or by a common mechanism converging toward IKK activation. The results show that IkB degradation after TNFα treatment was similar in MSCV-BCL11B and control cells (Fig. 5E), contrary to what we observed when cells were activated by PMA/ionomycin (Fig. 5A). These findings suggest that BCL11B acts specifically on TCR/CD28–mediated IKK activation.
Enhanced activation of NF-kB mediated by BCL11B occurs specifically in T lymphocytes
To further demonstrate that enhanced activation of NF-kB mediated by BCL11B involves downstream components of TCR/CD28 signaling pathway and occurs specifically in T lymphocytes, we used a HeLa cell line in which we stably expressed ectopic BCL11B (Fig. 5F, right panel). We chose HeLa cells because they do not express endogenous BCL11B (Fig. 5F, lower panel) and lack TCR/CD28 signaling pathway. However NF-kB can be activated by PMA/ionomycin treatment of these cells [24]. We conducted reporter assays with the consensus NF-kB-Luciferase construct. The results showed that the relative Luciferase activity was the same, regardless the presence or absence of BCL11B (Fig. 5F, left panel).
These results collectively demonstrate that the enhanced activation of NF-kB mediated by BCL11B is specific for T lymphocytes and involves the TCR/CD28 signaling pathway.
BCL11B upregulates the levels of Cot kinase mRNA
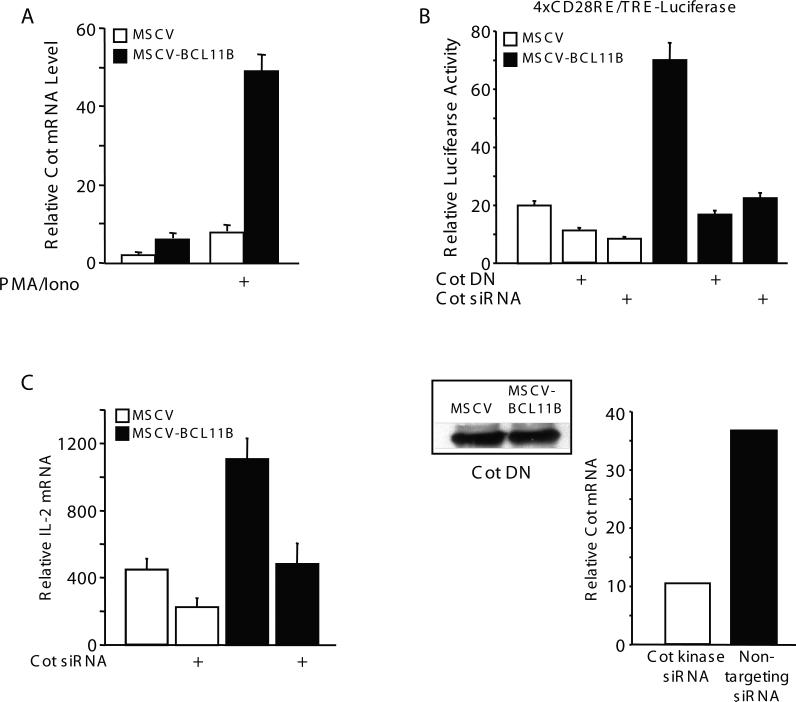
The results presented above indicate that BCL11B enhances NF-kB activation specifically in T lymphocytes, downstream of TCR/CD28 signaling pathway and upstream of IkB kinase. BCL11B is a transcription factor localized almost exclusively in the nucleus of Jurkat cells (data not shown). Therefore, it is unlikely that BCL11B is directly involved in IKK activation. Rather, it is possible that BCL11B regulates the expression of genes involved in TCR/CD28–triggered IKK activation. To test this hypothesis we investigated the expression levels of known genes implicated in TCR/CD28–mediated IKK activation in MSCV-BCL11B and MSCV control cells by quantitative RT-PCR, including IKKα, IKKβ, IKKγ, Vav, SLP-76, ZAP-70, LAT, Lck, PKCθ, Calcineurin, CARMA1, BCL10, MALT1, PI3K, Akt, Cot, NIK, PDK1, TRAF2, TRAF6, TAK1, RIP2, MLK3, βTrCP1, Cullin1, Rbx1, E2UbcH5, Rac1, Ubc13, Need8 and Caspase8 (Table 1). From all the genes tested, only Cot kinase mRNA levels were upregulated in MSCV-BCL11B compared to control cells (Table 1 and Fig. 6A). Cot kinase is known to be involved in the activation of IKK following T lymphocyte stimulation through TCR/CD28 [14]. Of note, Cot kinase mRNA levels were already higher even in the absence of TCR activation in cells overexpressing BCL11B (Fig. 6A).
