Abstract
Radicle emergence and reserves mobilization are two distinct programmes that are thought to control germination. Both programs are influenced by abscissic acid (ABA) but how this hormone controls seed germination is still poorly known. Phenotypic and microscopic observations of the embryo axis of Medicago truncatula during germination in mitotic inhibition condition triggered by 10 μM oryzalin showed that cell division was not required to allow radicle emergence. A suppressive subtractive hybridization showed that more than 10% of up-regulated genes in the embryo axis encoded proteins related to cell-wall biosynthesis. The expression of α-expansins, pectin-esterase, xylogucan-endotransglycosidase, cellulose synthase, and extensins was monitored in the embryo axis of seeds germinated on water, constant and transitory ABA. These genes were overexpressed before completion of germination in the control and strongly inhibited by ABA. The expression was re-established in the ABA transitory-treatment after the seeds were transferred back on water and proceeded to germination. This proves these genes as contributors to the completion of germination and strengthen the idea that cell-wall loosening and remodeling in relation to cell expansion in the embryo axis is a determinant feature in germination. Our results also showed that ABA controls germination through the control of radicle emergence, namely by inhibiting cell-wall loosening and expansion.
Keywords: ABA, cell-wall expansion, germination, Medicago truncatula, radicle emergence, XET
INTRODUCTION
In flowering plants, seed germination is the transition state between quiescent embryo and a new photo-synthetically active plant. This transition state itself can be divided into three phases. The first phase is short and is characterized by a sharp and passive increase in water content of the dry and mature seed, while the second phase, during which the water content remains constant, consists of the synthesis of new mRNAs and proteins. During the third and the last phase of germination, the embryo axis elongates and radicle emerges through surrounding tissue. A further increase in water uptake occurs afterwards during post-germination growth and seedling establishment (Bewley, 1997).
Seed germination has been proposed to be under the control of two distinct programs that can progress independently, one driving radicle emergence and the other driving mobilization of reserves (Pritchard et al., 2002). Carbon and nitrogen reserves’ mobilization during germination has been studied in various cereal, legume, and non-legume species (Fath et al., 2001; Garciarrubio et al., 1997; Glevarec et al., 2004; Gomez-Cadenas et al., 2001; Limami et al., 2002). It has been hypothesized that abscissic acid (ABA) inhibition of germination occurs by preventing reserve mobilization pointing out the dependence of germination upon reserves mobilization (Finkelstein and Lynch, 2000; Garciarrubio et al., 1997). This hypothesis that has been named, ‘the nutritional control of germination’, was supported by the fact that the inhibitory effects of ABA were strongly repressed in the presence of glucose, sucrose, or fructose; however, this repression was limited to the process of radicle emergence (Finkelstein and Lynch, 2000). This hypothesis has been, however, weakened by the experiments of Pritchard et al. (2002), who showed that ABA inhibition of Arabidopsis thaliana was not accompanied by an inhibition of lipid (triacylglycerol) mobilization; moreover, sucrose derived from lipids degradation accumulated in ABA-treated seeds (Pritchard et al., 2002).
Embryo axis elongation and radicle emergence that are turgor-driven processes require loosening of cell walls in the cells of embryo root axes that lie between the root cap and the base of the hypocotyls. Many cellular and metabolic changes that occur in non-dormant seeds before the completion of germination also occur in dormant seeds; but embryo axes of only non-dormant seeds elongate. Several studies suggest that the real inhibition of germination in dormant seeds occurs at the very last stage and that is the inability of cell walls of radicle cells to elongate (Bewley, 1997). Analysis of impact of ABA, a potent inhibitor of germination on Brassica napus seeds, showed that neither osmotic potential of embryo axis cells nor their ability to uptake water was affected by the presence of ABA, but rather cell-wall loosening was prevented, resulting in the inhibition of germination. In general, prevention of embryo radicle extension can be achieved by incubating mature seeds in solutions of ABA even when ABA is introduced late during germination. This strengthens the idea that ABA acts to prevent the third phase of germination, such as radicle cell-wall loosening. In sunflower, temporary application of ABA to embryos only prevents radicle extension while its removal leads to the completion of germination (Bewley, 1997). Altogether, these findings highlight the importance of cell-wall loosening in embryo axis cells that allows for cell elongation, probably a determinant event in embryo axis elongation during germination. However, the occurrence and control of this phenomenon, cell-wall loosening or radicle elongation, are poorly known.
Our strategy for improving knowledge of important and vital events of germination was based on a suppressive and subtractive hybridization (SSH) transcriptomics analysis of embryo axes for the identification of genes involved in germination completion. In the present study, the subtraction was carried out between two mRNA populations extracted at two imbibition stages, 6 hours (i.e. the beginning of the second phase of germination) and 23 h (i.e. 2 h after the end of the second phase of germination). It is important to note that new mRNAs are synthesized during the second phase of germination. The fact that this study revealed that more than 10% of up-regulated genes encode proteins related to cell-wall loosening and expansion spurred us to put forward the hypothesis that a late event during germination such as radicle cell-wall loosening, a prerequisite for cell expansion, might be a determinant in germination completion and that event might be controlled by ABA.
