Abstract
The genome of Arabidopsis thaliana encodes more than 600 receptor-like kinase (RLK) genes, by far the dominant class of receptors found in land plants. Although similar to the mammalian receptor tyrosine kinases, plant RLKs are serine/threonine kinases that represent a novel signaling innovation unique to plants and, consequently, an excellent opportunity to understand how extracellular signaling evolved and functions in plants as opposed to animals. RLKs are predicted to be major components of the signaling pathways that allow plants to respond to environmental and developmental conditions. However, breakthroughs in identifying these processes have been limited to only a handful of individual RLKs. Here, we used a Syngenta custom Arabidopsis GeneChip array to compile a detailed profile of the transcriptional activity of 604 receptor-like kinase genes after exposure to a cross-section of known signaling factors in plants, including abiotic stresses, biotic stresses, and hormones. In the 68 experiments comprising the study, we found that 582 of the 604 RLK genes displayed a two-fold or greater change in expression to at least one of 12 types of treatments, thereby providing a large body of experimental evidence for targeted functional screens of individual RLK genes. We investigated whether particular subfamilies of RLK genes are responsive to specific types of signals and found that each subfamily displayed broad ranges of expression, as opposed to being targeted towards particular signal classes. Finally, by analyzing the divergence of sequence and gene expression among the RLK subfamilies, we present evidence as to the functional basis for the expansion of the RLKs and how this expansion may have affected conservation and divergences in their function. Taken as a whole, our study represents a preliminary, working model of processes and interactions in which the members of the RLK gene family may be involved, where such information has remained elusive for so many of its members.
Keywords: Abiotic/environmental stress, hormone biology, receptors, transcriptome analysis, disease responses, Arabidopsis
INTRODUCTION
The emergence of multicellularity in eukaryotes represents a sophisticated achievement in evolutionary history. Although its exact origins remain obscure, multicellularity appears to have evolved independently in a variety of eukaryotic phyla, perhaps many times (Kaiser, 2001; Bonner, 1998; King, 2004). Despite these broad beginnings, a unifying requirement of multicellularity is the capacity for individual cells to sense environmental and developmental cues in order to coordinate their behavior with neighboring cells (Pires-daSilva and Sommer, 2003). Not surprisingly, the genomes of multicellular organisms encode a number of molecular mechanisms that confer such sensory capacity during environmental and developmental responses.
In particular, a number of classes of cell-surface receptors have evolved that service the extracellular signaling needs of multicellular organisms (Ben-Shlomo et al., 2003). These classes include seven-transmembrane receptors, such as the G-protein-coupled receptors (GPCRs); enzyme-linked receptors, such as the receptor tyrosine kinases (RTKs) and receptor serine/threonine kinases (RS/TKs); ion-channel-linked receptors; and two-component histidine kinase receptors. Of these classes, the ion-channel-linked receptors and two-component receptors appear in both prokaryotic and eukaryotic lineages, suggesting that these forms of signaling are of truly ancient origin (Koishi et al., 2004; Hoch, 2000; Stock et al., 2000).
However, a survey of eukaryotic-specific receptors found in plants reveals an interesting divergence from the animal kingdom in terms of receptor evolution. One surprising result from the analysis of the complete genome sequence of Arabidopsis thaliana is the apparent absence of receptor tyrosine kinases and the presence of only two putative GPCRs (Meyerowitz, 2002; Perfus-Barbeoch et al., 2004; Liu et al., 2007). Instead, the Arabidopsis genome contains an expansive set of receptor serine/threonine kinases, known as receptor-like kinases (RLKs), which are unique to the plant kingdom (Shiu and Bleecker, 2001a; Shiu et al., 2004). RLKs comprise a more than 600-member gene family in Arabidopsis, representing approximately 2% of the predicted protein coding genome. Like the animal RTKs and RS/TKs, the majority of Arabidopsis RLKs appear to consist of an intracellular kinase domain, a single-pass transmembrane helix, and an extracellular region containing signaling-related sequence motifs (Cock et al., 2002). However, phylogenetic analyses support their classification as a monophyletic group that evolved separately from animal receptor kinases (Shiu and Bleecker, 2001a).
Current experimental results from a variety of plant species indicate that genes encoding RLKs are involved in a range of environmental and developmental responses, including recognition of pathogens (Xa21, FLS2, EFR) (Song et al., 1995; Gomez-Gomez and Boller, 2000; Zipfel et al., 2006), symbionts (NORK, SYMRK, NFR1, NFR5) (Endre et al., 2002; Stracke et al., 2002; Madsen et al., 2003; Radutoiu et al., 2003), and self (SRK) (Stein et al., 1991; Kachroo et al., 2001); perception of the plant hormones brassinosteroid (BRI1, BAK1/AtSERK3, BRL1, BRL3) (Li and Chory, 1997; Wang et al., 2001; Kinoshita et al., 2005; Nam and Li, 2002; Li et al., 2002; Zhou et al., 2004; Cano-Delgado et al., 2004) and phytosulfokine (PSKR) (Matsubayashi et al., 2002), as well as abscisic acid responsiveness (RPK1) (Osakabe et al., 2005); cell proliferation (ER, ERL1, ERL2) (Torii et al., 1996; Shpak et al., 2004); cell fate (CR4, EXS/EMS1, SCM/SUB, SERK1, SERK2) (Becraft et al., 1996; Canales et al., 2002; Zhao et al., 2002; Kwak et al., 2005; Chevalier et al., 2005; Colcombet et al., 2005; Albrecht et al., 2005); cell expansion (WAK2, WAK4) (Lally et al., 2001; Wagner and Kohorn, 2001); meristem regulation (CLV1, BAM1-3, FON1, TD1) (Clark et al., 1997; DeYoung et al., 2006; Suzaki et al., 2004; Bommert et al., 2005); vascular development (PXY) (Fisher and Turner, 2007); and, root and nodule growth (HAR1, NARK, SUNN) (Nishimura et al., 2002; Searle et al., 2003; Schnabel et al., 2005).
Collectively, RLKs represent a novel signaling innovation in plants and, consequently, an excellent opportunity to understand how extracellular signaling evolved and functions in plants as opposed to animals. However, many basic questions regarding their evolution and function remain unresolved. For example, the sheer number of Arabidopsis RLKs and the breadth of motifs found in their extracellular domains suggest their involvement in a number of physiological processes. But, the nature and range of these processes have yet to be identified for all but a handful of the more than 600 Arabidopsis RLKs. Furthermore, phylogenetic analysis of the Arabidopsis RLKs supports their classification into 43 subfamilies, which correspond to the type and organization of motifs found in their extracellular domains. For this reason, it has been hypothesized that members of a given subfamily may respond to similar types of signals. But, this hypothesis has yet to be tested. Finally, the functional implications of the evolutionary expansion of RLKs in the Arabidopsis genome remain unclear. The large sizes of the RLK subfamilies may indicate functional redundancies among its members. On the other hand, this phenomenon may have allowed for functional divergences to have evolved among its members, thereby allowing for an expanded functional range of the RLKs as a whole.
To address these issues, we performed a systematic, large-scale analysis of the transcriptional response of the Arabidopsis RLK gene family to a host of known environmental and developmental stimuli in plants. Here, we report on the conditions under which RLK gene expression is modulated, both on an individual gene level and on the subfamily level. We also profile similarities and divergences in gene expression among the individual RLK genes, as well as within the framework of the RLK subfamilies. Finally, we present a comparative analysis of the associations between gene expression patterns and sequence distances and divergence rates in an attempt to understand the evolutionary basis for the expansion and retention of the RLK superfamily.
RESULTS
RLK Genes Respond to a Broad Set of Common Plant Signaling Factors
As sessile, autotrophic organisms, plants are particularly reliant on environmental factors to guide their physiological and developmental programs. A number of studies have embraced the use of expression profiling to elucidate the molecular components involved in the signaling pathways that coordinate such programs (Nemhauser et al., 2006; Denby and Gehring, 2005; Takahashi et al., 2004; Hazen et al., 2003; Rabbani et al., 2003; Chen et al., 2002; Cheong et al., 2002; Kreps et al., 2002). Additionally, studies of several of the Arabidopsis RLKs have revealed gene expression changes, as well as the emergence of RLK mutant phenotypes, under specific stress elicitors and hormones (Shiu and Bleecker, 2001b; Osakabe et al., 2005; Verica et al., 2003; Sivaguru et al., 2003; He et al., 1998). However, a comprehensive profile of the transcriptional response of Arabidopsis receptor-like kinase genes to known signaling factors had yet to be produced.
For this reason, we addressed whether, and to what extent, Arabidopsis RLK genes are responsive to a host of well studied environmental and developmental signaling factors in plants. Specifically, we used the Syngenta custom Arabidopsis GeneChip array (sySYNG002a, Affymetrix, Santa Clara, California, USA) to assay changes in RLK transcript levels after exposure to 12 categories of treatments, representing three basic, though complex, classes of plant physiological responses: abiotic stress, biotic stress, and hormonal/chemical. Collectively, 102 arrays representing 68 experiments were analyzed. The 12 specific treatments included cold (4°C), gravity (vertical rotation), high light (1500 μmol m−2 s−1), osmotic stress (200 mM mannitol), salt (100 mM NaCl), bacterial pathogens (Pseudomonas syringae strains DC3000 and ES4326), fungal pathogens (Botrytis cinerea, Erysiphe, Peronospora parasitica, Phytophthora porri), viral pathogens (beet curly top hybrigeminivirus, cauliflower mosaic virus, cucumber mosaic virus, oilseed rape mosaic virus, tobacco rattle virus, turnip mosaic virus, turnip vein-clearing virus), abscisic acid (1 μM, 10 μM), glucose (0.3%, 3.0%, 6.0%), auxin (0.1 μM), and jasmonic acid (45 and 50 μM). Given that stress, disease, and hormones have both immediate and long-term effects on gene expression, many of the treatments in this analysis represent data generated from more than one time point, in an attempt to capture a larger portion of the range of responses (Nemhauser et al., 2006; Kreps et al., 2002). These treatments include cold, high light, osmotic stress, salt, auxin, jasmonic acid, and most of the pathogens. However, data from some treatments (glucose, abscisic acid) represent changes seen at single time points under differing concentrations. Supplemental Table 1 contains a description of the individual experiments.