Table 1.
Expression of genes involved in TCR signaling in MSCV-BCL11B Jurkat cells
| Gene | Ratio Relative Expression Level MSCV-BCL11B/MSCV |
|---|---|
| IKKα | 1 |
| IKKβ | 1 |
| IKKγ | 1 |
| Vav | 1.1 |
| SLP-76 | 0.9 |
| ZAP-70 | 1 |
| LAT | 1 |
| Lck | 0.9 |
| PKCθ | 1 |
| Calcineurin | 1 |
| CARMA1 | 1 |
| BCL10 | 1.1 |
| MALT1 | 1.2 |
| PI3K | 1.2 |
| Akt | 1 |
| Cot kinase | 6 |
| NIK | 1 |
| PDK1 | 1 |
| TRAF2 | 0.9 |
| TRAF8 | 1 |
| TAK1 | 1.1 |
| RIP2 | 1 |
| MLK3 | 1 |
| βTrCP1 | 1 |
| Cullin1 | 1.1 |
| RBX1 | 1 |
| E2UbcH5 | 1.1 |
| Rac1 | 1 |
| Ubc13 | 1 |
| Caspase8 | 1.2 |
Figure 6. Enhanced activation of NF-kB by BCL11B is mediated through Cot kinase gene expression.
(A) MSCV (open bars) and MSCV-BCL11B (solid bars) Jurkat cells were treated or not with 50 ng/ml PMA and 1 μM ionomycin for 4 hrs. Cot mRNA expression was assessed by quantitative RT-PCR. The relative abundance of Cot kinase mRNA was normalized against actin in each sample. (B) (upper panel) MSCV (open bars) and MSCV-BCL11B (solid bars) Jurkat cells were transfected with the Cot dominant negative (DN) mutant construct, or Cot kinase siRNA, as indicated, and 4xRE/AP1-Luciferase and Renilla luciferase plasmids. 24 hours posttransfection the cells were stimulated with 50 ng/ml PMA and 1 μM ionomycin for 7 hrs and luciferase activity was determined. (lower panel, left) Expression of the Cot kinase dominant negative (DN) mutant construct detected by Western blot analysis in MSCV and MSCV-BCL11B Jurkat cells. (lower panel, right) Reduction in Cot kinase mRNA levels as a result of transfection with Cot kinase siRNA. Relative Cot kinase mRNA levels were evaluated by quantitative RT-PCR, as previously described [6, 7]. (C) MSCV (open bars) and MSCV-BCL11B (solid bars) Jurkat cells were transfected with Cot kinase siRNA or nontargeting siRNA, followed by treatment with 50 ng/ml PMA and 1 μM ionomycin. IL-2 mRNA expression was assessed by quantitative RT-PCRs. The relative abundance of IL-2 mRNA was normalized against actin in each sample.
A Cot kinase dominant negative mutant and knockdown of Cot kinase reduce BCL11B-mediated activation of NF-kB
To further demonstrate that BCL11B activates NF-kB through Cot kinase upregulation, MSCV and MSCV-BCL11B Jurkat cells were transfected with a Cot kinase dominant negative construct (DN) (Cot S400A/S413A) [15] or Cot kinase siRNA (Fig. 6B). Expression of the Cot kinase DN mutant, as well as knockdown of Cot kinase blocked the enhanced activation mediated by BCL11B on NF-kB-driven CD28RE/TRE reporter (Fig. 6B, upper panel), demonstrating that Cot kinase is implicated in BCL11B-mediated activation of NF-kB. The levels of expression of Cot kinase DN mutant were similar in MSCV and MSCV-BCL11B Jurkat cells (Fig. 6B, lower panel, left). Since the levels of endogenous Cot kinase are undetectable with the antibody used for the detection of the transfected mutant (data not shown), we used qRT-PCR to detect the reduction in the Cot kinase mRNA (Fig. 6B, lower panel, right).