RESULTS
Effect of Constant and Transitory Abscisic Acid Treatment on Germination and Post-Germination Growth of Medicago truncatula
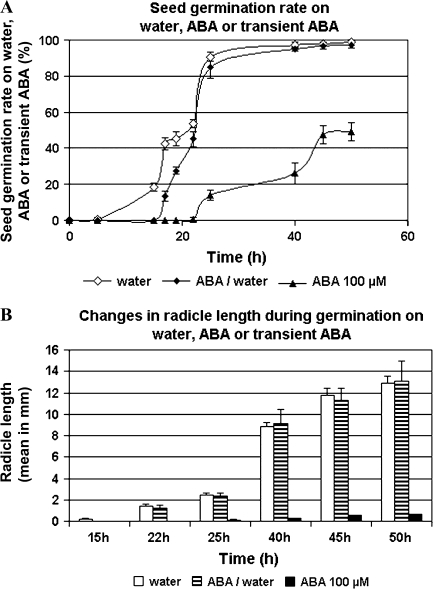
To determine the effects of a constant and transitory ABA treatment versus water (control) on germination and post-germination growth, germination rate, and radicle length were examined under the three conditions. In water condition, 50% germination (T50) was reached after 22 h of imbibition and 88% after 25 h (Figure 1A). This seed lot has a maximum germination rate of 98%. Non-germinating seeds were either not imbibed (hard seeds) or had broken radicle. Elongation of radicles was slow during the first hours of post-germination growth and accelerated markedly between 25 and 40 h of imbibition before decreasing (Figure 1B). In comparison with water condition, 100 μM ABA dramatically inhibited the germination rate by about 50% (Figure 1A) and blocked radicle elongation above 1 mm by mean (Figure 1B). In the transitory ABA treatment, seeds were imbibed on 100 μM ABA during 15 h and transferred on water. Under this condition, the first seeds started to germinate at 17 h, namely 2 h after stopping ABA treatment, and, after 22 h and later, germination rate and radicle length were comparable to that on water (Figure 1A and 1B).
Figure 1.
Effect of Permanent and Transient Abscisic Acid Treatment on Medicago truncatula Seed Germination and Radicle Growth.
Germination rates (A) and radicle length (B) are the mean of 150 measurements taken on 150 seeds germinated on water, 100 μM ABA or 100 μM ABA during 15 h before transfer of the seeds on water.
Microscopic Observation and Cell Size Determination in the Radicles of Medicago truncatula Embryo Axis throughout Germination
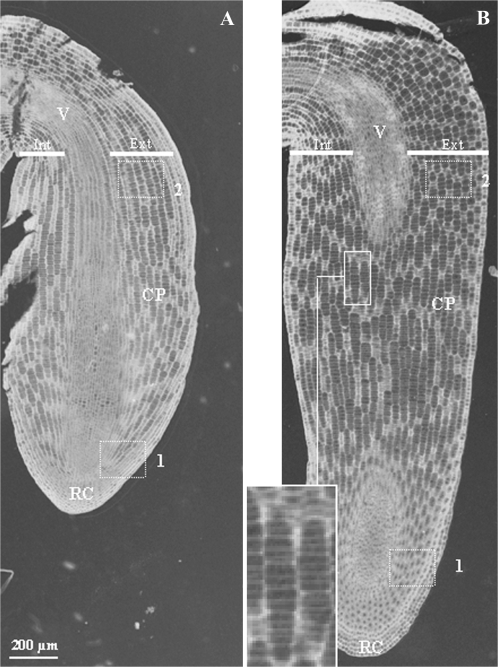
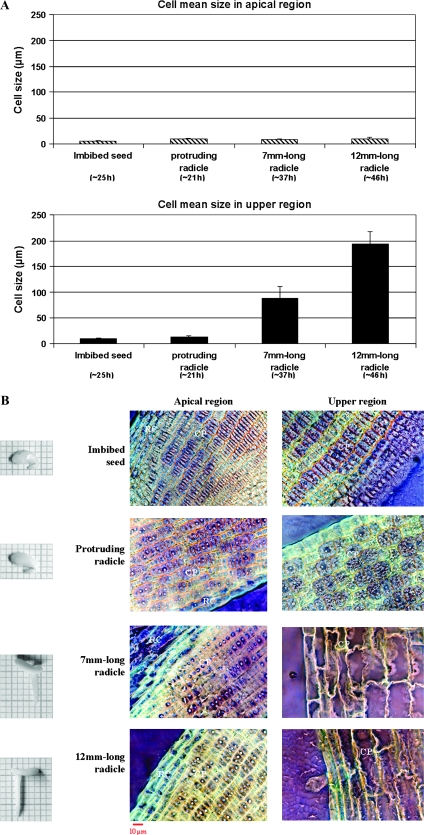
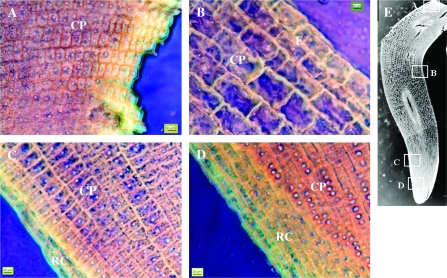
The cortical parenchyma cells were studied in conventional microscopy with meta-chromatic toluidine-blue coloration at several stages throughout germination and early post-germination (Figures 2 and 3). Longitudinal section of embryos in two states, before and after germination completion, namely ‘imbibed seeds’ and ‘protruding radicle’ (5 and 21 h of imbibition, respectively), shows that cortical parenchyma cells are clustered in spindles all along embryo axes (Figure 2). Transversal cell growth was asymmetrical. For instance, in the region just under the hypocotyl (sub-hypocotyl region), external cell layers were two-fold larger than internal ones. Both the apical and upper regions underwent axial cell growth (Figure 2). In the ‘imbibed seed’ state, cortical parenchyma cells were 6 mm in length in the apical region and 9 mm in length in the upper region, and all the cells were in undifferentiated state (Figure 3A). Then, they reached 10 and 13 mm, respectively (Figure 3A), leading to radicle protrusion through the seed coat. This first size increases affected cells overall and happened without differentiation of the cells and vacuole establishment (Figure 3B). Later, apical cells kept the same feature (10 mm, undifferentiated) even with radicle growth. In contrast, cells in the upper region were ∼90 mm in length, differentiated with large vacuoles in 7-mm-length radicles, and ∼190 mm in 10-mm-length radicles (Figure 3).