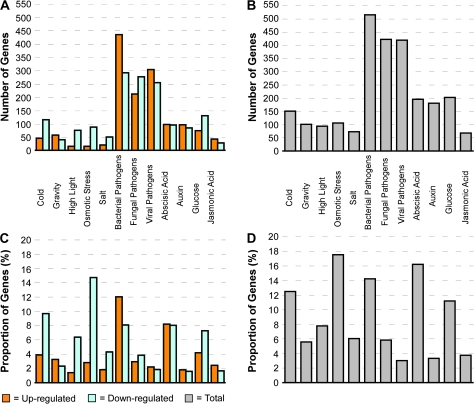
Based on our analysis, we identified responses equal to or greater than a two-fold difference in expression from 582 of the 604 Arabidopsis RLK genes in at least one of the 12 treatments in our study (Figure 1A). Interestingly, of the 12 treatment groups, the bacterial, fungal, and viral pathogens elicited the largest proportion of differentially expressed RLK genes (60%), compared to the abiotic stresses (18%) and hormone/chemical treatments (22%) (Figure 1B). The hormone and chemical treatments also had a relatively large number of responses, in comparison with the abiotic stresses. In some treatment categories, individual RLK genes registered an up-regulated response in one experiment and a down-regulated response in another. In the case of time-course-based experiments, this result would suggest the temporal nature of the transcriptional activity of the given RLK gene. However, in experiments in which the nature of the treatment differed, such as with the pathogens, the diverse transcriptional response would indicate a disparate role in signaling activities related to the different treatments comprising that category. As a result of these dynamic responses, the total of number of respondents for a given treatment in some cases was smaller than the summation of up-regulated and down-regulated genes observed for that category of treatment.
Figure 1.
Distribution of Differentially Expressed RLK Genes per Treatment Type.
Genes were classified as differentially expressed based on a two-fold-change or greater threshold.
(A) Total number of up-regulated and down-regulated RLK genes by treatment.
(B) Total number of differentially expressed genes by treatment.
(C) Proportion of up and down-regulated RLK genes by treatment. Proportions were calculated by scaling the total number of up-regulated or down-regulated RLK genes per treatment by the total number of RLK genes and the number of experiments used in the given treatment: number up/down-regulated genes/(number of all RLK genes × number experiments).
(D) Proportion of differentially expressed genes by treatment: number DE genes/(number of all RLK genes × number experiments).
Though it is tempting to speculate that the large number of pathogenesis-related responses may be due to an evolutionary process, the response proportions may be reflective of the proportion of experiments performed per treatment class: 61% biotic stresses, 14% abiotic stresses, and 25% hormone/chemical. To better understand this distribution, we also calculated the number of differentially expressed RLK genes per treatment, scaled by the number of experiments used in each treatment and the number of genes analyzed (Figure 1C and 1D). Based on this analysis, relatively large numbers of differentially expressed RLK genes were registered for the osmotic stress, abscisic acid, bacterial pathogen, cold, and glucose treatments. Table 1 lists the RLK genes with the top increased and decreased expression levels per treatment.
Table 1.
Top Up-Regulated and Down-Regulated RLK Genes per Treatment.
| Treadment | Accession | Subfamily | Signal log Radio | Treatment | Accession | Subfamilly | Signal log Radio |
| Abscisic Acid | AT3G05140 | RLCK VI | 2.99 | Auxin | AT1G21250 | WAKL | 4.00 |
| AT3G47580 | LRR XII | 2.90 | AT2G19190 | LRR I | 2.99 | ||
| AT3G68420 | CR4L | 2.64 | AT2G13800 | LRRII | 2.83 | ||
| AT3G51990 | CR4L | 2.45 | AT1G21240 | WAKL | 2.57 | ||
| AT2G25220 | RLCK X | 2.29 | AT4G23180 | DUF26 | 2.48 | ||
| AT4G02410 | L-LECTIN | -3.75 | AT3G51740 | LRR III | -2.61 | ||
| AT4G11460 | L-LECTIN | -3.67 | AT3G45430 | L-Lectin | -2.07 | ||
| AT4G11460 | DUF26 | -3.20 | AT5GD1060 | RLCK II | -2.03 | ||
| AT1G15110 | WAKL | -2.95 | AT4G32000 | RLCK | -2.00 | ||
| AT5G02070 | WAKL | -2.69 | AT5G54380 | CrRLK1L-1 | -1.99 | ||
| Bacterial | AT1G79680 | WAKL | 5.99 | Cold | AT3G59350 | RLCK VIII | 4.50 |
| AT5G25930 | LRR XI | 5.18 | AT5G61350 | CrRLK1L-1 | 3.20 | ||
| AT4G25390 | RLCK XI | 4.91 | AT5G60900 | SD-2 | 2.59 | ||
| AT5G46080 | UC | 4.69 | AT4G27290 | SD-1 | 2.34 | ||
| AT1G51370 | SD 1 | 4.11 | AT5G66790 | WALK | 2.19 | ||
| AT4G39400 | LRR X | -4.11 | AT5G03140 | L-Lectin | -3.00 | ||
| AT2G30740 | RLCK VIII | -2.89 | AT4G11460 | DUF26 | -2.70 | ||
| AT5G51770 | RLCK XI | -2.88 | AT4G04960 | L-Lectin | -2.63 | ||
| AT4G08850 | LRR XII | -2.40 | AT1G79670 | WAKL | -2.61 | ||
| AT3G03770 | LRR VI | -2.34 | AT4G11500 | DUF26 | -2.59 | ||
| Gravity | AT5G03140 | L-LECTIN | 1.93 | High Light | AT1G21250 | WAKL | 2.02 |
| AT1G79680 | WAKL | 1.84 | AT1G51550 | SD-1 | 1.45 | ||
| AT1G28390 | CR4L | 1.73 | AT1GD7560 | LRR I | 1.41 | ||
| AT1G51620 | LRR I | 1.71 | AT4G00960 | DUF26 | 1.38 | ||
| AT3G53840 | WAKL | 1.64 | AT5G81350 | CrRLK1L-1 | 1.35 | ||
| AT1G55610 | LRR X | -2.05 | AT3G47580 | LRR XII | -2.99 | ||
| AT3G28450 | LRR X | -1.96 | AT5GD3140 | L-Lectin | -1.98 | ||
| AT1G25390 | LRK10L-1 | -1.96 | AT1G56720 | TAXL | -1.93 | ||
| AT2G41890 | SD-3 | -1.92 | AT1G51940 | LysM | -1.91 | ||
| AT4G11890 | DUF26 | -1.71 | AT5G37790 | RLCK VI | -1.85 | ||
| Salt Strees | AT5G53890 | LRR X | 2.49 | Viral | AT4G39110 | CrRLK1L-1 | 4.99 |
| AT4G02420 | L-Lectin | 1.48 | AT1G76370 | RLCK VII | 4.85 | ||
| AT5G35960 | RLCK VI | 1.47 | AT3G02810 | RLCK VII | 4.78 | ||
| AT1G15120 | WAKL | 1.38 | AT2G07040 | LRR III | 4.70 | ||
| AT3G08870 | L-Lectin | 1.25 | AT4G11890 | DUF26 | 4.31 | ||
| AT3G57710 | RLCK III | -2.02 | AT1G65800 | SD-1 | -3.36 | ||
| AT4G11460 | DUF26 | -1.91 | AT2G05940 | RLCK VII | -3.30 | ||
| AT1G11330 | SD-1 | -1.75 | AT4G23290 | DUF26 | -3.15 | ||
| AT1G17540 | RLCK IX | -1.70 | AT4G23300 | DUF25 | -2.70 | ||
| AT1G51850 | LRR I | -1.69 | AT4G23180 | DUF25 | -2.70 | ||
| Furgal | AT5G10250 | RLCK VI | 4.24 | Glucose | AT3G24240 | LRR XI | 3.14 |
| AT1G79680 | WAKL | 4.06 | AT3G58690 | Extensin | 2.95 | ||
| AT4G21390 | SD-1 | 3.74 | AT5G20480 | LRR XII | 2.73 | ||
| AT4G04490 | DUF26 | 3.66 | AT2G33580 | LysM | 2.67 | ||
| AT1G81370 | SD-1 | 3.59 | AT5G47070 | RLCK VII | 2.54 | ||
| AT3G49670 | LRRXI | -4.01 | AT3G49570 | LRR XI | -4.49 | ||
| AT5G67280 | LRR III | -2.68 | AT3G28040 | LRR VII | -4.09 | ||
| AT1G30570 | CrRLK1L-1 | -2.58 | AT1G48480 | LRR III | -3.08 | ||
| AT1G28440 | LRR XI | -2.34 | AT1G28440 | LRR XI | -2.93 | ||
| AT5G16590 | LRR III | -2.31 | AT4G20270 | LRR XI | -2.90 | ||
| Jasmonic Acid | AT3G05140 | RLCK VI | 2.21 | Osmotic Strees | AT4G27300 | SD-1 | 3.82 |
| AT3G47580 | LRR XII | 2.18 | AT1G51850 | LRR I | 3.80 | ||
| AT3G59420 | CR4L | 1.87 | AT1G51800 | LRR I | 3.28 | ||
| AT3G51990 | CR4L | 1.77 | AT4G23290 | DUF26 | 2.56 | ||
| AT2G25220 | RLCK X | 1.73 | AT4G11460 | DUF26 | 1.94 | ||
| AT4G02410 | L-Lectin | -1.93 | AT4G11890 | DUF26 | -3.67 | ||
| AT4G28350 | L-Lectin | -1.83 | AT5G24080 | SD-2 | -3.62 | ||
| AT4G11460 | DUF26 | -1.53 | AT1G69790 | RLCK VII | -2.85 | ||
| AT1G15110 | WAKL | -1.47 | AT3G59350 | RLCK VIII | -2.73 | ||
| AT5G02070 | WAKL | -1.44 | AT1G35710 | LRR XII | -2.46 |
The signal log2 ratio represents a log2 transformation of the fold-change difference between treated and control signal readings exhibited by the designated gene under a given treatment category. Each treatment category comprised multiple experiments, with the exception of the osmotic stress assay. The top respondents listed reflect the highest signal log2 ratios registered by any experiment within a given treatment category.
The results of the absolute and scaled analyses indicate that a large number of RLK genes are responsive to known signaling factors, and that the responses cover a range of plant physiological programs, including the response to abiotic and biotic stresses, as well as hormone and chemical-related processes. Given that mutant phenotypes in Arabidopsis often present themselves only under the appropriate, functionally relevant screening conditions, the specific treatments and RLK gene responses identified here serve as a resource for the targeted screening of individual RLK genetic mutants. The full list of differentially expressed RLK genes for all 68 experiments can be viewed in Supplemental Table 2. For a detailed description of the data analysis, see Methods.
