Abstract
Mitochondrial (mt) genomic study may reveal significant insight into the molecular evolution and several other aspects of genome evolution such as gene rearrangements evolution, gene regulation, and replication mechanisms. Other questions such as patterns of gene expression mechanism evolution, genomic variation and its correlation with physiology, and other molecular and biochemical mechanisms can be addressed by the mt genomics. Rare genomic changes have attracted evolutionary biology community for providing homoplasy free evidence of phylogenetic relationships. Gene rearrangements are considered to be rare evolutionary events and are being used to reconstruct the phylogeny of diverse group of organisms. Mt gene rearrangements have been established as a hotspot for the phylogenetic and evolutionary analysis of closely as well as distantly related organisms.
Keywords: gene expression, evolution, mitochondrial gene, genomic variation, replication, gene regulation
Background
Genes and genomes are the product of complex processes of evolution, influenced by mutation, random drift, and natural selection. The inference of genome rearrangement events such as duplication, inversion, and translocation, is crucial in multiple genome comparisons. During the past three decades, DNA and protein sequence data has increasingly become the main asset to infer the general phylogeny; however, certain difficulties and limits have been found with the use of nucleotide sequences in phylogeny, in particular evolutionary rate differences between taxa, non-homogenous base composition, and multiple substitutions. “Rare genomic changes” have attracted great interest because of their potential to provide homoplasy free evidence of phylogenetic relationships [1]. Gene rearrangements are considered to be rare evolutionary events, and as such the existence of a shared derived gene order between taxa is often indicative of common ancestry [2].
Enormous numbers of phylogenetic studies using mt gene sequences have been carried out and reported, some dealing with the use of mitochondrial genes in the establishment of different levels of phylogenetic relationships. The success of mt DNA in molecular systematics lies in its prevalent characteristics, such as maternal inheritance, rapid rate of evolution, and haploid nature. Different parts of mt genome with different functional constraints are expected to evolve at different rates. Thus, comparative mt genomics promises to offer a comprehensive study of distinct patterns and processes of molecular evolution [3–6].
Description
Mitochondria are the power house of the cell. They are present in virtually every cell in body. They play an imperative role in metabolism, apoptosis, disease and aging. They are the site of oxidative phosphorylation, essential for the production of ATP, as well as for other biochemical functions. Mitochondria have a genome separate from the nuclear genome referred to as mitochondrial DNA (mtDNA). Animal mt DNA is a small (~16 kb), compact, economically-organized circular molecule, composed of 37 genes coding for 22 tRNAs, 2 rRNAs and 13 proteins, with few exceptions specifically in invertebrates [2]b The thirteen proteins are mainly involved in electron transport and oxidative phosphorylation of the mitochondria.
Recent advances in sequencing techniques have made available a great deal of data on whole genome basis. Complete mt genome sequences are available for thousands of organisms. The order of the genes in the mitochondrial DNA molecule in a wide variety of organisms has begun to be disclosed during last two decades. The gene order is highly conserved in vertebrates (Figure 1b) except for the region around the control region (D-loop), which is more prone to gene rearrangement. Maximum variability has been found in the gene order in invertebrates. The control region of the mitochondrial genome is frequently used in population studies due to the high variability in its nucleotide sequence, while protein-coding genes, such as cytochrome b (Cyt b), are generally used for phylogenetic analysis of taxa above the species level [7,8]b.
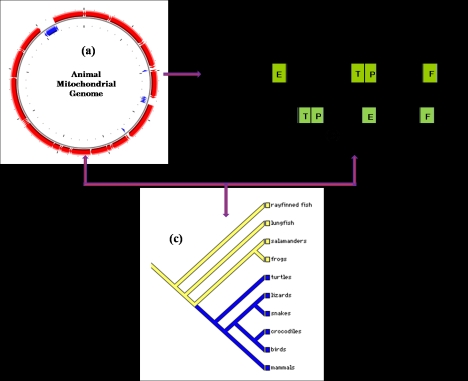
Figure 1.
Schematic representation of mt genome, mt gene order and its narrative role in elucidating evolutionary information. (a) Circular animal mt genome. (b) Universal mt gene order in vertebrates and one derived form in birds as ancestral gene order. The gene order near CR (control region) are shown which is the characteristics of higher organisms and more prone to gene rearrangements. (c) A schematic phylogenetic tree to indicate the generation of evolutionary information form mt gene order and its rearrangements.