Figure 2.
Microscopic Observation of Longitudinal Sections of Medicago truncatula Embryos at Two Developmental Stages.
Imbibed seed (5 h) before germination completion (A) and after germination completion and radicle emergence (21 h) (B). ×4 enlargement shows details of cells clustered in spindles. Boxes correspond to the apical region (1) and the upper region (2) of the radicle. Int, internal cell layers; Ext, external cell layers; CP, cortical parenchyma; V, vascular tissues; RC, root cap.
Figure 3.
Changes in Cortical Parenchyma Cell Length (A) and Anatomy (B) in the Apical and Upper Region of Medicago truncatula Radicle at Various Developmental Stages, Before and After Germination Completion.
Pictures are presented on highly saturated inverted colors. ×40. CP, cortical parenchyma; RC, root cap.
Effect of Oryzalin, an Inhibitor of Mitosis Metaphase on Germination and Radicle Elongation of Medicago truncatula
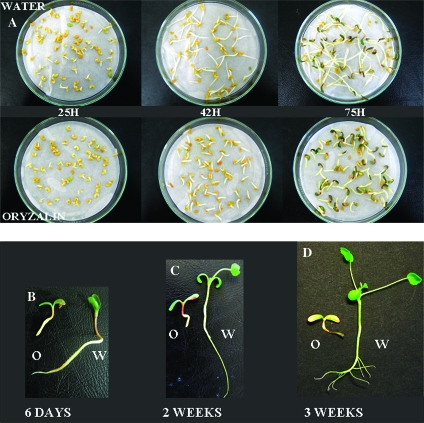
In order to determine the role of both mitosis and cell elongation in Medicago seed germination, oryzalin was used to inhibit mitosis metaphase (de-polymerization of cellular microtubules). No difference in germination rates and kinetics can be seen during the overall experiment between water (control) and 10 μM oryzalin condition (data not shown) and, after 25 h, radicle growth was identical in both conditions (Figure 4). After 42 h and later, radicles of seeds soaked on oryzalin reached approximately 10 mm and were stopped at this stage whereas seeds soaked on water continued to develop normally. On nutritive solution and under light conditions, seedlings subjected to oryzalin kept short and swollen radicles but followed their developing program by root hair growth, hypocotyl rising, and cotyledon greening and opening (6 d seedlings, Figure 4). After 2 weeks, in comparison with control, hypocotyls of seedlings on oryzalin were shorter and had a red coloration. Epicotyl and the first leaves failed to elongate completely (Figure 4). Later, seedlings on oryzalin bleached and started to decompose (3 weeks seedlings, Figure 4).
Figure 4.
Effect of 10 μM Oryzalin on Medicago truncatula Seed Germination and Seedling Post-Germination Growth.
O, oryzalin condition; W, water condition.
Longitudinal sections of 4-mm-length radicles from seeds germinated on oryzalin (Figure 5) were used for conventional microscopic observation. This experiment showed that cells were not synchronized for axial growth, since they had variable extent, depending on position. In hypocotyls (A) and apical (D) zones, cells were very small (approximately 7 mm) with dense cytoplasm and visible nuclei, similar to cells observed in the radicle apex of seeds germinated on water (Figure 3). Because of oryzalin treatment, these cells were definitely not formed from the early mitotic process during germination, but formed during embryo development. Between these positions, cells were gradually longer and more differentiated. For this experiment, the maximum length observed was approximately 50 mm with developed vacuoles.
Figure 5.
Microscopic Observation of a Longitudinal Section of Medicago truncatula Embryo Axis Cells after Germination Completion and Radicle Emergence after 25 h of Oryzalyn (10 μm) Treatment.
Hypocotyl (A), upper region (B), sub-apical region (C), and apical region (D) of the radicle. Pictures are presented on highly saturated inverted colors. ×40. RC, root cap; CP, cortical parenchyma; E, epidermis.
Expression of Up-Regulated Genes Involved in Cell-Wall Loosening and Expansion in Medicago truncatula Embryo Axis throughout Germination
In order to identify genes involved in the control of Medicago truncatula germination, a cDNA library was constructed by suppressive hybridization. For this purpose, cDNA libraries of genes specifically expressed in the embryo axis have been generated by a PCR-based Suppressive Subtractive Hybridization (SSH) technique on mRNAs of populations of embryo axes obtained at two imbibition times, namely 6 h (i.e. the beginning of the second phase of germination) and 23 h (i.e. 2 h after radicle protrusion). Nine hundred clones were generated that revealed after sequencing 343 unique ESTs with 276 ESTs of up-regulated genes and 67 ESTs of down-regulated genes. A comprehensive list of these clones as well as a functional classification are presented as Supplementary Data (two tables and one figure). Among the clones obtained by this subtraction, about 10% of up-regulated genes were identified as genes encoding cell-wall biosynthetic and architecture-modifying enzymes and structural proteins in the embryo axis (Table 1). No genes implicated in cell cycle or proliferation were detected in this library.
Table 1.