Individual RLK Genes Respond to Multiple Treatments
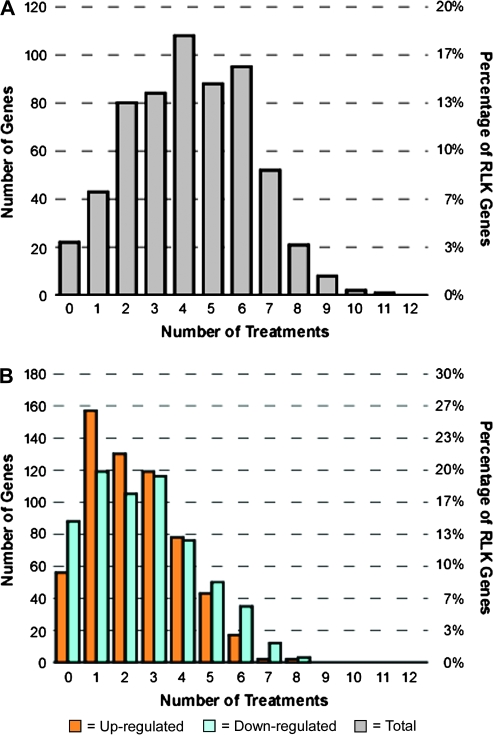
Previously, it had been discovered that individual RLKs might be responsive to more than one signaling factor and thereby participate in multiple signaling pathways (Johnson and Ingram, 2005; Morris and Walker, 2003; Montoya et al., 2002). To investigate this phenomenon, we calculated the number of treatments to which individual RLK genes were differentially expressed, as well as the overlap in responses among the different conditions. First, we looked at the distribution of the number of overall responses registered by individual RLK genes across the 12 treatments (Figure 2A). Of the 604 RLK genes, 22 (4%) displayed no response to any of the experimental conditions tested, and 43 (7%) displayed an increase or decrease in expression to exactly one condition. The remaining number of respondents displayed an increase or decrease in expression to more than one condition. We also found a large number of genes that showed an increase or decrease in expression to more than one condition when looking at up-regulated responses and down-regulated responses separately (Figure 2B).
Figure 2.
Distribution of Number of Treatments in which a Given RLK Gene Is Differentially Expressed.
An RLK gene is said to be differentially expressed for a given treatment if it is differentially expressed at the two-fold-change or greater level in at least one experiment for this treatment. Note that experiments within a treatment tested for responses to a range of treatment-related conditions and are not experimental replicates.
(A) Distribution of total number of treatments in which a given RLK gene is differentially expressed. Twenty-two (4%) of the RLK genes did not exhibit a change in expression level to any treatment while 43 RLK genes (7%) responded to exactly one treatment.
(B) Distribution of the number of treatments in which a given RLK gene is differentially expressed separated into up-regulated and down-regulated responses.
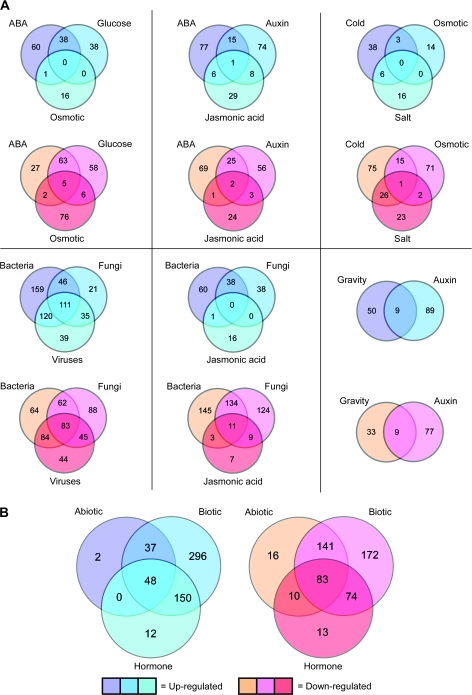
These results support the idea that single RLK genes may be involved in several physiological processes or in a common downstream process initiated by more than one condition. To identify those conditions under which individual RLK genes display multiple responses, we performed a Venn diagram-based analysis. Specifically, we looked for relationships among sets of responding RLK genes under conditions that are known to produce related physiological responses in plants (Figure 3A). In looking at groups of three conditions, we detected only small overlaps among the abiotic and hormone/chemical groupings. For example, abscisic acid, glucose, and osmotic stress had no overlapping respondents with increased expression and five shared respondents (1%) among those with decreased expression levels. Similarly, among the hormones abscisic acid, auxin, and jasmonic acid, one respondent (0.5%) had increased expression levels to all three treatments, and two (1%) had decreased expression. Cold, salt, and osmotic stress had no overlapping respondents with increased expression, and only one (0.5%) with decreased expression. However, the bacterial, fungal, and viral pathogens showed larger proportions of overlap with 111 (21%) shared respondents with increased expression levels and 83 (18%) with decreased levels.
Figure 3.
Comparison of Treatment Types with Overlapping Sets of Differentially Expressed RLK Genes.
(A) Distribution of overlaps in physiologically related treatment types.
(B) Distribution of overlaps according to the three major treatment classes: abiotic stress, biotic stress, and hormone/chemical.
Comparisons of sets of two treatments that are known to induce related physiological processes revealed more overlaps in RLK gene responses. We found that of the RLK genes with increased expression levels to abscisic acid and glucose, 38 (28%) of them responded to both. For those RLK genes with decreased expression levels to either glucose or abscisic acid, 68 (42%) displayed decreased levels for both. Gravity and auxin had nine (6%) shared respondents with increased expression, and nine (8%) with decreased expression. Abscisic acid and osmotic stress are also known to have cooperative effects in plant development (Zhu, 2002). Interestingly, we found that only one of the RLK genes (0.9%) displayed increased expression levels under both osmotic stress and abscisic acid. For genes with decreased expression levels to either abscisic acid or osmotic stress, seven (6%) had responses to both conditions. Plant responses to osmotic stress, salt stress, and cold are known to be intricately related (Mahajan and Tuteja, 2005). Surprisingly, osmotic stress and salt stress had no overlapping increased response, and an overlap of three (1%) RLK genes with decreased expression levels. Salt stress and cold had more shared responses, with six (10%) RLK genes with increased expression levels and 27 (19%) with decreased levels. Cold and osmotic stress had three shared respondents (5%) with increased expression, and 16 (8%) with decreased expression. Given the cross-talk that can occur in the signaling pathways involving abscisic acid, salt, and osmotic stress, the little overlap seen here may indicate that these genes are partitioned into condition-specific responses outside of those cross-talk networks. For example, it is known that the plant response to osmotic stress involves multiple signaling pathways, some of which are abscisic acid-dependent and some of which are abscisic acid-independent (Zhu, 2002). However, it also possible that temporal and tissue-specific differences may be influencing these results.
Among the biotic stress-related treatments, bacterial and viral pathogens shared more respondents than either bacterial–fungal pathogens or viral–fungal pathogens. The bacterial and viral pathogens had 231 (45%) shared respondents with increased expression levels, and 167 (44%) with decreased expression. In contrast, bacterial and fungal pathogens had 157 (32%) and 145 (34%) with increased and decreased expression levels, respectively, while fungal and viral pathogens had 146 (39%) increased and 128 (32%) decreased. Plant resistance to fungal pathogens is known to be related to jasmonic acid signaling (Kunkel and Brooks, 2002). Here, we found that fungal and jasmonic acid treatments produced 19 shared respondents (8%) with increased expression levels, and 20 shared respondents (7%) with decreased expression.
Overall, in looking at the three major classes of treatments—abiotic stress, biotic stress, and hormones/chemicals—we found that 48 RLK genes (9%) with increased expression levels and 83 (16%) with decreased expression levels were shared among the three (Figure 3B). While the results presented here have highlighted overlaps seen in responses to related treatments, it should be noted that this analysis additionally reveals those groups of RLK genes with distinct responses to specific treatments. Supplemental Table 3 lists all up-regulated and down-regulated RLK genes per treatment.
RLK Subfamilies Display Wide Signal Coverage
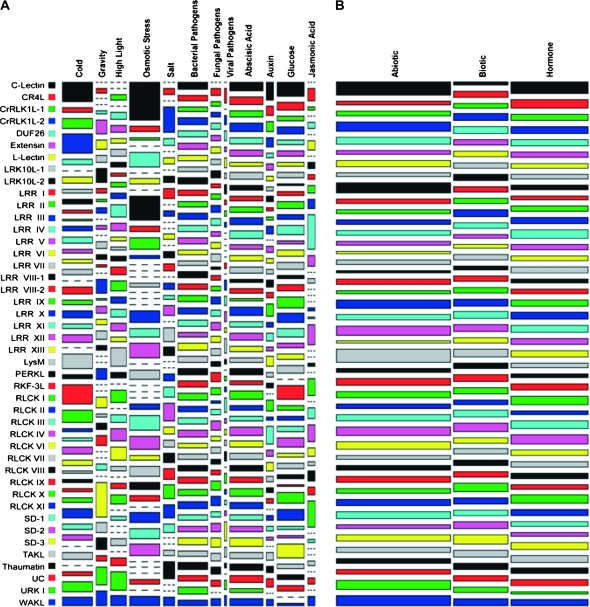
Molecular phylogenetic analysis of the amino acid sequences encoding the Arabidopsis RLK kinase domains has led to the classification of 43 RLK subfamilies plus one grouping of unclassified RLKs (UC), with most subfamilies also bearing distinct types and organization of sequence motifs in their extracellular domains (Shiu and Bleecker, 2001a). An appealing hypothesis is that each subfamily may service a specific, related set of physiological responses in plants (Shiu and Bleecker, 2001b). To investigate this question, we grouped the results of the expression analysis by subfamily and compared them across treatments (Figure 4A). Note that we also counted the responses found among the unclassified RLKs.
Figure 4.
Differentially Expressed RLK Genes by Subfamily and Treatment.
Area of tile indicates joint distribution of differentially expressed genes by subfamily and treatment. Tile width represents marginal distribution of differentially expressed genes per treatment or treatment class. Tile length reflects marginal distribution of differentially expressed genes per subfamily.
(A) Proportion of differentially expressed genes per subfamily and treatment. Tile area calculated by taking the number of differentially expressed genes for a given subfamily under a given treatment and scaling by the subfamily size and number of experiments within the treatment category: number DE genes/(number gene members × number experiments).
(B) Proportion of differentially expressed RLK genes per subfamily and major class of treatment. Tile area calculated by taking the number of differentially expressed genes exhibited by a subfamily and scaling by the subfamily size and number of experiments falling under the major treatment class of abiotic stress, biotic stress, or hormone/chemical: number DE genes/(number gene members × number experiments in treatment class).