To further demonstrate that Cot kinase is responsible for BCL11B-mediated activation of NF-kB, and further effects on downstream genes, such as IL-2, we knocked down Cot kinase and evaluated IL-2 mRNA levels in MSCV and MSCV-BCL11B. The reduction in IL-2 mRNA level in MSCV-BCL11B was more pronounced compared to MSCV (2.5 versus 1.9 fold) (Fig. 6C), demonstrating the contribution of Cot kinase in the upregulation of IL-2 mediated by BCL11B through NF-kB. Still the levels of IL-2 mRNA remained higher in MSCV-BCL11B compared to MSCV cells after knockdown of Cot kinase, supporting our previous observation that BC11B also controls IL-2 gene expression directly [7].
These results taken together demonstrate that Cot kinase is responsible for the BCL11B-mediated activation of NF-kB, and for further downstream effects.
BCL11B modulates expression of NF-kB-dependent genes in response to T cell activation
NF-kB was previously demonstrated to regulate expression of genes encoding cytokines, chemokines and several cytokine receptors, including IL-2 [25], TNFα [26], TNFβ [27], IFNγ [28], IL-8 [29], lymphotoxin β [30], macrophage inflamatory protein-1 (Mip1 α/β) [31], IL-2Rα [32] and IL-7Rα [33]. As BCL11B enhances NF-kB activation in lymphocytes, we measured the mRNA levels for genes previously demonstrated to be regulated by NF-kB. Quantitative RT-PCR assays demonstrated that the expression of the NF-kB-dependent genes, but not the expression of IL-4, was upregulated in MSCV-BCL11B cells (Table 2).
Table 2.
Cytokine and cytokine receptor genes are upregulated in MSCV-BCL11B Jurkat cells
| Gene | Ratio Relative mRNA Levels MSCV-BCL11B/MSCV |
|---|---|
| TNFα | 12 |
| TNFβ | 3.6 |
| IFNγ | 2 |
| IL-2Rα | 22 |
| IL-7Rα | 11 |
| Lymphotoxin B | 10 |
| IL-4 | 1 |
| MIP1a | 3 |
| IL-8 | 3 |
To demonstrate that endogenous BCL11B plays a role in the activation of the NF-kB-dependent genes we knocked down BCL11B, which resulted in the reduction of mRNA levels of the majority of NF-kB-dependent genes (Table 3), demonstrating that endogenous BCL11B plays a role in regulation of their expression. As expected, Cot kinase and IL-2 mRNA levels were also downregulated (Table 3).
Table 3.
Cytokine and cytokine receptor genes expression is downregulated in Jurkat cells transfected with BCL11B siRNA
| Gene | Ratio Relative mRNA Levels siRNA BCL11B/control |
|---|---|
| TNFα | 0.25 |
| TNFβ | 0.7 |
| IFNγ | 0.4 |
| IL-2Rα | 0.3 |
| IL-7Rα | 0.3 |
| Lymphotoxin B | 0.12 |
| IL-4 | 1 |
| MIP1a | 0.5 |
| IL-8 | 1 |
| Cot kinase | 0.25 |
| IL-2 | 0.2 |
BCL11B associates with Cot kinase gene intron 2
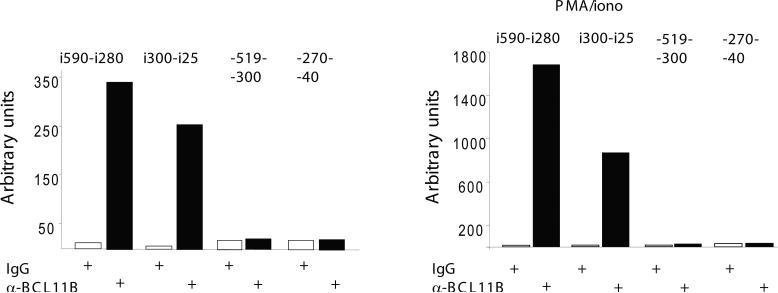
We further investigated whether BCL11B associates with the Cot kinase promoter and found that there was no binding above background within the 519 bp upstream of mRNA start site (Fig. 7), which suggest that BCL11B is unlikely to bind to the promoter. It has been reported that Cot kinase gene has 9 exons, out of which the first 2 correspond to the 5’ untranslated region [34, 35]. Regulatory elements can be also located within intronic regions, often within the first introns. We therefore investigated whether BCL11B binds to the intron 2, which precedes the first ATG. Our results show that BCL11B bound within this intron to a region upstream to the 5’splice site. Interestingly, the binding was enhanced in response to PMA/ionomycin treatment (Fig. 7). These results suggest that BCL11B regulates expression of Cot kinase gene by associating to a region in intron 2. Our results also indicate that cot kinase gene intron 2 potentially contains regulatory elements responsive to PMA/ionomycin, in addition to those previously demonstrated to be present in the promoter.