List of ESTs of cell wall metabolism related genes revealed by SSH approach during germination of Medicago truncatula
| Gen Bank Accession number | Medicago trucatula Gene index TC number | Identity of the clone |
| Up regulated | ||
| ES481863 | TC104491 | Cellulose synthase |
| ES466859 | TC96054 | Alpha-expansin |
| ES466858 | TC100994 | Expansin precursor |
| EL563604 | AL372570 | Extensin-like protein (1) |
| EL563664 | TC94245 | Extensin-like protein (2) |
| EL563549 | TC106582 | Extensin-like protein (3) |
| ES466854 | TC100493 | Extensin-like protein (4) |
| ES466861 | TC95797 | F-actin capping protein, alpha subunit |
| EL563565 | TC100262 | MtPPRDI |
| ES466860 | AJ501848 | Pectinesterase |
| EL563643 | TC103769 | Phosphatidylinositolglycan class N |
| EL563674 | TC97045 | Plant lipid transfer (1) |
| EL563532 | TC94138 | Plant lipid transfer (3) |
| EL563610 | TC100736 | Plant lipid transfer / prolin-rich protein |
| EL563654 | TC100266 | Plant lipid transfer (2) |
| EL563668 | TC93943 | Root cap protein-like |
| ES466857 | TC94949 | T2L5.6 protein [heparanase like, (endoglucosidase)] |
| EL563527 | AW687357 | Tubulin / FtsZ protein (1) |
| EL563516 | CX541346 | Tubulin / FtsZ protein (2) |
| ES466856 | TC106434 | Tubulin / FtsZ protein (3) |
| EL563544 | TC100408 | Tubulin beta chain |
| EL563500 | TC106341 | UDP-glucose:protein transglucosylase |
| EL563503 | TC106650 | Weakly similar to Lycopersicon esculentum beta galactosidase |
| EL563647 | TC103297 | Beta-galactosidase |
| EL563663 | TC97248 | Copper amine oxidase |
| EL563606 | TC109732 | L-ascorbate oxidase |
| ES466811 | TC95190 | Auxin-induced beta-glucosidase |
| EL563657 | AW697240 | Mob1-like protein / phocein family |
| Down regulated | ||
| ES408378 | TC95684 | Mob 1-like protein / phocein family |
| ES408368 | TC100580 | Plant lipid transfer |
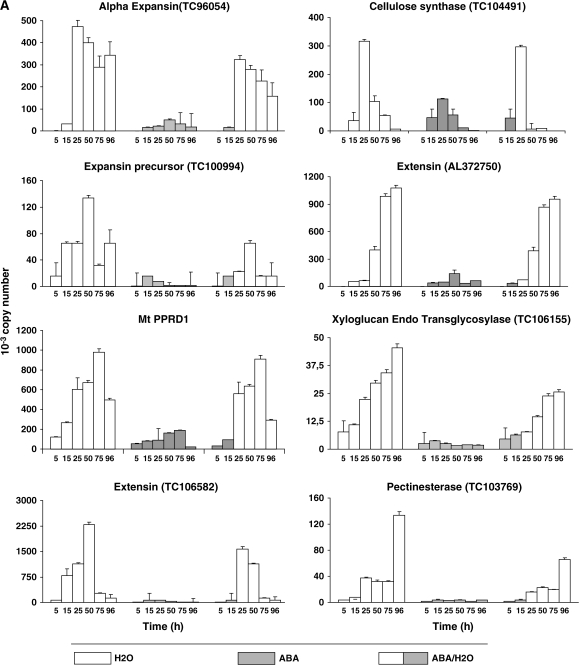
To find links between events of radicle cells (elongation and division) and transcription activity, we studied the expression of genes encoding enzymes related to cell-wall loosening or enzymes responsible for cell-wall expansion and the synthesis of cell-wall constituents: α-expansins (TC96054 and TC104491), a pectin-esterase (TC103769), xylogucan endotransglycosidase (XET, TC106155), cellulose synthase (TC104491), extensins (TC106582 and AL373750), and MtPPRD (protein with proline-rich domain). They were studied on water and in the presence of constant or transitory ABA and in seeds treated with oryzalin (Figure 6). All these genes were increasingly expressed on water (from 5 h measure to maximum) and strongly inhibited in ABA condition (Figure 6A). Transitory ABA inhibited gene expression until seeds were back to water (after 15 h), although later transcript abundance was in some cases lower as compared to control (Figure 6A).
Figure 6.
Expression of Genes Involved in Cell-wall Biosynthesis and Architecture Modification and Cell Cycle in Medicago truncatula Embryo Axis under Various Conditions of Germination: Water, Permanent, and Transient (100 μM) ABA and 10 μM Oryzalin Treatment.
The results are the mean ± SE of at least two biological replicates with duplicated PCR reaction for determining Ct values. Expression levels are presented in arbitrary units.
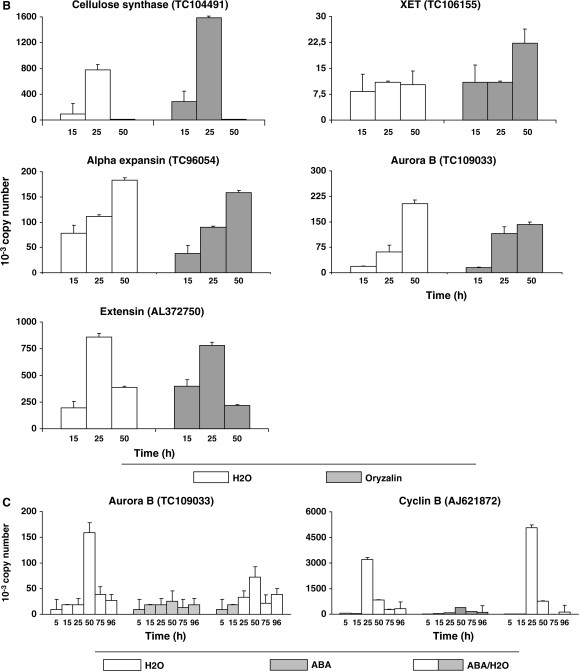
Expression of four of the above-mentioned genes was checked out under oryzalin conditions in comparison with the control (Figure 6B). Expression profiles of the four genes namelyXET, α-expansin, extensin, and cellulose synthase, were similar under both conditions, allowing us to conclude that oryzalin did not affect the expression of key genes related to cell-wall processing.