The mosaic plot depicted in Figure 4A is based on the number of differentially expressed RLK genes for a given subfamily under the specified treatment, scaled by the number of genes in the subfamily and the number of experiments related to the treatment. The area of each tile in the plot represents the joint distribution of differentially expressed genes for a given combination of subfamily and treatment. The lengths of each tile indicate the marginal distribution of differentially expressed genes per subfamily (vertical length) or treatment (horizontal length). Here, we find that the bacterial pathogen and abscisic acid treatments induced responses across all subfamilies. To a lesser extent, the fungal and viral pathogens, as well as glucose, had near widespread effects. It is also interesting to note the amount of distortion in the tiles for the abiotic stress-related treatments and for jasmonic acid, which suggests associations between subfamilies and these treatments in terms of the number of differentially expressed members.
Figure 4B depicts the proportion of each subfamily's total number of responses according to the three major classes of treatments: abiotic stress, biotic stress, and hormones/chemicals. The plot in Figure 4B reveals that, in general, a slightly larger proportion of each subfamily's responses are found under abiotic stress and hormone/chemical treatments, with a smaller proportion induced by the biotic stress conditions. However, for certain subfamilies, such as PERKL, SD-2, and Thaumatin, the effects were more evenly distributed.
In absolute terms, the number of responses varied for each subfamily, with the DUF26 and LRR III subfamilies each registering the largest number at 207, while the C-Lectin and SD-3 subfamilies each had the smallest number at six. We found that overall, every RLK subfamily displayed responses to a broad range of treatments, with the smallest number of treatments being five (the RKF3L subfamily) and 13 of the subfamilies showing at least one differentially expressed member to all 12 of the treatments. Based on these results, it is clear that each RLK subfamily contains members that are responsive to different types of signals, as opposed to a specific type of treatment.
RLK Subfamilies Exhibit Different Levels of Divergence in their Expression Patterns
In the previous analysis, we found that each RLK subfamily had members that were differentially expressed under different types of treatments. Next, we investigated the extent to which expression profiles diverge or overlap among the RLK genes, particularly within the 43 subfamilies. Dissimilar expression patterns may be indicative of functional divergences among closely related RLK members, while similar expression patterns may indicate the presence of functional overlaps. Such redundancies have been reported for several of the RLKs (Shiu and Bleecker, 2001b; Johnson and Ingram, 2005; Dievart et al., 2003). For example, three members of the LRR XIII subfamily have been shown to have functional overlaps through the analysis of a triple mutant (Shpak et al., 2004). The presence of such overlaps was also offered as a plausible explanation for the difficulty often encountered in generating identifiable phenotypes in single loss-of-function RLK mutants.
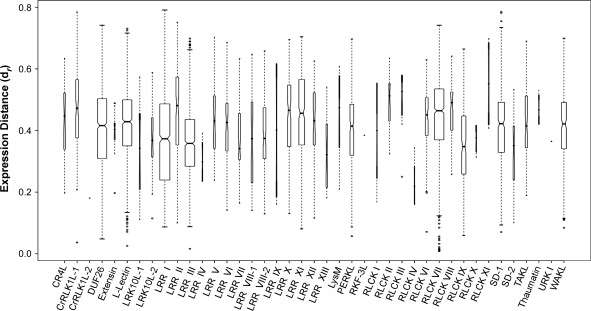
To analyze whether expression patterns appear to be conserved or divergent within each subfamily, we measured the distance between RLK gene expression profiles across the arrays used in our study in pairwise fashion, using a metric based on the Pearson correlation coefficient (r) (see Methods). The final gene expression distance for a subfamily represents the median of the pairwise distances (dr) among all of its members, using a normalized scale of 0 to 1, with 1 being most distant. Based on this analysis, we found that the level of expression divergence varied widely among the RLK subfamilies (Figure 5). The largest divergence, seen in the RLCK XI subfamily (median dr = 0.55), was approximately three times that of the smallest divergence, registered by the CrRLK1L-2 subfamily (median dr = 0.18). Note that these results apply only to those subfamilies with two or more members and therefore exlude the C-Lectin and SD-3 subfamilies.
Figure 5.
Pairwise Gene Expression Distances (dr) by Subfamily.
Expression distances were calculated based on the Pearson correlation coefficient (r) for each pairwise comparison of expression vectors using the maximum r value found across all subfamilies as a normalization factor: dr = 1 – (rij/max(r)), where i and j represent the expression vectors being compared. Markers within boxes represent median dr values for each subfamily. Notches along boxes that do not overlap are evidence the medians differ for the given subfamilies. Box widths reflect the number of pairwise calculations performed for a given subfamily. Box lengths represent interquartile ranges. Ranges of each plot cover observations that extend to 1.5 times the interquartile range. Outliers for each subfamily are designated as individual points. Note that the single-member subfamilies, C-Lectin and SD-3, are excluded.
However, we also found that, for the most part, the majority of subfamilies exhibited a broad range of distances among their expression profiles, which we calculated by subtracting the lowest pairwise dr value from the highest for each subfamily. The smallest dr range was 0.10, found in the RLCK X subfamily, and the CrRLK1L-1 subfamily had the largest dr range at 0.75. The average dr range for all subfamilies was 0.48. To ensure that the subfamily expression distances were not an artifact of membership size, we checked the correlation of the median expression distance of each subfamily to its membership size and found no significant correlation (r = 0.05).
We also looked at the expression profile distances among the three LRR XIII subfamily members with known functional overlaps (Shpak et al., 2004). We found that their expression profiles were indeed similar, with pairwise distances of dr = 0.18, dr = 0.21, and dr = 0.32 among the three. In a ranking of all distances generated in our analysis from shortest to longest, these distances fell into the 97th, 96th, and 85th percentiles, respectively. Supplemental Dataset 1 contains a list of all the pairwise distances between subfamily members for each subfamily.
RLK Expression Profiles Transcend Subfamily Classifications
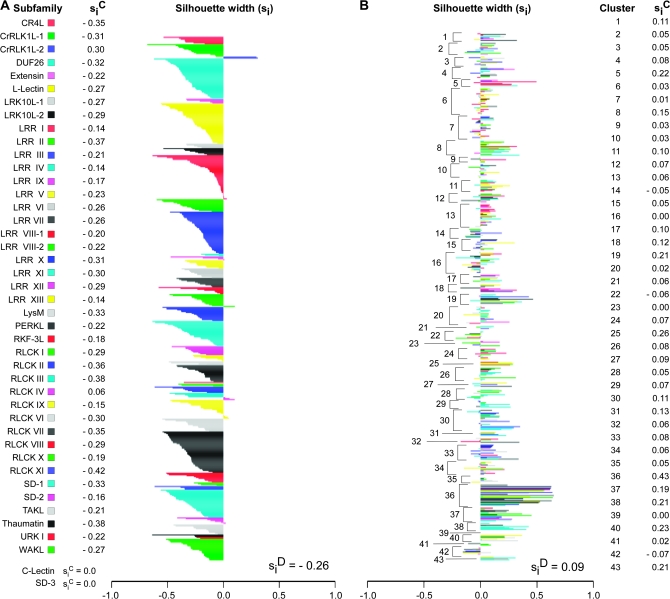
The broad range of distances found among the expression profiles within each subfamily led us to ask whether associations existed between expression profiles of different subfamilies. On a functional level, interactions between different subfamily members have been shown to occur, for instance, with members of the LRR II and LRR X subfamilies (Nam and Li, 2002; Li et al., 2002). To perform this analysis, we used the subfamily classification scheme as an a priori clustering of the expression profiles and calculated the silhouette widths (si) for all members of each subfamily (Figure 6A) (Kaufman and Rousseeuw, 1990). In this case, the silhouette width measures how closely associated an individual expression profile is to those found in its own subfamily, compared to profiles from other subfamilies.
Figure 6.
Clustering and Silhouette Width Analysis of RLK Expression Profiles.
Silhouette plots indicate strength of expression profile cluster structures derived from (A) an a priori clustering based on subfamily classification and (B) Partitioning Around Medoids (PAM) clustering with the number of clusters (K) = 43. Each bar represents the silhouette width of an individual expression profile. Bar widths indicate strength of assignment of a given expression profile to its designated cluster, with larger values representing better classification. Silhouette width values for a cluster (siC) represent how well a cluster is separated from neighboring clusters, with values closer to 1 indicating better separation. The silhouette width value for the entire cluster structure (siD) reflects the strength of support for the overall clustering of the data, with larger values reflecting better support. In (A), subfamilies are color-coded as designated. In (B), expression profiles from different subfamilies within a given cluster are represented by different colors.
Our results show that the majority of RLK genes had associations outside of their own subfamily, as represented by the negative silhouette width scores in Figure 6A. Furthermore, the silhouette width average for the entire a priori clustering was siD = –0.26. These results show that the expression profiles do not cluster well according to subfamily. For the most part, each subfamily had widely disparate expression profiles, so much so that they appear to have associations outside of their subfamily to a greater degree than within their own subfamily. This result is consistent with the notion that the activity of the genes in each subfamily are related to divergent physiological processes, as suggested in Figure 4A and 4B.
Next, we attempted to separate the data into groups of conserved expression profiles, regardless of subfamily membership. To do so, we used a clustering approach called Partitioning Around Medoids (PAM), and calculated the silhouette widths for the resulting cluster structure. PAM clusters data by first identifying the best K medoids in the data, and then assigning all other observations to its nearest medoid (Kaufman and Rousseeuw, 1990). In this case, we used PAM to find the best 43 clusters (K = 43), matching the number of RLK subfamilies minus the unclassified grouping. In the resulting cluster structure, we found no support for the PAM-derived clusters based on an overall silhouette width average of siD = 0.09 (Figure 6B). However, two interesting points emerged from the silhouette plot of the PAM clustering. First, the composition of each cluster was typically a heterogeneous mix of subfamily types. This result suggests the possibility of widespread interactions among different RLK genes. Second, the results show that, for the conditions tested, the responses to a particular treatment may involve the activity of an assortment of RLK genes.
We also tested a range of cluster numbers (K = 2 to K = 50) in our PAM analysis but the results indicated that no ideal number could produce silhouette width measures that strongly supported the clustering (siD > 0.5) (Supplemental Figure 1). Finally, we used the Hierarchical Ordered Partitioning and Collapsing Hybrid (HOPACH) algorithm as a second attempt to cluster the expression profiles but found a similar lack of support for the HOPACH clustering (25 clusters, siD = 0.06, Supplemental Figure 2) (van der Laan and Pollard, 2003).