Figure 7. Endogenous BCL11B binds to Cot kinase gene intron 2.
Chromatin immunoprecipitation of Jurkat cells, stimulated or not with PMA/ionomycin, with anti-BCL11B antibodies or IgG. Eluted DNA was analyzed by quantitative PCR using primers for Cot kinase gene promoter (−519−300 and −270−40) or intron 2 (i590-i280 and i300-i25).
DISCUSSION
Our previous studies demonstrated that BCL11B participates in IL-2 gene expression in response to T cell activation through direct binding of the US1 site in the IL-2 promoter [7]. Here we show that BCL11B also participates in the activation IL-2 gene expression through an indirect mechanism, as a result of modulation of NF-kB activity, through enhanced Cot kinase gene expression. In the first mechanism, as a consequence of T cell activation through TCR/CD28 stimulation or PMA/ionomycin treatment, BCL11B binds and directly activates the IL-2 promoter through the US1 site [7]. In the second, indirect mechanism, BCL11B enhances NF-kB activity, which further binds and activates the IL-2 promoter. Interestingly, we found that BCL11B upregulates Cot gene expression even before T cell stimulation. Thus, it is expected that NF-kB activation is enhanced immediately after T cell stimulation in cells expressing ectopic BCL11B. Indeed, it is sufficient to stimulate the cells for only one hour in order to detect stronger translocation of NF-kB into the nucleus and more DNA-NF-kB complex formation. Of note that at this early time point of activation, p65 (RelA)-p50 complex is the major NF-kB complex present on IL-2 promoter [19]. In support that Cot kinase plays a role in BCL11B-mediated NF-kB activation comes the fact that a dominant negative mutant of Cot kinase, as well as Cot kinase siRNA inhibit BCL11B-mediated NF-kB activation. Enhanced Cot kinase expression in cells with higher levels of BCL11B results in elevated IKK activity, and enhanced IkBα/β phosphorylation and degradation. In addition, our data suggest that BCL11B-mediated NF-kB activation involves pathways specific to T lymphocytes and is triggered by TCR/CD28 signals or PMA/ionomycin, but not by TNFα or UV radiation.
The NF-kB family of transcription factors has a key role in coordinating expression of a wide variety of genes that control both innate and adaptive immune responses [23]. NF-kB-activating signaling pathways converge to IKK complex activation [23, 36]. IKK-mediated IkB phosphorylation triggers subsequent proteolytic destruction of the inhibitory complex, thus allowing the NF-kB to translocate to the nucleus and activate target genes [37]. Activation of NF-kB pathway by IL-1, lipopolysaccharide and TNFα has been well characterized. Recently, much progress has been made in understanding the biochemical mechanisms involved in NF-kB activation triggered by the TCR/CD28 pathways (reviewed in [23, 36]). TCR engagement leads to activation of several transcription factors including NFAT, AP1 and NF-kB [8]. Stimulation of the TCR complex alone efficiently activates NF-AT, while activation of NF-kB requires costimulatory signals from CD28 [16]. Several studies demonstrated that CD28 costimulation is required for a potent NF-kB activation and a sustained IL-2 expression in T lymphocytes [12, 38, 39]. Here we demonstrate that the enhanced activation of CD28RE/TRE response elements by BCL11B requires CD28 costimulatory signals. Interestingly, it has been demonstrated that Cot kinase is required for TCR/CD28-induced, but not TNFα-induced NF-kB activation in lymphocytes [14, 15], which supports our observations that BCL11B enhances IkBα/β degradation after TCR-, but not after TNFα-mediated stimulation of Jurkat cells.