Expression of Up-Regulated Genes Involved in Activation of Cell Division in the Medicago truncatula Embryo Axis throughout Germination
Activation of cell division in the Medicago truncatula embryo axis throughout germination was studied using the genes related to mitosic activity revealed by SSH. Studied genes were the mitotic Cyclin B-1,2 (AJ621872), known to be present only in G2 phase and early M phase, the Aurora B (TC109033), known to mediate the regulation of ‘centromere–kinetochore–microtubule’ linkage in mitotic cells, and the Mob-1-like protein/phocein family (TC95684), known to be involved in cell proliferation and localized in the cell division plane during cytokinesis. The three genes were transiently overexpressed during germination but expression peak occurred after germination completion and radicle protrusion, namely 25 h for Cyclin B-1,2 and after 50 h for aurora B and Mob-1-like. The expression of all three genes was inhibited by 100 μM ABA treatment. Back to water, the genes were expressed again with a global profile of expression similar to that in the control with, however, lower levels of mRNA, except for Cyclin B-1,2, whose level was greater (Figure 6C).
DISCUSSION
Radicle Protrusion in Germinated Medicago truncatula Seeds Is Driven by Cell Extension and Does Not Require Cell Division
In the present work, it is shown that cell division is not required to allow radicle protrusion in Medicago truncatula seed germination. Under conditions of mitosic inhibition triggered by oryzalin treatment and until radicle reached 10 mm, seeds germinated with the same rate and aspect as observed in control. This lack of early phenotype implies that mitosis occurs after radicle protrusion. This is corroborated by the results of the expression of cell-cycle-related genes and particularly aurora B. Aurora B is a key-protein for chromosome alignment during metaphases (Francis, 2007). At 25 h of imbibition, its expression did not rise, yet, over the basal level, even though the majority of seeds had undergone radicle protrusion. This suggests that cells did not reach metaphases when the radicle protrudes through the seed coat. This observation is concordant with Faria et al. (2005), who noticed, in germinating Medicago truncatula embryo axes, that mitotic microtubules did not appear before the radicles protruded and reached 2–3 mm in length (Faria et al., 2005).
In order to have more information at the tissue level about the early events of Medicago truncatula germination, morphological modifications between longitudinal sections of radicles at the ‘imbibed seed state’ (5 h) and ‘protruding radicle state’ (21 h) were studied. Asymmetrical cell expansion was observed along the radicles, resulting in a radical change in cell structure. Between both states, all cells were implicated in growth but enlargement was more important in the central region of the radicles, suggesting that growth starts at this point and spreads to up and downstairs neighboring cells. In order to challenge this idea, cell elongation was studied along the embryo axis during germination when mitosis was inhibited by 10 μM oryzalin. The microscopic observations showed that not only cells in the central region were the largest observed in the longitudinal section of 4-mm-long embryos, but also neighboring cells on both sides were smaller and smaller as they were closer to the hypocotyl and apical region. In other words, the first enlargement is observed for cells at a distal region from the apex. For root growth, cells are sequentially newly produced from root meristem and elongate while pushing down the root apex through substrate, while, in embryo, it seems that radicle cells did not elongate after division during the maturation phase and the oldest ones were the first to engage in enlargement during germination. These observations also confirmed that cell division is not required to allow radicle protrusion in Medicago truncatula germinating seeds.
Altogether, these results support the idea that cell-wall loosening and expansion, a prerequisite for cell extension, might be a determinant feature in germination completion. Consistently, more than 10% of the up-regulated genes in the embryo axis of germinating Medicago truncatula were identified as encoding cell-wall biosynthetic and architecture-modifying enzymes and structural proteins. Cell-wall-related genomic approaches in different physiological contexts have revealed that many genes encoding enzymes of cell-wall biosynthesis and architecture modification and structural proteins are regulated at the transcriptional level (Holmes et al., 2008; Milioni et al., 2001; Schrader et al., 2004). Quantitative real-time RT–PCR was used to confirm the level of expression of 10 transcripts from the up-regulated genes related to cell-wall loosening and expansion. The transcripts analyzed were chosen based on their demonstrated role in angiosperms. For cell-wall loosening, genes studied were two expansins, β-1,3-glucanase and pectin-esterase. For cell-wall expansion and strengthening, genes studied were xylogucan endotransglucosylase (XET), cellulose synthase, arabinogalactan protein, two extensins, and PPRD. In embryo axes of non-dormant Medicago truncatula seeds germinating on water (control batch), the expression of all these genes increased throughout germination and post-germination. More importantly, genes known to play key role in cell-wall loosening and expansion, such as expansins, 1-3 glucanase, and XET, were significantly expressed in the embryo axis as early as 5 or 15 h of imbibition, namely before germination completion and radicle extension. The primary cell wall is composed of cellulose microfibrils embedded in hydrated matrix of hemicelluloses (xyloglucan in non-gramineous), pectins, and glycoproteins. Hemicelluloses form non-covalent bonds with cellulose, allowing adjacent cellulose microfibrils to tie (Lerouxel et al., 2006). During turgor-driven cell-wall expansion, the xyloglycan–cellulose network needs to be disrupted in a controlled manner to allow the necessary slippage of cellulose microfibrils in growing cell walls without losing their integrity (Marga et al., 2005). Expansins and XET have been shown to be the key players in this process (Sampedro and Cosgrove, 2005; Vissenberg et al., 2005a, 2005b). Expansins disturb the hydrogen bonds between xyloglycan and the cellulose microfibrils, allowing the slippage of microfibrils while XET allow the re-organization of xyloglycan within the growing cell wall by incorporating newly synthesized xyloglycan into the network. XET is thought to exhibit a hydrolase activity along with its endotransglucosylase activity (Vicky et al., 2007). The enzyme would then break and rejoin tethering xyloglucan with another xyloglucan.