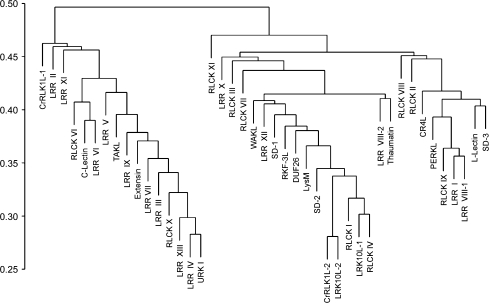
Given that our analysis found a high level of associations between expression profiles from different subfamilies, as seen in Figure 6A, we used hierarchical clustering to view subfamily relationships according to the average expression profile distances among them (Figure 7). The clustering itself was based on average linkage between subfamilies and resulted in a dendogram with three major clusters. Interestingly, nine of the 14 LRR subfamilies are found in the first cluster, and the 10 RLCK subfamilies appear to be distributed across the clusters. Taken together, the clustering and silhouette width analyses reveal a high level of diversity in the RLK transcriptional response, with complex associations found among different groupings of profiles and across RLK subfamilies. Supplemental Dataset 2 contains a list of gene expression distances across subfamilies, as well as a summary of the clustering and silhouette width analyses, including cluster associations, nearest neighboring clusters, and silhouette width measures for each RLK.
Figure 7.
Average Linkage Hierarchical Clustering of RLK Subfamilies Based on Average Linkage Gene Expression Distance.
Pearson correlation coefficient (r) was used as the basis for a distance measure between gene expression profiles. Branches represent the average gene expression profile distance between each clustering of subfamilies. Initial distances between each subfamily were based on the average expression profile distances between all members of each subfamily being compared. The scale represents the expression profile distance calculated as 1 – r, normalized to fit in the 0–1 range.
Opposing Trends in Sequence and Expression Distance among the RLK Subfamilies
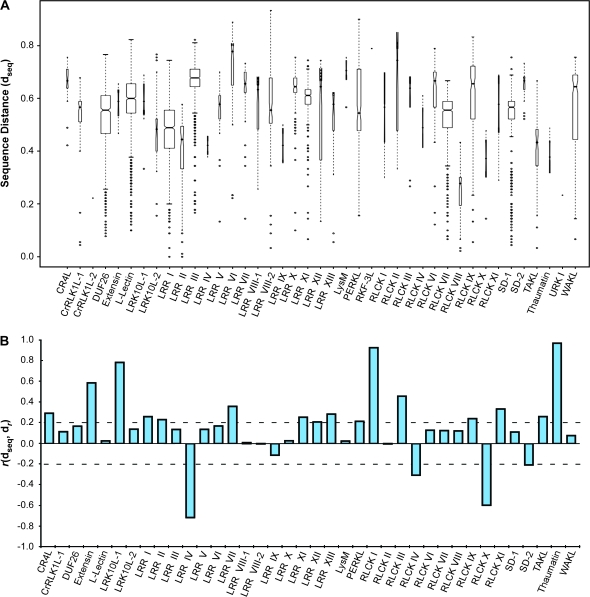
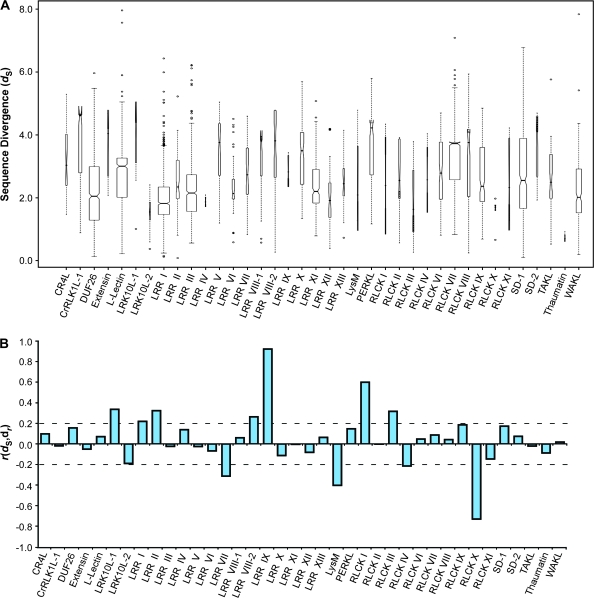
To investigate whether there was a sequence basis for the levels of expression profile divergence seen within each RLK subfamily, we asked whether subfamily members with similar sequences exhibited similar expression profiles, and, conversely, whether less similar sequences had more divergent expression profiles. In order to answer this question, we performed all-against-all pairwise alignments of RLK amino acid sequences, using ClustalW and the Gonnet series of scoring matrices (Thompson et al., 1994; Gonnet et al., 1992). From each pairwise alignment, we calculated a distance based on the normalized alignment score. As with the expression profile distances, the sequence distances (dseq) fall between 0 and 1, with 1 being most distant. Overall, the median sequence distances for all within-subfamily comparisons varied (Figure 8A).
Figure 8.
Correlation between Sequence Distances and Expression Distances by Subfamily.
(A) Pairwise sequence distances by subfamily. Distances between amino acid sequences within a subfamily (dseq) were based on pairwise alignment scores (sij) using the Gonnet substitution matrices, with the maximum score (max(sij)) found across all subfamilies as a normalization factor. Distances were calculated as: dseq = 1 – (sij/max(s)). Boxes represent the interquartile range with medians for each subfamily indicated by marks. Ranges extend to data that fall within 1.5 times the boxed interquartile range, with outliers marked as individual points. Box widths represent the proportion of observations within a given subfamily and notches that do not overlap among subfamilies provide evidence of differences in their medians. Note that the single-member subfamilies C-Lectin and SD-3 are excluded.
(B) Pearson correlation between sequence distance and expression distance (r(dseq, dr)) for each pairwise comparison of subfamily members within each subfamily. Final r value for a subfamily represents the average of all observations within that subfamily. Subfamilies with fewer than three members (C-Lectin, CrRLK1L-2, RKF3L, SD-3, and URK I) were excluded.
Next, for each subfamily, we extracted the sequence and expression profile distances for all within-subfamily pairwise comparisons and calculated the correlation between the two sets of values. We found that of the 38 subfamilies, 20 (53%) exhibited a correlation (r ≥ 0.2 or r ≤ –0.2) between sequence similarity score and expression profile distance (Figure 8B). Note that five subfamilies (C-Lectin, CrRLK1L-2, RKF3L, SD-3, and URK I) had two or fewer members and could not be included in the analysis. In addition, we did not include the unclassified RLKs.
Within these 20 subfamilies, we found two distinct, though vastly different, sets of results. Sixteen of the subfamilies exhibited a positive correlation (r ≥ 0.2), indicating that the closer the sequence similarity among subfamily members, the more conserved their expression profiles. Perhaps more intriguingly, four of the subfamilies displayed a negative correlation (r ≤ –0.2) between sequence and expression. In this case, the more similar the sequences within the subfamily, the more distant their expression profiles. We also checked whether the correlation between sequence distance and expression profile distance was affected by subfamily size and found no such correlation (r = –0.03). In addition, we found no overall correlation between sequence distance and expression profile distance for the RLK superfamily as a whole (r = –0.02).
RLK Subfamilies Demonstrate Signs of Differential Evolution of Sequence and Expression
Based on our analysis of amino acid sequence and expression profile distances, it appeared that different evolutionary forces might be related to the divergence of expression patterns seen within the RLK subfamilies. For this reason, we formally investigated whether a link existed between RLK sequence evolution and expression divergence. Recent studies have paired molecular phylogenetic methods with expression profiling to investigate the evolutionary and functional implications of gene expression divergences (Nuzhdin et al., 2004; Akashi, 2001; Wagner, 2000). In particular, studies have looked at measures of sequence divergence and selective pressure among both orthologous and paralogous genes and compared them to similarities seen in their expression patterns, in order to make initial assessments of functional evolution among the sequences (Good et al., 2006; Oakley et al., 2005; Lemos et al., 2005).
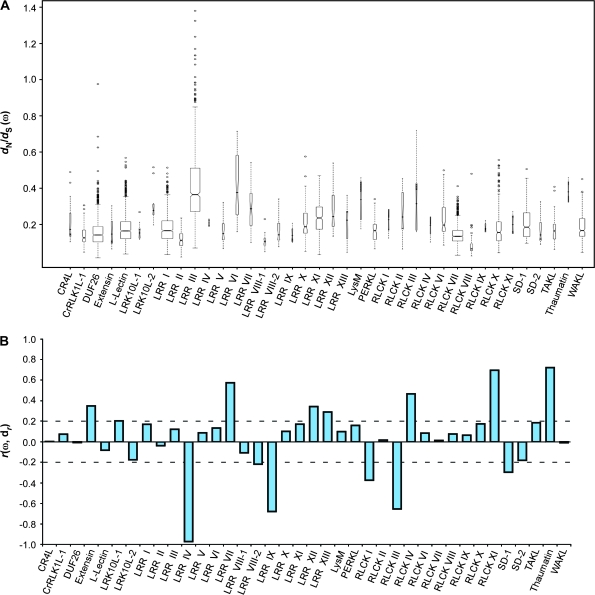
In our case, to discern whether evolutionary events might have had an effect on RLK subfamily expression profiles, we first attempted to establish whether members of the subfamilies displayed signs of selective pressure. To do so, we calculated the ratio of non-synonymous-to-synonymous substitution rates, the so-called dN/dS ratio, among the sequences using the method of Yang and Nielsen (2000). The dN/dS ratio (ω) provides a measure of whether the molecular sequences in question appear to be under negative (purifying) selection or positive (diversifying) selection. For each subfamily, we created multiple amino acid sequence alignments of its members, and used these alignments to guide the alignment of their coding DNA (see Methods). From the aligned DNA sequences, we calculated the dN/dS ratios for all pairwise comparisons within each subfamily using the yn00 program in PAML (Yang, 1997). We then calculated the median dN/dS ratio for each subfamily, as well as the range of all ratios found within each subfamily (Figure 9A).
Figure 9.
Correlation between dN/dS (ω) Ratios and Expression Distances (dr) by Subfamily.
(A) Pairwise dN/dS ratios by subfamily. Ratios of non-synonymous-to-synonymous substitution rates (dN/dS) were calculated based on alignments of coding sequences among all members within each subfamily. Boxes represent the interquartile range with medians for each subfamily indicated by marks. Ranges extend to data that fall within 1.5 times the boxed interquartile range, with outliers marked as individual points. Box widths represent the proportion of observations within a given subfamily and notches that do not overlap among subfamilies provide evidence of differences in their medians. Note that the single-member subfamilies C-Lectin and SD-3 are excluded.
