Abstract
A bacterial strain, designated AcBz01, was isolated from a water sample collected from Gomti River, Lucknow, India, and identified using a molecular approach. On the basis of the bacterial 16S rRNA gene sequence phylogeny and comparison of this gene sequence with sequences in Ribosomal Database project II, evidence given in this study, it is proposed that isolate is closely related to members of the genus Acinetobacter. Identification and annotation of regulatory elements in the 16S rRNA gene and characterization of their interaction with the respective transcription factor can provide basis for better understanding of the mechanism of network of gene interaction of functionally related genes. The identification of such sites is relevant for locating promoter boundary of a gene and it also allows the prediction of specific gene expression pattern and response to disturbances in a known signaling pathway. Computational identification of regulatory elements and Transcription Factor with their binding sites in 16S rRNA gene of Acinetobacter sp. was performed using BPROM tool. We predicted the regulatory elements are TSS, -10 box, -35 box and three Transcription Factor (narP, ompR and fadR) with their binding sites in the upstream region of 16S rRNA gene of Acinetobacter sp. AcBz01. The GenBank accession number for 16S rRNA gene of Acinetobacter sp. AcBz01 is EU931637.
Keywords: RNA polymerase binding site, 16S ribosomal RNA gene, transcription factor, promoter region, Acinetobacter, PCR
Background
Gomti, one of the rivers in India, represents a unique niche for many animals and plants in Lucknow, which is one of the cities of India. In recent years, it has received great attention from the public, due to its potential for biodiversity and biological conservation. We initiated a systematic screening programme to catalogue the microbial composition of Gomti river water in Lucknow. The traditional identification of bacteria on the basis of phenotypic characteristics is generally not as accurate as identification based on genotypic methods. Comparison of the bacterial 16S rRNA gene sequence has emerged as a preferred genetic technique [1]. In this work, we reported the isolation and identification of a previously unknown bacterium from Gomti, for which the name Acinetobacter sp. AcBz01 is proposed with the help of 16S rRNA gene sequence analysis.
Acinetobacter species have been attracting interest in both environmental and biotechnological applications: they are known to be involved in biodegradation of a number of different pollutants and in the extra and intracellular production of a number of economically valuable products [2]. In 16S ribosomal gene, transcription process begins with binding of RNA polymerase to DNA sequence in the promoter region, which is located immediate upstream of the transcription start site (TSS). A typical prokaryotic promoter region is thought to comprise of two hexameric motifs TATAAT and TTGACA centered at or near -10 and -35 positions relative to the TSS [3]. These sequence motifs were identified based on the analysis of a large number of promoter sequences and represent consensus sequences. It has been seen that a wide variety of sequences similar to these representative motifs are present in promoter. Today, how RNA polymerase locates these binding sites in the non-promoter DNA region remains a field of intensive investigation.
Several computational approaches have been reported for the identification of regulatory elements during the last decade [4]. The regulatory elements TSS, -10 box, -35 box and known Transcription Factor (TF) with their binding sites in the upstream sequence of bacterial 16S rRNA gene of Acinetobacterr sp. AcBz01 were predicted by using BPROM [5] tool sited at Softberry.
Methodology
Culturing of bacteria
Water sample collected from Gomti River in Lucknow, India was serially diluted and spread onto peptone/Beef extract/Nacl/Agar-Agar palates and kept for incubation at 30°C under anaerobic conditions. Different single colonies of bacterial strains were picked and further grown and subcultured several times to obtain a pure culture.
DNA isolation of bacteria
Pure culture of the target bacteria was grown overnight in liquid NB medium for the isolation of genomic DNA, which was done as per the method described by Hiney and colleagues [6] with some modification.