Under the condition of mitosis inhibition triggered by oryzalin treatment, expression of four genes selected on the basis of their roles in cell-wall loosening (expansin), expansion (XET, cellulose synthase), and structural protein (extensin) was determined. The results showed that these genes were expressed in the embryo axis as in the control before germination completion.
ABA-Driven Inhibition of Germination of Medicago truncatula Involves the Inhibition of the Expression of Genes Involved in Cell-Wall Loosening and Expansion
Endogenous abscisic acid (ABA) is responsible for the inhibition of germination in dormant seeds, despite every condition being suitable (Baskin and Baskin, 1998). In mature non-dormant seeds, prevention of embryo radicle extension can be achieved by incubation in solutions of ABA. This inhibition can occur even when ABA is introduced later during germination; conversely, early application of ABA followed by its removal later during the germination process does not prevent germination completion. This is illustrated by Figure 1, which shows the inhibitory effect of 100 μM ABA treatments on germination of Medicago truncatula seed, while its removal after 15 h of treatment resulted in a slight delay in germination kinetics and did not impact radicle growth. Analysis of the impact of ABA, a potent inhibitor of germination on Brassica napus seeds, showed that neither osmotic potential of embryo axis cells nor their ability to take up water was affected by the presence of ABA, but rather cell-wall loosening was prevented, resulting in inhibition of germination (Bewley, 1997).
Taken together, the available literature and the present results led us to suppose that cell-wall loosening and expansion are targets of ABA control of germination. In order to challenge this hypothesis, the expression of the above-studied genes was monitored by quantitative real-time RT–PCR in embryo axes of germinating Medicago truncatula in response to our two ABA treatments: constant and transitory application of 100 μM ABA. All the genes were strongly inhibited by ABA in the ABA constant treatment. This inhibition was abolished in the ABA transitory treatment once the seeds were transferred back on water. These results validate the recruitment of these genes as major contributors to the completion of the germination process and strengthens the idea that changes in cell-wall loosening and remodeling in relation to cell expansion in the embryo axis is a determinant feature in germination.
It is important to note that the inhibitory effect of ABA was selective. Several genes were expressed in the embryo axis of ABA-treated seeds at the same level as in the control or even higher, in the absence of radicle emergence and germination completion, such as the gene encoding S-adenosylmethionine synthase (SAMS, TC106563). The SAMS protein, which catalyses the synthesis of S-adenosylmethionine from methionine and ATP, absent in the dry mature seed, was found to accumulate in the embryo axis concomitantly to radicle emergence. S-adenosylmethionine synthetase was proposed to play a fundamental role in the control of the metabolism during the transition from a quiescent to a highly active state during seed germination (Bouton et al., 2005; Gallardo et al., 2003). Among genes whose expression was not inhibited in ABA-treated seeds were genes encoding aquaporin (TC100308), hexokinase (109111), methionine synthase (TC106598), methyltransferase (TC100717), γ-aminobuturate transaminase (TC109262), and methylene tetrahydrofolate reductase (TC94425) (data not shown).
Furthermore, our results shed light on the way ABA control of germination is exerted. It has been established that ABA plays a crucial role in embryo development, seed maturation, dormancy, and germination. The inhibition of germination by ABA has been used as a bioassay for screening mutants for ABA responses allowing for the discovery of several intermediates in ABA perception and signal transduction, such as the family of ABI (ABA-Insensitive) proteins (Finkelstein et al., 2002). However, the mechanism by which ABA inhibits germination is still poorly understood. Several evidences support an ABA control of germination through mobilization of reserves, such as the ABA antagonistic effect on Gibberrellic acid (GA)-induced starch mobilization in cereal grains (Bethke and Jones, 2001; Lovegrove and Hooley, 2000) and the antagonistic effect of exogenous sugar on ABA-induced inhibition of germination (Garciarrubio et al., 1997). The latter results suggested that ABA inhibition of germination was induced by nutritional deficiency caused by the inhibition of mobilization of reserves by exogenous ABA. However, a purely nutritional effect of ABA on germination was counteracted by several experiments. It has been shown that glyoxilate cycle is not essential for germination in isocitrate lyase-deficient Arabidopsis mutant (Eastmond et al., 2000). Furthermore, Arabidopsis seeds were shown to mobilize lipid reserves in the presence of ABA and absence of germination (Pritchard et al., 2002). The present results are in favor of the alternative hypothesis of an ABA control of germination through the control of radicle emergence. Our results strongly suggest that by inhibiting the expression of genes encoding enzymes of cell-wall biosynthesis and architecture modification and structural proteins, ABA inhibits cell-wall loosening and expansion and consequently inhibits radicle emergence and germination.
METHODS
Plant Material
For all conditions, 50 seeds of Medicago truncatula cv. Parragio were germinated at 20°C in darkness in 9-cm-diameter Petri dishes on Whatman paper soaked with 3.5 ml solution. Solutions were: ultra-pure water, 100 μM Abscisic acid or 10 μM oryzalin (3,5-dinitro-N4,N4-dipropylsulfanilamide).
As scarified and non-scarified seeds soaked on ultra-pure water and 10 μM oryzalin showed the same rates of germination, it was concluded that the seed coat of Medicago truncatula cv. Paragio might not block the absorption of oryzalin (data not shown). Oryzalin has been shown to be absorbed by the seeds of several populations of Acacia dealbata and Acacia mangium.