(B) Correlation between dN/dS ratios and expression distances by subfamily. Pairwise dN/dS ratios were normalized using the maximum value found across all subfamilies and correlated with matching pairwise comparisons of expression distances for all members within each subfamily. Final r value for a subfamily represents the average of all observations within the subfamily. Subfamilies with fewer than three members (C-Lectin, CrRLK1L-2, RKF3L, SD-3, and URK I) were excluded.
Based on this analysis, we found that none of the subfamilies as a whole showed signs of positive selection (ωmed > 1). In fact, the median values for all subfamilies were less than ωmed = 0.40, indicating that the majority of the sequences were under varying degrees of negative selection. Interestingly, one of the subfamilies, LRR III, contained 13 members that showed signs of positive selection (ω > 1), although the median ratio for the subfamily was ωmed = 0.37. Similarly, the DUF26 subfamily had a pair of members with a ratio nearing ω = 0.98, where ω = 1 is often interpreted as selectively neutral conditions.
Although the RLK subfamilies appear to have evolved under negative selection, variations in the strength of selection might have had an impact on expression divergence within each subfamily. For this reason, we tested whether the degree of selective pressure as measured by ω had any relationship to the amount of expression divergence seen within each subfamily. To test for this, we calculated the correlation between all pairwise dN/dS ratios and their corresponding pairwise expression profile distances within each subfamily (Figure 9B). Of the 38 subfamilies analyzed, we found that 14 (37%) had correlations of either r ≥ 0.2 or r ≤ –0.2. Eight of the 14 subfamilies (Extensin, LRK10L-1, LRR VII, LRR XII, LRR XIII, RLCK IV, RLCK XI, and Thaumatin) show a positive correlation between dN/dS and their expression divergence. That is, the more relaxed the negative selection, the more divergent their expression profiles. In contrast, the remaining six subfamilies (LRR IV, LRR VIII-2, LRR IX, RLCK I, RLCK III, and SD-1) show a negative correlation in which sequences under greater negative selective pressure had greater divergence in their expression profiles.
Next, we used the synonymous substitution rate (dS) as a measure of divergence time found among RLK subfamily members, and calculated the correlation of this measure with expression divergence (Figure 10A and 10B) (Gu et al., 2002). We found that of the 38 subfamilies, 11 (29%) had correlations of either r ≥ 0.2 or r ≤ –0.2. Seven of the subfamilies (LRK10L-1, LRR I, LRR II, LRR VIII-2, LRR IX, RLCK I, and RLCK III) had a positive correlation between divergence time and expression divergence among its members. Four of the subfamilies (LRR VII, LysM, RLCK IV, and RLCK X) registered a negative correlation between divergence time and expression divergence. We also checked to ensure that subfamily size did not influence the correlation between dN/dS and expression divergence and found no such effect (r = –0.01), and found only a slight association (r = 0.10) between dS and family size.
Figure 10.
Correlation between Estimates of Sequence Divergence (dS) and Expression Distances (dr) by Subfamily.
(A) Pairwise dS measurements by subfamily. Divergences between coding sequences were calculated based on the rates of synonymous substitution found between each pairwise comparison of all members within a given subfamily. Coding sequence alignments were based on a multiple amino acid sequence alignment of subfamily members. Boxes represent the interquartile range with medians for each subfamily indicated by marks. Ranges extend to data that fall within 1.5 times the boxed interquartile range, with outliers marked as individual points. Box widths represent the proportion of observations within a given subfamily and notches that do not overlap among subfamilies provide evidence of differences in their medians. Note that the single-member subfamilies C-Lectin and SD-3 are excluded.
(B) Correlation between sequence divergence and expression profile distance for each RLK subfamily. Pairwise dS measurements were normalized using the maximum value found across all subfamilies. Final r value for a subfamily represents the average of all observations within the subfamily. Subfamilies with fewer than three members (C-Lectin, CrRLK1L-2, RKF3L, SD-3, and URK I) were excluded.
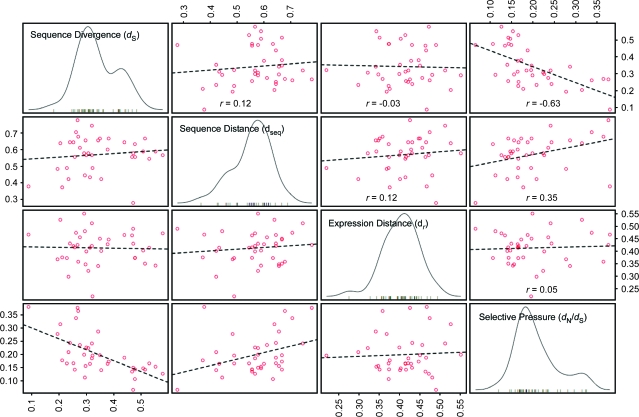
Finally, to look at the general trends in the relationships among divergence time (dS), sequence distance (dseq), expression distance (dr), and selective pressure (dN/dS) at the subfamily level, we plotted the median values for each subfamily in a four-way scatter plot matrix (Figure 11). Measured at the level of the subfamily, it appeared that divergence time and sequence distance had a very weak association (r = 0.12), as did sequence distance and expression distance (r = 0.12). Divergence time and expression distance as a whole appeared unrelated (r = –0.03), as did selective pressure and expression distance (r = 0.05). Finally, sequence distance and selective pressure had a positive association (r = 0.35), and, as expected, divergence time and selective pressure were negatively associated (r = –0.63).
Figure 11.
Associations among Median Values of Sequence Divergence (dS), Sequence Distance (dseq), Expression Profile Distance (dr), and Selective Pressure (dN/dS) for Each RLK Subfamily.
For each measure, median values for a given subfamily were calculated based on all pairwise comparisons of members within that subfamily. Off-diagonal panels contain scatter plots of median values for each subfamily according to the measures indicated. Dashed lines represent linear best fits, with the overall correlation among observations given (r). On-diagonal panels depict the distribution of median values among all RLK subfamilies for each measure noted.
DISCUSSION
Signaling-Related Changes in the RLK Transcriptome
The genome of Arabidopsis thaliana contains more than 600 receptor-like kinase genes, the great majority of which have yet to be investigated experimentally. Here, we have created a detailed expression profile of 604 receptor-like kinase genes after exposure to a cross-section of major developmental and environmental signaling factors in plants, including abiotic stresses, biotic stresses, and hormones. Our results show that RLK gene activity is regulated at the transcriptional level in response to many of these factors and suggest that a great number of RLKs may be functionally relevant components of a broad spectrum of essential signaling processes in plants. Furthermore, these results specifically identify those signaling conditions that elicit changes in RLK transcriptional levels on a gene-by-gene basis, as well as similarities and divergences in expression among closely related RLK genes.
These results are significant in that, prior to the detection of the first receptor-like kinase in maize, it was believed that the primary mechanism for signaling in plants was the symplastic transit of molecules through the intracellular tubules known as plasmodesmata (Dievart and Clark, 2004; Walker and Zhang, 1990). The discovery of RLKs as large gene families in land plants changed this conception and provided a new focus for research into plant extracellular signaling (Shiu and Bleecker, 2001a). However, although ongoing experimental analyses have bolstered the hypothesis that RLKs are involved in signaling pathways that service fundamental plant physiological processes, breakthroughs in identifying these pathways have been limited to a relatively small subset of individual RLKs. By taking a functional genomics approach to investigate this hypothesis, we have generated a large body of initial evidence as to RLK functionality in Arabidopsis for the complete genomic inventory of RLKs. Additionally, the preponderance of changes that we found in the RLK transcriptome further supports the widely held belief that RLKs are major contributors to the processing of a vast array of plant developmental and environmental cues.
Our results also extend the initial observation that transcriptional regulation may serve as an important mechanism for controlling RLK-mediated responses. Previous reports have implicated post-translational events at the biochemical and cellular level as possible forms of regulating RLK activity. For instance, receptor oligomerization and hyperphosphorylation have been demonstrated in several of the well studied RLKs, including BRI1, BAK1, and SYMRK (Nam and Li, 2002; Li et al., 2002; Wang et al., 2005; Yoshida and Parniske, 2005). In addition, receptor internalization and dephosphorylation were exhibited by ACR4, SERK3, and BRI1 (Russinova et al., 2004; Gifford et al., 2005). A third possible form of regulation was speculated to occur at the transcriptional level. Initial evidence demonstrated clear changes in the gene expression of several RLKs in response to signaling factors such as abscisic acid, salicylic acid, oxidative stress, and light (Shiu and Bleecker, 2001b; Becraft, 2002). Our results greatly expand on this evidence and indicate that transcriptional regulation may be an important form of controlling the activity of a large number of RLK genes in a wide variety of signaling events. As an obvious advancement of this result, it will be critical to establish the relationship between the transcriptional response of a given RLK gene and the functional response provided by its gene products.
RLKs and the Capacity for Complex Signaling
Our study also revealed that the transcriptional levels of many RLK genes are affected by more than one signaling factor. These results support current models of RLK versatility that have been reported recently. In one study, an RLK homologue in tomato, tBRI1/SR160, appeared to be capable of sensing both steroid and peptide hormones, and subsequently functioning in distinct signaling pathways (Montoya et al., 2002). A second form of versatility was demonstrated in BRI1, an Arabidopsis RLK, which was able to either homodimerize or interact with another RLK, BAK1, under differing conditions (Wang et al., 2005; Russinova et al., 2004; Wang et al., 2005). The ERECTA RLKs also appear to be capable of differential interactions in response to different ligands (Johnson and Ingram, 2005; Shpak et al., 2005). Given that most of the insights into RLK mechanics revolve around a few individual RLKs, the extent that such versatility exists among the large contingent of unexplored RLKs is unknown. However, in light of these models, our results highlight a number of RLK genes that may be involved in multiple signaling pathways, and suggest that such functional versatility may be a widespread feature of the RLKs.
Another explanation for the multiplicity of responses seen among the RLKs is that an individual RLK may be a component of a secondary response that exists downstream of separate signaling pathways. This interpretation is particularly appealing for those RLKs whose transcriptional levels were regulated by treatments known to induce physiologically related responses in plants. In particular, it would explain the widespread response, as well as the great degree of overlap, seen in the bacterial, fungal, and viral pathogen assays. Although the initial signaling events differ among these stimuli, they are known to target a number of related downstream pathways within the infected host, including those involved in the hypersensitive response, systemic acquired resistance, and the de-novo generation of additional signaling molecules, such as hydrogen peroxide and nitrous oxide (Kunkel and Brooks, 2002). This interpretation, although likely, remains to be demonstrated. In addition, the forms of RLK signaling versatility mentioned here are not mutually exclusive; these mechanisms may all be represented in the functional repertoire of the RLK superfamily.