PCR amplification 16S rDNA gene
PCR reaction was performed in a gradient thermal cycler (Eppendorf, Germany). The universal primers (Forward primer 5'- AGAGTTTGATCCTGGCTCAG -3' and reverse primer 5'- CTTGTGCGGGCCCCCGTCAATTC-3') were used for the amplification of the 16S rDNA gene fragment. The reaction mixture of 50 μl consisted of 10 ng of genomic DNA, 2.5 U of Taq DNA polymerase, 5 μl of 10 X PCR amplification buffer (100 mM Tris-HCl, 500 mM KCl pH-8.3) , 200μM dNTP, 10 p moles each of the two universal primers and 1.5mM MgCl2. Amplification was done by initial denaturation at 94°C for 3 minutes, followed by 30 cycles of denaturation at 940C for 30 second, annealing temperature of primers was 55°C for 30 second and extension at 72°C for 1 minute. Final extension was at 72°C for 10 minutes
Agarose gel electrophoresis
10 μl of the reaction mixture was then analyzed by submarine gel electrophoresis using 1.0 % agarose with ethidium bromide at 8V/cm and the reaction product was visualized under Gel doc/UV transilluminator
Purification of PCR product
PCR product was purified by Qiagen gel extraction kit using the following protocol: Excise the DNA fragment from the agarose gel with a clean sharp scalpel. Weigh the gel slice in an eppendorf. Add 3 volumes of buffer QG to 1 volume of gel (100 mg ∼ 100 μl). Incubate at 50°C for 10 minutes. To help dissolve gel, mix by vortexing the tube every 2-3 min during the incubation. After the gel slice has dissolved completely, check that the colour of the mixture is yellow. Add 1 gel volume of Iso-propanol to the sample and mix. Place a QIAquick spin column in a provided 2 ml collection tube. To make the DNA bind to the column bind, apply the sample to the QIAquick column, and centrifuge for one minute. Discard flow-through and place QIAquick column back in the same collection tube. To wash, add 0.75 ml of buffer PE to QIAquick column and centrifuge for 1 minute. Discard the flow-through and centrifuge the QIAquick column for an additional 1 minute at 10,000 × g. Place QIAquick column into a clean 1.5 ml eppendorf. To elute DNA, add 50 μl of buffer EB (10mM Tris-Cl, pH 8.5) to the center of the QIAquick membrane and centrifuge the column for 1 min.
DNA sequencing of the 16S rDNA fragment
A total 100ng concentration of 16S rDNA amplified PCR product was used for the sequencing with the single 16S rDNA 27F Forward primer: 5'- AGAGTTTGATCCTGGCTCAG-3` by ABI DNA sequencer (Applied Biosystem Inc).
Computational analysis
Identification of Acinetobacterr sp.
A comparison of the 16S rRNA sequence of test strain with sequences in the non-redundant collection (Genbank, DDBJ, EMBL & PDB) using BLAST [7,8] and Ribosomal Database Project II [9,10] databases, has been done. The 16S rRNA gene sequence of the test strain was aligned with selected members of the genus Acinetobacter; sequences were obtained from BLAST hits, using multiple sequence alignment programme ClustalW [11]. A phylogenetic tree was drawn, using neighbor-joining method for this alignment by using ClustalW [11].
Regulatory elements analysis
We identified upstream DNA sequence region (17883 nt to 18443 nt) of 16S rRNA gene from the complete genome of Acinetobacter sp. strain ADP1 [12] as a input sequence for the regulatory elements analysis because this strain has maximum similarity (99% identity) with proposed Acinetobacter sp. AcBz01 at 16S rRNA gene sequence level. Bacterial promoter prediction program, BPROM [5] identified position of promoter i.e. Transcription start site (TSS), -10 box and -35 box in input sequence as well as predicts know Transcription Factor (TF) with their binding sites.
Discussion
rRNA-based analysis is a central method in microbiology, used not only to explore microbial diversity but also as a method for bacterial strain identification. The genomic DNA was extracted from isolated bacterial strain AcBz01 and universal primers 27F and 939R were used for the amplification and sequencing of the 16S rRNA gene fragment. A total of 805 bp of the 16S rRNA gene was sequenced and used for the identification of isolated bacterial strain. Ribosomal Database Project II (RDP) Classifier [9,10] tool with confidence threshold of 95% has classified that the test strain belongs to genus Acinetobacter. A 16S rRNA based phylogenetic tree showing the relationships between strain AcBz01 and representatives of the family Acinetobacter is given in Figure 1. It is evident from phylogenetic analysis of 16S rRNA gene that isolate AcBz01 represents a novel genomic species in the genus Acinetobacter.