For germination curves, seeds transiently on ABA were washed after 15 h and placed on Whatmann paper soaked with 3 mL ultra-pure water. For real-time quantitative PCR analysis, embryo axes were immediately frozen in liquid nitrogen after collection. For photography, seeds were germinated during 42 h on water condition and then placed under periodic light conditions (16 h/24 h) at 21°C on nutritive solution (1 mM KH2PO4; 1 mM MgSO4; 0.25 mM CaCl2; 5 mM (NH4)2SO4; 5 mM KNO3; 50 μM KCl; 25 μM Na2Fe–EDTA; 1.44 mg L−1 H3BO3; 0.04 mg L−1 CuCl2(2H2O); 0.9 mg L−1 MnCl2(4H2O); 0.12 mg L−1 ZnCl2; 0.02 mg L−1 MoO4) with or without oryzalin addition.
Determination of Germination Rate and Radicle Length
Measures were made on 150 seeds (three Petri dishes). Germination rate was determined by the radicle appearance, according to the definition of Bewley (1997). Radicle length at tx was calculated as follows: (total radicle length at tx)/(total germinated seeds), where tx represents time (h) measure. Measures were repeated three times.
RNA Extraction
For one biological measure, 50 embryo axes were disrupted under mortar and pestle in liquid nitrogen. RNA extraction was made with TriReagent (Ambion, Austin, TX, USA) following the manufacturer's instructions for plant material. RNA quantity was evaluated by ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA), and quality was checked by agarose gel electrophoresis.
Subtractive Suppressive Hybridization (SSH) Library Construction and Differential Screening
Suppression subtractive hybridization was carried out between mRNAs of embryo axes corresponding to two imbibition times, 6 and 23 h, using a Smart PCR cDNA synthesis kit and a PCR-Select Subtractive Hybridization kit (Clontech, Palo Alto, California, USA). For the construction of the 6 and 23 h imbibition libraries, driver and tester cDNAs were synthesized using 2 μg of polyA RNA purified from the total RNA with poly-A tract System 1000 (Promega, Madison, Wisconsin, USA). For the 6 h library, PolyA RNA from embryos axes imbibed 6 h were used for the synthesis of the tester cDNA and PolyA RNA from embryos axes imbibed for 23 h were used for the synthesis of the driver cDNA. For the 23 h library, PolyA RNA from embryos axes imbibed for 23 h were used for the synthesis of the tester cDNA and PolyA RNA from embryos axes imbibed for 6 h were used for the synthesis of the driver cDNA. The efficiency of the subtraction was estimated by comparing the abundance of a gene known to be overexpressed during germination (Mt PPRD1, Bouton et al., 2005) and a constitutive gene, MSC27, before and after subtraction by real Q–PCR (data not shown).
PCR products were purified using the QIAquick PCR purification kit, polyA 3’ elongated with Promega Taq Polymerase and then were ligated in PGEM-T Easy vector (Promega, Madison, Wisconsin, USA). Escherichia coli one shot® Top 10F Chemically Competent Cells (invitrogen, Breda, The Netherlands) were transformed with ligated plasmids. Clones selection was performed by hybridization as described in Merchan et al. (2007). The selected positives clones (355) were sequenced and ESTs annotated on the basis of TIGR, NCBI, and MENS (Medicago EST Navigation System) databases using BLASTN.
Reverse Transcription
cDNAs were obtained by retro-transcription of 2 μg of total RNA using 200 units of RT–MMLV (Invitrogen, Breda, The Netherlands), 2 μg of random primer (Invitrogen) in the presence of 40 units of Recombinant Rnasin, Ribonuclease Inhibitor (Promega, Madison, WI, USA). Reaction occurred 1 h at 37°C in total volume of 50 μL.
Real-Time Quantitative PCR
Reaction was performed on the light cycler ABI Prism 7000 SDS (Applied Biosystems, Foster city, CA, USA). Each reaction was performed on 3 μL of a 1:2 (v/v) dilution of the first cDNA strands using SYBR Green PCR Master Mix (Applied Biosystems) following the manufacturer's instructions with 200 nM of each primer in a total reaction of 25 μL. The reaction was incubated for 2 min at 50°C and 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 58°C. The specificity of the PCR amplification procedure was checked with a heat-dissociation protocol (from 65 to 95°C) after the final cycle of PCR. Each measure was done with at least two biological replicates with duplicated PCR reaction for determining Ct values. Amplified fragments of each gene were cloned into the pGEM-T Easy vector (Promega, Madison, Wisconsin, USA). The plasmids were diluted several times to generate templates ranging from 105 to 100 copies used for standard curves for the estimation of copy numbers in total cDNA. The results are expressed as copy number times 10−3. The gene encoding the transcription factor MSC27 (Medicago sativa cDNA 27) was used as a constitutive control (data not shown).
Microscopy
Samples were harvested and dissected in 3.5% v/v glutaraldehyde in phosphate buffer, then infiltrated under vacuum in fresh fixative during 2 h, on ice. After three washes in phosphate buffer and three washes in distilled water, samples were dehydrated through a graded series of ethanol and embedded in resin (Technovit 7100 resin, Heraeus Kulzer GmbH, Wehrheim, Germany) following the manufacturer's instructions. Samples were cut longitudinally to 8–10-mm sections, settled in water drops on glass slides, and dried at 40°C overnight. Tissues were stained with 0.5% w/v aqueous toluidine-blue solution and mounted in Entellan® (ProSciTech, Thuringowa, Australia) prior to observations.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was funded by grants from the EU FP6 project FOOD-CT-2004-506223, ‘Grain Legumes Integrated Project (GLIP)’ and COSAVE Contract with Région Pays de la Loire, France. No conflict of interest declared.