The potential for complex signaling responses seen among individual RLKs is further enhanced when considered at the subfamily level. Previously, it had been speculated that, due to the differential configurations of their extracellular domains, members of each subfamily might be involved in similar signaling pathways. Here, we have shown that each subfamily exhibited transcriptional responses to a diverse set of signal types. The targeting of signals, such as hormones, to different members of the same gene family is a known phenomenon in plants (Nemhauser et al., 2006). In the case of RLKs, it may be that, within a given subfamily, individual members have roles in disparate signaling events. However, the diversity of responses seen within each subfamily may also indicate a mechanism by which different types of signals may be perceived by specific members of the same subfamily, yet induce similar downstream signaling events. This mechanism would allow for the integration of multiple signaling inputs in forming a finely tuned physiological response to complex environmental conditions.
Redundancy and Novelty in the Expansion of the RLK Superfamily
As noted in the literature, a major problem in determining RLK functionality has been the frequent inability to establish phenotypes in single loss-of-function genetic lines (Johnson and Ingram, 2005). One factor possibly contributing to this problem was the lack of physiologically relevant screening conditions (Shiu and Bleecker, 2001b). However, a second factor cited was the likely existence of functional redundancies among similar RLKs (Shiu and Bleecker, 2001b; Dievart and Clark, 2004; Shpak et al., 2003). A common use of microarray data is to predict functional similarities and interactions among genes by looking for similarities in their expression patterns (Walker et al., 1999; Hughes et al., 2000). This concept has been the basis for active areas of research in clustering and classification methods, as well as in terms of biological discovery, such as inferring regulatory networks and signaling pathway components.
The results of our work provide a detailed expression profile of each RLK gene not only in response to a range of screening conditions, but in relationship to genes both within its own subfamily and across other RLK subfamilies. The conserved expression profiles within each subfamily—particularly from genes with high sequence similarity—are obvious candidates for functional redundancies. Curiously, we also found a great degree of expression similarity between members from different subfamilies. This result suggests that functional relationships may also be common among members of separate subfamilies. Such a phenomenon has been experimentally demonstrated in the interaction of a member of the LRR X subfamily, BRI1, with a member of the LRR II subfamily, SERK1II/BAK1 (Nam and Li, 2002; Russinova et al., 2004). It should be noted, however, that our data do not capture tissue-specific divergences that may exist among the expression patterns. Although functional overlaps may exist among those RLKs with similar expression patterns, they may not always result in redundancy if expression is restricted to discrete domains. Nonetheless, the datasets we provide allow future experimental designs to account for possible complementarity or interactions among such similarly expressed RLK genes.
The diversity of expression profiles we found within each subfamily may also explain the expansion and retention of RLK genes within the Arabidopsis genome. Previous analyses support the hypothesis that gene duplication played a major role in the appearance of the multi-member RLK subfamilies in Arabidopsis (Shiu et al., 2004; Morillo and Tax, 2006; Becraft, 2002). It is thought that one key to retaining such duplicates is through functional diversification, and that divergence in gene expression may be a primary step in this process (Gu et al., 2002; Ganko et al., 2007). Here, we found a high level of divergent expression among RLK subfamilies, and we investigated possible evolutionary dynamics underlying this divergence.
In our analysis of selective pressure on the RLKs, we found that all of the subfamilies appear to be under some degree of purifying selection when considering the ratio of non-synonymous-to-synonymous substitution rates for full-length sequences within each subfamily. It should be noted that others have reported finding signs of positive selection at specific sites within the extracellular domains of some RLK subgroups (Shiu et al., 2004; Verica et al., 2003; Strain and Muse, 2005; Zhang et al., 2006). However, we did find that the strength of purifying selection varied among the subfamilies and that about a third of the subfamilies displayed some degree of association between selective pressure and expression divergence. The positive association found among eight of the subfamilies may reflect a dynamic in which relaxed purifying selection may have allowed for greater divergence in expression to evolve (Good et al., 2006; Gu et al., 2002; Zhang, 2003). Interestingly, we found that six of the subfamilies had an inverse association between selective pressure and expression divergence. In these cases, even those sequences under greater selective pressure relative to other members of its subfamilies were able to diverge in expression.
Finally, we compared expression divergence to two measures of the amount of sequence evolution found among subfamily members: sequence distance (dseq) and sequence divergence time (dS). In terms of expression distance, we found 20 of the subfamilies (53%) displayed some association with sequence distance, and 11 of the subfamilies (29%) showed an association with sequence divergence time. Given that seven of the subfamilies overlapped between these results, 24 of the 38 subfamilies (63%) we analyzed, representing 251 of the 604 RLK genes (42%), showed signs of an association between sequence evolution and expression divergence. Eighteen of the subfamilies (47%), representing 220 RLK genes (36%), had a positive association in which more diverged sequences had higher levels of expression divergence. Interestingly, six of the subfamilies (16%) representing 31 RLK genes (5%) had an inverse association in which the more closely related sequences had the more divergent expression patterns. In either case, it may be that the great amount of expression divergence seen within the individual RLK subfamilies served as a primary mechanism for maintaining such an expansive number of RLK genes in the Arabidopsis genome (Gu et al., 2002; Ganko et al., 2007). However, as our results suggest, although many paralogs within the RLK subfamilies have been retained, it appears that distinctly different evolutionary processes might have determined their fates.
METHODS
Microarray Experiments, Data Preparation, Signal Log2 Ratios
The data presented in this study were generated from 68 individual experiments coordinated by Syngenta, Inc., using the Syngenta custom Arabidopsis GeneChip array (sysSYNG002a, Affymetrix, Santa Clara, California, USA). Note that data produced in these experiments passed manual quality examination and datasets representing different areas of focus from the one presented here have been published (see Supplemental Table 1). Experimental parameters from these experiments, including details of treatments, dosages, and tissues, are described in Supplemental Table 1. Subsequent cDNA synthesis and hybridization were performed as described by Zhu et al. (2001).
CEL files from each experiment were processed by a uniform algorithm (Zhu and Wang, 2000) and signal readings were globally scaled across each array to an arbitrarily defined target value (Zhu et al., 2001). The expression indices of 604 known and predicted RLK genes (Shiu and Bleecker, 2001a) represented on the GeneChip array were extracted and paired as control and treated samples according to experimental design. For each experiment, any gene with an average difference (signal reading) of less than 5 in either or both the control or treatment sample were disregarded (Chen et al., 2002; Zhu et al., 2001). Signal log2 ratios were generated by taking the ratio of the treatment signal reading to the control signal reading and performing a log2 transformation (Chen et al., 2002).
Note that, in each experiment, the samples were pooled from at least eight individual plants receiving the same treatment (or mock treatment) in order to control for biological variation. Consequently, the detected mRNA levels represent an average reading of the biological replicates. To ensure data quality and stability in the analyses, the biological reproducibility of the samples was tested by performing 11 experiments with biological replicates. The results showed a high level of correlation among the replicated experiments, with each compared experimental results having a correlation of r > 0.90.
Because the complexity and diversity of experimental designs represented in the dataset did not allow for explicit modeling of variation among biological replicates across all conditions, we were restricted to using a heuristic approach to identifying changes in gene expression. Consequently, we sought to impose relatively stringent criteria for classifying differentially expressed genes. In this case, we chose a two-fold cut-off based on a preliminary analysis of false-positive rates, in which the false-positive rate at a two-fold threshold was demonstrated to be 0.2% (Zhu and Wang, 2000). Finally, an exhaustive, secondary confirmation of all gene expression changes under all conditions in this study was not tractable. However, an independent set of experiments based on RNA gel analysis have confirmed the GeneChip data for a subset of genes and conditions presented here (Zhu et al., 2001).
Gene Expression Profile Distances
To measure similarity among expression profiles, we concatenated 104 arrays from our study and created a matrix of normalized signal readings for the 604 RLK genes. An expression profile for a given gene was represented by its vector of signal readings across the 104 arrays. To calculate gene expression distances between any two genes, we first computed the mean-centered Pearson product-moment correlation coefficient (r) between the expression vectors X and Y of the genes, where i represents the array number and N the total number of arrays:
where
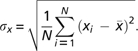 |
The distance in gene expression between any two genes i and j was then calculated as:
In the process of determining correlation values, if a signal reading for either gene from a particular array was below 5, we removed the array for that particular comparison.
Once the distances were computed for all pairwise comparisons within the matrix (182 106 comparisons in total), we normalized the distances by dividing each distance between any two genes i and j by the largest value found in the resulting dataset:
This normalization transformed the distances to a scale of 0 to 1, with 1 being most distant. In parsing the dataset into subfamily-based comparisons, we followed the subfamily classification scheme designated by Shiu and Bleecker (2001a).
Clustering Methods and Silhouette Width Analysis
The RLK gene expression profiles were clustered in two ways for analysis. The first clustering consisted of the manual separation of the 604 expression profiles into 43 a priori clusters based on the noted subfamily classification scheme (Shiu and Bleecker, 2001b). The second clustering was performed using the R implementation of the Partitioning Around Medoids (PAM) algorithm (Kauffman and Rousseeuw, 1990). Given a set of K medoids, PAM clusters observations by assigning each observation to its nearest medoid. The K medoids selected by the algorithm minimize the sum of the distances of each observation from its nearest medoid. Forty-three clusters were specified for the PAM clustering to correspond with the number of subfamily clusters.
Silhouette widths were calculated for the a priori clustering and the PAM clustering (K = 43). The same distance matrix based on the Pearson correlation (dr) was used to calculate silhouette widths for each clustering approach. The silhouette width (si) of a given observation i is defined as:
where ai represents the average distance of i to all other observations in its cluster, and bi represents the average distance of i to the observations in its nearest neighboring cluster. The average silhouette width for a cluster (siC) is the average of the silhouette widths for all members of a cluster. The average silhouette width for a cluster structure (siD) is the average of all silhouette widths in the dataset.
The Hierarchical Ordered Partitioning and Clustering Hybrid (HOPACH) algorithm was also used as a potential clustering method. HOPACH combines hierarchical clustering with partitioning, creating a data-adaptive tree that is composed of partitions at each level K. The optimal number of child clusters in partitioning steps, as well as the optimal number of clusters to be joined in collapsing steps, rely on the mean or median split silhouette measure of cluster heterogeneity (van der Laan and Pollard, 2003). For a given number of clusters k, a partitioning clustering method (in this case, PAM) is applied to each cluster. The split silhouette is defined as the maximum average silhouette width when the partitioning clustering method is applied to the elements of cluster k only. The mean split silhouette is the average of the split silhouettes over all clusters.