Genome level character comparison can address a large number of evolutionary branch points. A small number of such comparisons have provided strong resolution of some evolutionary relationships which were controversial earlier. This suggests the reliability and confidence in their usage as markers [9]. Mitochondrial gene rearrangements are considered to be rare evolutionary events. In principle, ’rare genomic changes‘ [1], such as changes in gene order, can retain phylogenetic information for long periods of time, even when primary sequence data must have become randomized due to the involvement of long time periods.
Differences in mitochondrial gene order have been useful for phylogenetic resolution of some groups of species, for example Arthropoda being monophyletic, and within this Crustacea grouping with Hexapoda to the exclusion of Myriapoda and Onychophora. The ’universal‘ gene order for living vertebrates (Figure 1b) is not followed by birds [10]. In some recent studies based on complete mt genomes in birds, it has been suggested that gene orders evolved independently more than once. Also mapping of these gene orders on the avian phylogenetic tree suggested that one of these gene orders is an ancestral while the other is a derived form. The most acceptable model for the mt gene rearrangements till date is the tandem duplication followed by the deletion [4,9, 11,12].
Conclusion
With the increasing amount of mt and nuclear sequence data there will be new outcomes for phylogenetic studies at several levels and will soon emerge as the knowledge base for molecular evolution. As the understanding of the secondary structure of the control region, non coding region and ribosomal genes and their role in the mt genome will improve, these sequences will be helpful for use in phylogeny. Several studies and debate are still in progress on the structure of tRNAs and their specific role in gene regulation and evolution. Gene regulation may also serve as a substrate for evolutionary change, since control of the timing, location, and amount of gene expression can have an insightful effect on the functions and existence of the genes in the organism. Mt gene rearrangements are likely to reveal structural, functional and evolutionary aspects of the biology distinct from those evident from homologous sequence comparisons. It will also help in the prediction and reconstruction of homoplasy free phylogenetic relationships, and will also be used to develop models for the phylogenetic analysis of remotely related organisms and to better understand the evolutionary history of organisms.
References
- 1.Rokas, Holland PWH. Trends Ecol Evol. 2000;15:454. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- 2.Boore JL. Nucleic Acids Res. 1999;27:1767. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumazawa Y, Nishida M. Mol Biol Evol. 1995;12:759. doi: 10.1093/oxfordjournals.molbev.a040254. [DOI] [PubMed] [Google Scholar]
- 4.Mindell DP, et al. Proc Natl Acad Sci USA. 1998;95:10693. doi: 10.1073/pnas.95.18.10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nachman MW. Genetica. 1998;102/103:61. [PubMed] [Google Scholar]
- 6.Rand D. Trends Ecol Evol. 2001;16:2. doi: 10.1016/s0169-5347(00)02036-x. [DOI] [PubMed] [Google Scholar]
- 7.Boore JL. Trends Ecol Evol. 2006;8:439. doi: 10.1016/j.tree.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Desjardins P, Morais R. J Mol Biol. 1990;212:599. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- 9.Bensch S, Härlid A. Mol Biol Evol. 2000;17:107. doi: 10.1093/oxfordjournals.molbev.a026223. [DOI] [PubMed] [Google Scholar]
- 10.Slack KE, et al. Mol Phylogenet Evol. 2007;42:1. doi: 10.1016/j.ympev.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Singh TR, et al. Mol Biol Evol. 2008;25:475. doi: 10.1093/molbev/msn003. [DOI] [PubMed] [Google Scholar]
- 12.Rawlings TA, et al. Mol Biol Evol. 2001;18:1604. doi: 10.1093/oxfordjournals.molbev.a003949. [DOI] [PubMed] [Google Scholar]
- 13.Dowton M, et al. J Mol Evol. 2003;56:517. doi: 10.1007/s00239-002-2420-3. [DOI] [PubMed] [Google Scholar]
- 14.Smith MJ, et al. J Mol Evol. 1993;36:545. doi: 10.1007/BF00556359. [DOI] [PubMed] [Google Scholar]
- 15.Boore JL, et al. Nature. 1995;376:163. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- 16.Phillips MJ, Penny D. Mol Phylogenet Evol. 2003;28:171. doi: 10.1016/s1055-7903(03)00057-5. [DOI] [PubMed] [Google Scholar]
- 17.Zardoya R, Meyer A. Genetics. 1996;142:1249. doi: 10.1093/genetics/142.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]