Supplementary Material
References
- Baskin C, Baskin J. Seeds: Ecology, Biogeography, and Evolution of Dormancy and Germination. San Diego: Academic Press; 1998. [Google Scholar]
- Bethke PC, Jones RL. Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. Plant J. 2001;25:19–29. doi: 10.1046/j.1365-313x.2001.00930.x. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton S, Viau L, Lelievre E, Limami AM. A gene encoding a protein with a proline–rich domain (MtPPRD1), revealed by suppressive subtractive hybridization (SSH), is specifically expressed in the Medicago truncatula embryo axis during germination. J. Exp. Bot. 2005;56:825–832. doi: 10.1093/jxb/eri077. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA. Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl Acad. Sci. U S A. 2000;97:5669–5674. doi: 10.1073/pnas.97.10.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria JM, Buitink J, van Lammeren AA, Hilhorst HW. Changes in DNA and microtubules during loss and re–establishment of desiccation tolerance in germinating Medicago truncatula seeds. J. Exp. Bot. 2005;56:2119–2130. doi: 10.1093/jxb/eri210. [DOI] [PubMed] [Google Scholar]
- Fath A, Bethke PC, Jones RL. Enzymes that scavenge reactive oxygen species are down-regulated prior to gibberellic acid-induced programmed cell death in barley aleurone. Plant Physiol. 2001;126:156–166. doi: 10.1104/pp.126.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol. 2000;122:1179–1186. doi: 10.1104/pp.122.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;(14 Suppl):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D. The plant cell cycle––15 years on. New Phytol. 2007;174:261–278. doi: 10.1111/j.1469-8137.2007.02038.x. [DOI] [PubMed] [Google Scholar]
- Gallardo K, Le Signor C, Vandekerckhove J, Thompson RD, Burstin J. Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol. 2003;133:664–682. doi: 10.1104/pp.103.025254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciarrubio A, Legaria JP, Covarrubias AA. Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta. 1997;203:182–187. doi: 10.1007/s004250050180. [DOI] [PubMed] [Google Scholar]
- Glevarec G, Bouton S, Jaspard E, Riou MT, Cliquet JB, Suzuki A, Limami AM. Respective roles of the glutamine synthetase/glutamate synthase cycle and glutamate dehydrogenase in ammonium and amino acid metabolism during germination and post-germinative growth in the model legume Medicago truncatula. Planta. 2004;219:286–297. doi: 10.1007/s00425-004-1214-9. [DOI] [PubMed] [Google Scholar]
- Gomez-Cadenas A, Zentella R, Walker-Simmons MK, Ho TH. Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell. 2001;13:667–679. [PMC free article] [PubMed] [Google Scholar]
- Holmes P, Goffard N, Weiller GF, Rolfe BG, Imin N. Transcriptional profiling of Medicago truncatula meristematic root cells. BMC Plant Biol. 2008;8:21. doi: 10.1186/1471-2229-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel O, Cavalier DM, Liepman AH, Keegstra K. Biosynthesis of plant cell wall polysaccharides—a complex process. Curr. Opin. Plant Biol. 2006;9:621–630. doi: 10.1016/j.pbi.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Limami AM, Rouillon C, Glevarec G, Gallais A, Hirel B. Genetic and physiological analysis of germination efficiency in maize in relation to nitrogen metabolism reveals the importance of cytosolic glutamine synthetase. Plant Physiol. 2002;130:1860–1870. doi: 10.1104/pp.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove A, Hooley R. Gibberellin and abscisic acid signalling in aleurone. Trends Plant Sci. 2000;5:102–110. doi: 10.1016/s1360-1385(00)01571-5. [DOI] [PubMed] [Google Scholar]
- Marga F, Grandbois M, Cosgrove DJ, Baskin TI. Cell wall extension results in the coordinate separation of parallel microfibrils: evidence from scanning electron microscopy and atomic force microscopy. Plant J. 2005;43:181–190. doi: 10.1111/j.1365-313X.2005.02447.x. [DOI] [PubMed] [Google Scholar]
- Merchan F, de Lorenzo L, Gonzalez Rizzo S, Niebel A, Manyani H, Frugier F, Sousa C, Crespi M. Identification of regulatory pathways involved in the reacquisition of root growth after salt stress in Medicago truncatula. Plant J. 2007;51:1–17. doi: 10.1111/j.1365-313X.2007.03117.x. [DOI] [PubMed] [Google Scholar]
- Milioni D, Sado PE, Stacey NJ, Domingo C, Roberts K, McCann MC. Differential expression of cell-wall-related genes during the formation of tracheary elements in the Zinnia mesophyll cell system. Plant Mol. Biol. 2001;47:221–238. [PubMed] [Google Scholar]
- Pritchard SL, Charlton WL, Baker A, Graham IA. Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J. 2002;31:639–647. doi: 10.1046/j.1365-313x.2002.01376.x. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. The expansin superfamily. Genome Biol. 2005;6:242. doi: 10.1186/gb-2005-6-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandberg G. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell. 2004;16:2278–2292. doi: 10.1105/tpc.104.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicky ST, Van S, Herman S, Yves G, Jean-Pierre V, Kris V. XET activity is found near sites of growth and cell elongation in bryophytes and some green algae: new insights into the evolution of primary cell wall elongation. Ann. Bot. (Lond.) 2007;99:39–51. doi: 10.1093/aob/mcl232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Fry SC, Pauly M, Hofte H, Verbelen JP. XTH acts at the microfibril–matrix interface during cell elongation. J. Exp. Bot. 2005a;56:673–683. doi: 10.1093/jxb/eri048. [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Oyama M, Osato Y, Yokoyama R, Verbelen JP, Nishitani K. Differential expression of AtXTH17, AtXTH18, AtXTH19 and AtXTH20 genes in Arabidopsis roots. Physiological roles in specification in cell wall construction. Plant Cell Physiol. 2005b;46:192–200. doi: 10.1093/pcp/pci013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.