In response to TCR activation, NF-kB controls expression of genes encoding multiple cytokines, chemokines and cytokine receptors [23]. Here we show that BCL11B, in addition to participating in the activation of IL-2 gene expression, also plays a role in activation of expression of other NF-kB target genes, including TNFα and TNFβ, lymphotoxin β, Mip1α, as well as IL-2Rα and IL-7Rα. In conclusion, the data presented here demonstrate that BCL11B enhances NF-kB activation through upregulation of Cot kinase expression and, consequently, by promoting the TCR/CD28 triggered IkB kinase activation.
In addition to the important role of NF-kB in the transcriptional control of genes encoding cytokines, chemokines, cytokine receptors in response to TCR/CD28-triggered T lymphocyte activation (reviewed in [36]), several reports demonstrated that NF-kB is constitutively activated in T cell leukemia, but the molecular mechanism remains poorly defined [40, 41]. Also, Cot kinase was found to be constitutively activated in several human T cell leukemia cell lines [42], and BCL11B was recently identified as a potential oncogene for ATL [43]. Thus, our data may also suggest an interesting functional link between enhanced expression of BCL11B and the NF-kB constitutive activation in the adult T cell leukemia/lymphoma cells.
Acknowledgements
We greatly acknowledge Dr. Arthur Weiss (University of California, San Francisco) for constructs, and Dr. Gary Nolan (Stanford University) for Phoenix A and E packaging cells. We thank Hong-Mei Chen and Jennifer Gecewicz for technical assistance, Adrian Avram for graphical presentation, and Debbie Moran for secretarial assistance.
Supported by grants to Dorina Avram from the National Institute of Health/NIAID (R01AI078273) and the American Cancer Society (ACS-RSG-04-265-01-MGO).
Abbreviations
- BCL11B
B-cell leukemia/lymphoma 11B
- TCR
T cell receptor
- IL-2
interleukin 2
- US1
upstream site 1
- Cot
cancer Osaka thyroid, oncogene
- Tpl2
tumor progression locus 2
- MAP3K8
mitogen activated protein kinase kinase kinase 8
- NF-kB
nuclear factor of kappa light polypeptide gene enhancer in B-cells
- NFAT
nuclear factor of activated T-cells
- IkBα/β
NF-kappa-B inhibitor alpha/beta
- IKKα/β
IkB kinase alpha/beta
- siRNA
small interference RNA
- PMA
phorbol-12-myristate-13-acetate
- GFP
green fluorescence protein
- EMSA
electrophoretic mobility shift assay
REFERENCES
- 1.Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, Aizawa S, Kominami R. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 2.Albu DI, Feng D, Bhattacharya D, Jenkins NA, Copeland NG, Liu P, Avram D. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med. 2007;204:3003–3015. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J Biol Chem. 2000;275:10315–10322. doi: 10.1074/jbc.275.14.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem J. 2002;368:555–563. doi: 10.1042/BJ20020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 2005;24:6753–6764. doi: 10.1038/sj.onc.1208904. [DOI] [PubMed] [Google Scholar]
- 7.Cismasiu VB, Ghanta S, Duque J, Albu DI, Chen HM, Kasturi R, Avram D. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood. 2006;108:2695–2702. doi: 10.1182/blood-2006-05-021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwashima M. Kinetic perspectives of T cell antigen receptor signaling. A two-tier model for T cell full activation. Immunol Rev. 2003;191:196–210. doi: 10.1034/j.1600-065x.2003.00024.x. [DOI] [PubMed] [Google Scholar]
- 9.Loh C, Shaw KT, Carew J, Viola JP, Luo C, Perrino BA, Rao A. Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. J Biol Chem. 1996;271:10884–10891. doi: 10.1074/jbc.271.18.10884. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Hu J, Vita R, Sun B, Tabata H, Altman A. SPAK kinase is a substrate and target of PKCtheta in T-cell receptor-induced AP-1 activation pathway. Embo J. 2004;23:1112–1122. doi: 10.1038/sj.emboj.7600125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serfling E, Avots A, Neumann M. The architecture of the interleukin-2 promoter: a reflection of T lymphocyte activation. Biochim Biophys Acta. 1995;1263:181–200. doi: 10.1016/0167-4781(95)00112-t. [DOI] [PubMed] [Google Scholar]