Figure 1.
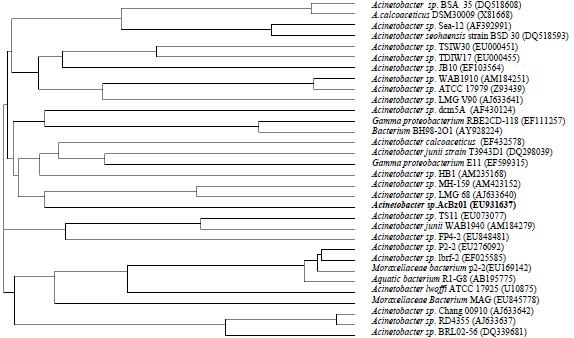
A Cladogram, using neighbor-joining method of selected 16S rRNA gene sequences of the genus Acinetobacter, obtained from BLAST hits, showing relationships between strain AcBz01 and some closely related representative members.
Genomic regulatory elements are frequently represented by DNA motifs. As such these representations are general and can be used to describe any class of short DNA sequence elements. Here we primarily discuss the transcriptional regulatory elements, more specifically, promoter elements and DNA binding sites that are bound by the TFs. In the non coding upstream sequences of Acinetobacter sp. AcBz01, the 16s rRNA gene promoter contains three signals, Transcription Start Site (TSS), -10 box (GAGTAACAT) and -35 box (TTGACT) at 321 bp, 306 bp, 287 bp and three TF binding sites at 302 bp, 308 bp, 317 bp relative to the start position of the input sequence, respectively shown in Figure 2 (see supplementary material). The detail of the three known TF narP, ompR and fadR are listed in table 1 (see supplementary material).
Conclusion
All bacterial species possess at least one copy of the 16S rRNA gene, which contains highly conserved as well as hyper variable nucleic acid sequences. Therefore the PCR targeting conserved nucleic acid sequence of 16S rRNA gene of bacterial isolate as a molecular tool is used to identify the presence of Acinetobacter sp. in water sample from Gomti River. Sequencing of 16S rRNA gene may also explain the discrepancy between phenotypic identification and the antibiotic susceptibility profile of a microorganism. Upstream region of 16S rRNA gene of Acinetobacter sp. AcBz01 containing promoter region, have three signals, TSS, -10box (GAGTAACAT), -35box (TTGACT) and three known TF narP, ompR and fadR with their binding sites.
It can be stated that these data provide a backbone to understand the basis of transcription regulation mechanism of 16S rRNA gene in Acinetobacter Sp. AcBz01.
Supplementary material
Acknowledgments
The present work was supported by a joint venture of the laboratory facilities at Chromous Biotech Pvt. Ltd. Bangalore, India and Biotechnology and Bioinformatics Division, BIOBRAINZ, Lucknow, U.P., India.
References
- 1.Jill E, III Clarridge. Clinical Microbiology Reviews. 2004;17:840. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdel-El-Haleem Desouky. African Journal of Biotechnology. 2003;2:71. [Google Scholar]
- 3.Harley CB, et al. Nucleic Acid Res. 1987;15:2343. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown CT. Methods Cell Biol. 2008;87:337. doi: 10.1016/S0091-679X(08)00218-5. [DOI] [PubMed] [Google Scholar]
- 5. http://www.softberry.com/berry.html.
- 6.Hiney M, et al. Appl Environ Microbiol. 1992;58:1039. doi: 10.1128/aem.58.3.1039-1042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. http://blast.ncbi.nlm.nih.gov/Blast.cgi.
- 8.Zhang Z, et al. J Comput Biol. 2000;7:203. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, et al. Appl Environ Microbiol. 2007;73:5261. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. http://rdp.cme.msu.edu/
- 11. http://www.ebi.ac.uk/Tools/clustalW.
- 12.Barbe V, et al. Nucleic Acids Res. 2004;32:5766. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