- 12.Lai JH, Tan TH. CD28 signaling causes a sustained down-regulation of I kappa B alpha which can be prevented by the immunosuppressant rapamycin. J Biol Chem. 1994;269:30077–30080. [PubMed] [Google Scholar]
- 13.Tsatsanis C, Patriotis C, Tsichlis PN. Tpl-2 induces IL-2 expression in T-cell lines by triggering multiple signaling pathways that activate NFAT and NF-kappaB. Oncogene. 1998;17:2609–2618. doi: 10.1038/sj.onc.1202460. [DOI] [PubMed] [Google Scholar]
- 14.Lin X, Cunningham ET, Jr., Mu Y, Geleziunas R, Greene WC. The proto-oncogene Cot kinase participates in CD3/CD28 induction of NF-kappaB acting through the NF-kappaB-inducing kinase and IkappaB kinases. Immunity. 1999;10:271–280. doi: 10.1016/s1074-7613(00)80027-8. [DOI] [PubMed] [Google Scholar]
- 15.Kane LP, Mollenauer MN, Xu Z, Turck CW, Weiss A. Akt-dependent phosphorylation specifically regulates Cot induction of NF-kappa B-dependent transcription. Mol Cell Biol. 2002;22:5962–5974. doi: 10.1128/MCB.22.16.5962-5974.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapiro VS, Truitt KE, Imboden JB, Weiss A. CD28 mediates transcriptional upregulation of the interleukin-2 (IL-2) promoter through a composite element containing the CD28RE and NF-IL-2B AP-1 sites. Mol Cell Biol. 1997;17:4051–4058. doi: 10.1128/mcb.17.7.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verweij CL, Geerts M, Aarden LA. Activation of interleukin-2 gene transcription via the T-cell surface molecule CD28 is mediated through an NF-kB-like response element. J Biol Chem. 1991;266:14179–14182. [PubMed] [Google Scholar]
- 18.Baeuerle PA, Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-kappa B transcription factor. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- 19.Himes SR, Coles LS, Reeves R, Shannon MF. High mobility group protein I (Y) is required for function and for c-Rel binding to CD28 response elements within the GM-CSF and IL-2 promoters. Immunity. 1996;5:479–489. doi: 10.1016/s1074-7613(00)80503-8. [DOI] [PubMed] [Google Scholar]
- 20.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 21.Li N, Karin M. Ionizing radiation and short wavelength UV activate NF-kappaB through two distinct mechanisms. Proc Natl Acad Sci U S A. 1998;95:13012–13017. doi: 10.1073/pnas.95.22.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bender K, Gottlicher M, Whiteside S, Rahmsdorf HJ, Herrlich P. Sequential DNA damage-independent and -dependent activation of NF-kappaB by UV. Embo J. 1998;17:5170–5181. doi: 10.1093/emboj/17.17.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 24.Nelsen B, Hellman L, Sen R. The NF-kappa B-binding site mediates phorbol ester-inducible transcription in nonlymphoid cells. Mol Cell Biol. 1988;8:3526–3531. doi: 10.1128/mcb.8.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serfling E, Barthelmas R, Pfeuffer I, Schenk B, Zarius S, Swoboda R, Mercurio F, Karin M. Ubiquitous and lymphocyte-specific factors are involved in the induction of the mouse interleukin 2 gene in T lymphocytes. Embo J. 1989;8:465–473. doi: 10.1002/j.1460-2075.1989.tb03399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shakhov AN, Collart MA, Vassalli P, Nedospasov SA, Jongeneel CV. Kappa B-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor alpha gene in primary macrophages. J Exp Med. 1990;171:35–47. doi: 10.1084/jem.171.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul NL, Lenardo MJ, Novak KD, Sarr T, Tang WL, Ruddle NH. Lymphotoxin activation by human T-cell leukemia virus type I-infected cell lines: role for NF-kappa B. J Virol. 1990;64:5412–5419. doi: 10.1128/jvi.64.11.5412-5419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sica A, Tan TH, Rice N, Kretzschmar M, Ghosh P, Young HA. The c-rel protooncogene product c-Rel but not NF-kappa B binds to the intronic region of the human interferon-gamma gene at a site related to an interferon-stimulable response element. Proc Natl Acad Sci U S A. 1992;89:1740–1744. doi: 10.1073/pnas.89.5.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunsch C, Rosen CA. NF-kappa B subunit-specific regulation of the interleukin-8 promoter. Mol Cell Biol. 1993;13:6137–6146. doi: 10.1128/mcb.13.10.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuprash DV, Osipovich OA, Pokholok DK, Alimzhanov MB, Biragyn A, Turetskaya RL, Nedospasov SA. Functional analysis of the lymphotoxin-beta promoter. Sequence requirements for PMA activation. J Immunol. 1996;156:2465–2472. [PubMed] [Google Scholar]
- 31.Grove M, Plumb M. C/EBP, NF-kappa B, and c-Ets family members and transcriptional regulation of the cell-specific and inducible macrophage inflammatory protein 1 alpha immediate-early gene. Mol Cell Biol. 1993;13:5276–5289. doi: 10.1128/mcb.13.9.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ballard DW, Bohnlein E, Lowenthal JW, Wano Y, Franza BR, Greene WC. HTLV-I tax induces cellular proteins that activate the kappa B element in the IL-2 receptor alpha gene. Science. 1988;241:1652–1655. doi: 10.1126/science.241.4873.1652. [DOI] [PubMed] [Google Scholar]
- 33.Vacca A, Felli MP, Palermo R, Di Mario G, Calce A, Di Giovine M, Frati L, Gulino A, Screpanti I. Notch3 and pre-TCR interaction unveils distinct NF-kappaB pathways in T-cell development and leukemia. Embo J. 2006;25:1000–1008. doi: 10.1038/sj.emboj.7600996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoki M, Hamada F, Sugimoto T, Sumida S, Akiyama T, Toyoshima K. The human cot proto-oncogene encodes two protein serine/threonine kinases with different transforming activities by alternative initiation of translation. J Biol Chem. 1993;268:22723–22732. [PubMed] [Google Scholar]
- 35.Sanchez-Gongora E, Lisbona C, de Gregorio R, Ballester A, Calvo V, Perez-Jurado L, Alemany S. COT kinase proto-oncogene expression in T cells: implication of the JNK/SAPK signal transduction pathway in COT promoter activation. J Biol Chem. 2000;275:31379–31386. doi: 10.1074/jbc.M000382200. [DOI] [PubMed] [Google Scholar]
- 36.Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 38.Harhaj EW, Sun SC. IkappaB kinases serve as a target of CD28 signaling. J Biol Chem. 1998;273:25185–25190. doi: 10.1074/jbc.273.39.25185. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro VS, Mollenauer MN, Weiss A. Nuclear factor of activated T cells and AP-1 are insufficient for IL-2 promoter activation: requirement for CD28 up-regulation of RE/AP. J Immunol. 1998;161:6455–6458. [PubMed] [Google Scholar]
- 40.Sun SC, Ballard DW. Persistent activation of NF-kappaB by the tax transforming protein of HTLV-1: hijacking cellular IkappaB kinases. Oncogene. 1999;18:6948–6958. doi: 10.1038/sj.onc.1203220. [DOI] [PubMed] [Google Scholar]
- 41.Hironaka N, Mochida K, Mori N, Maeda M, Yamamoto N, Yamaoka S. Tax-independent constitutive IkappaB kinase activation in adult T-cell leukemia cells. Neoplasia. 2004;6:266–278. doi: 10.1593/neo.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babu G, Waterfield M, Chang M, Wu X, Sun SC. Deregulated activation of oncoprotein kinase Tpl2/Cot in HTLV-I-transformed T cells. J Biol Chem. 2006;281:14041–14047. doi: 10.1074/jbc.M512375200. [DOI] [PubMed] [Google Scholar]
- 43.Oshiro A, Tagawa H, Ohshima K, Karube K, Uike N, Tashiro Y, Utsunomiya A, Masuda M, Takasu N, Nakamura S, Morishima Y, Seto M. Identification of subtype-specific genomic alterations in aggressive adult T-cell leukemia/lymphoma. Blood. 2006;107:4500–4507. doi: 10.1182/blood-2005-09-3801. [DOI] [PubMed] [Google Scholar]