Hierarchical Clustering and Gene Expression Linkage Analysis
We created an initial distance matrix containing the normalized expression distances (dr) for all pairwise comparisons among 585 RLK genes, with the 19 unclassified RLK genes removed from the dataset. Then, for each comparison between subfamilies, we averaged the expression distances found between their members to create a 43-by-43 subfamily distance matrix. We performed hierarchical clustering on the subfamily distance matrix using the hclust function provided by the stats package in R (version 2.6.2, www.r-project.org). hclust relies on an agglomerative algorithm in which each initial object (here, subfamilies) is assigned to its own cluster. The two most similar clusters in the matrix as measured by average gene expression distance are then joined and distances between the new cluster and all remaining objects are re-calculated. In our implementation, we used an average linkage method to update distances, in which the new distances are calculated based on the average distances between members of separate clusters. Consequently, the dendogram depicted in Figure 7 represents an average-linkage hierarchical clustering of subfamilies based on average-linkage gene expression distances.
Pairwise Sequence Alignments and Distances
We extracted amino acid sequences representing the 604 Arabidopsis RLKs from the NCBI GenBank protein database (www.ncbi.nlm.nih.gov) using the AGI identifiers provided by Shiu and Bleecker (2001a). Supplemental Table 5 contains a map of the AGI and GI identifiers and subfamily classification for all RLKs used in this study. In determining distances among these sequences, we first used sequence alignment scores (si,j) as a measure of similarity. The alignment scores were computed by performing all-against-all pairwise alignments (182 106 in total) among the set of 604 RLK sequences. The alignments and scores were generated by ClustalW (version 1.83) and the Gonnet series of substitution matrices, with gap open and extension penalties of 10.0 and 0.1, respectively (Thompson et al., 1994; Gonnet et al., 1992).
To convert the alignment score between any two sequences i and j into a sequence distance, we normalized each score using the maximum value found among all observations and subtracted it from 1:
Correlations between sequence distances and expression distances were calculated for each subfamily by comparing dseq and dr for matching sets of pairwise comparisons over all comparisons within each subfamily, using the Pearson product-moment correlation coefficient.
Multiple Sequence Alignments, dN/dS Ratios, and Sequence Divergence Estimates
We retrieved nucleotide sequences representing coding DNA for 585 RLKs from the Arabidopsis Information Resource website (TAIR, www.arabidopsis.org), excluding those RLK genes that remain unclassified. We divided the RLK protein and nucleotide coding sequences into 43 subfamilies based on the classification scheme designated by Shiu and Bleecker (2001b). For each subfamily, we produced a multiple protein sequence alignment using the program MUSCLE (version 3.6) (Edgar, 2004). Based on the multiple protein sequence alignment, we then created a multiple sequence alignment of the coding DNA for all members of each subfamily. In some cases, extremely long or short sequences caused the inclusion of a high number of gaps in the protein sequence alignment, which resulted in very short DNA alignments. When this occurred, the long/short sequences were removed from further analysis and the protein and nucleotide sequences were re-aligned. Overall, three subfamilies were affected by this modification: LRK10L-2 (two of eight sequences removed), LRR VIII-2 (five of 14 sequences removed), and PERKL (four of 19 sequences removed).
For each subfamily, dS and dN/dS were calculated for each pairwise comparison of codon alignments within the subfamily using the yn00 program in the PAML package (Yang and Nielsen, 2000; Yang, 1997). Codons aligned to gapped regions in the alignment were removed during this process. When normalizing these measures, we divided each value by the highest measurement found across all subfamilies. For each type of measurement, correlations with normalized expression profile distances were calculated for matching sets of pairwise comparisons over all comparisons within each subfamily, using the Pearson product-moment correlation coefficient.
Mosaic, Box, and Scatter Plots
The mosaic and box plots depicted in the manuscript were created using the mosaicplot and boxplot functions provided by the graphics package in R (version 2.6.2). The scatter plot matrix was produced in R using the car package (version 1.2-7). The silhouette plots were generated using the plot function on silhouette objects created both by custom R code and by the silhouette function found in the cluster package in R (version 1.11.10).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
Supplemental Table 1. Description of Microarray Experiments and List of Related Publications.
Supplemental Table 2. List of Differentially Expressed RLK Genes for All 68 Experiments, Grouped by Experiment.
Supplemental Table 3. Table of Differentially Expression RLK Genes per Treatment, Separated into Up-Regulated and Down-Regulated Responses.
Supplemental Table 4. Index Listing the AGI and GI Identifiers and Subfamily Classification for Each of the 604 RLKs Analyzed in the Study.
Supplemental Dataset 1. Normalized Pairwise Expression Distances between Subfamily Members for Each Subfamily.
Supplemental Dataset 2. Files Containing Normalized Expression Profile Distances across Subfamilies and Summaries of the Clustering and Silhouette Width Analyses.
Supplemental Figure 1. Average Silhouette Widths for PAM Clustering of RLK Expression Profiles across Different Values of K.
Supplemental Figure 2. Silhouette Plot for HOPACH Clustering of RLK Expression Profiles.
FUNDING
Funding for this work was provided by a grant from the National Science Foundation (award to S.L.). L.C. is also supported in part by funding from the California Agricultural Experimental Station (to S.L. and Steven Brenner).
Supplementary Material
References
- Akashi H. Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 2001;11:660–666. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell. 2005;17:3337–3349. doi: 10.1105/tpc.105.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW. Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol. 2002;18:163–192. doi: 10.1146/annurev.cellbio.18.012502.083431. [DOI] [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR. CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science. 1996;273:1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- Ben-Shlomo I, Hsu SY, Rauch R, Kowalski HW, Hsueh AJW. Signaling receptome: a genomic and evolutionary perspective of plasma membrane receptors involved in signal transduction. Science STKE. 2003;187:RE9. doi: 10.1126/stke.2003.187.re9. [DOI] [PubMed] [Google Scholar]
- Bommert P, et al. Thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development. 2005;132:1235–1245. doi: 10.1242/dev.01671. [DOI] [PubMed] [Google Scholar]
- Bonner JT. The origins of multicellularity. Integr. Biol. 1998;1:27–36. [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 2002;12:1718–1727. doi: 10.1016/s0960-9822(02)01151-x. [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A, et al. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- Chen W, et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, et al. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier D, et al. STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2005;102:9074–9079. doi: 10.1073/pnas.0503526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Cock JM, Vanoosthuyse V, Gaude T. Receptor kinase signalling in plants and animals: distinct molecular systems with mechanistic similarities. Curr. Opin. Cell Biol. 2002;14:230–236. doi: 10.1016/s0955-0674(02)00305-8. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell. 2005;17:3350–3361. doi: 10.1105/tpc.105.036731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby K, Gehring C. Engineering drought and salinity tolerance in plants: lessons from genome-wide expression profiling in Arabidopsis. Trends Biotechnol. 2005;23:547–552. doi: 10.1016/j.tibtech.2005.09.001. [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, et al. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- Dievart A, Clark SE. LRR-containing receptors regulating plant development and defense. Development. 2004;131:251–261. doi: 10.1242/dev.00998. [DOI] [PubMed] [Google Scholar]
- Dievart A, et al. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell. 2003;15:1198–1211. doi: 10.1105/tpc.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, et al. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- Fisher K, Turner S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 2007;17:1061–1066. doi: 10.1016/j.cub.2007.05.049. [DOI] [PubMed] [Google Scholar]
- Ganko EW, Meyers BC, Vision TJ. Divergence in expression between duplicated genes in Arabidopsis. Mol. Biol. Evol. 2007;24:2298–309. doi: 10.1093/molbev/msm158. [DOI] [PubMed] [Google Scholar]
- Gifford ML, Robertson FC, Soares DC, Ingram GC. ACR4 function, internalization and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell. 2005;17:1154–1166. doi: 10.1105/tpc.104.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Gonnet GH, Cohen MA, Benner SA. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- Good JM, Hayden CA, Wheeler TJ. Adaptive protein evolution and regulatory divergence in Drosophila. Mol. Biol. Evol. 2006;23:1101–1103. doi: 10.1093/molbev/msk002. [DOI] [PubMed] [Google Scholar]
- Gu Z, Nicolae D, Lu HH, Li WH. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 2002;18:609–613. doi: 10.1016/s0168-9525(02)02837-8. [DOI] [PubMed] [Google Scholar]
- Hazen SP, Wu Y, Kreps JA. Gene expression profiling of plant responses to abiotic stress. Funct. Integr. Genomics. 2003;3:105–111. doi: 10.1007/s10142-003-0088-4. [DOI] [PubMed] [Google Scholar]
- He ZH, He D, Kohorn BD. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 1998;14:55–63. doi: 10.1046/j.1365-313x.1998.00092.x. [DOI] [PubMed] [Google Scholar]
- Hoch JA. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Johnson KL, Ingram GC. Sending the right signals: regulating receptor kinase activity. Curr. Opin. Plant Biol. 2005;8:648–656. doi: 10.1016/j.pbi.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Kachroo A, Schopfer CR, Nasrallah ME, Nasrallah JB. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science. 2001;293:1824–1826. doi: 10.1126/science.1062509. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Building a multicellular organism. Annu. Rev. Genet. 2001;35:103–123. doi: 10.1146/annurev.genet.35.102401.090145. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw P. New York: Wiley; 1990. Finding Groups in Data: An Introduction to Cluster Analysis. [Google Scholar]
- King N. The unicellular ancestry of animal development. Dev. Cell. 2004;7:313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- Koishi R, Xu H, Ren D, Navarro B, Spiller BW, Shi Q, Clapham DE. A superfamily of voltage-gated sodium channels in bacteria. J. Biol. Chem. 2004;279:9532–9538. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Kwak SH, Shen R, Schiefelbein J. Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science. 2005;307:1111–1113. doi: 10.1126/science.1105373. [DOI] [PubMed] [Google Scholar]
- Lally D, Ingmire P, Tong HY, He ZH. Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell. 2001;13:1317–1331. doi: 10.1105/tpc.13.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of proteinprotein interactions. Mol. Biol. Evol. 2005;22:1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Liu X, Yue Y, Li B, Nie Y, Li W, Wu WH, Ma L. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- Madsen EB, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science. 2002;296:1470–1472. doi: 10.1126/science.1069607. [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM. Plants compared to animals: the broadest comparative study of development. Science. 2002;295:1482–1485. doi: 10.1126/science.1066609. [DOI] [PubMed] [Google Scholar]
- Montoya T, et al. Cloning the tomato Curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell. 2002;14:3163–3176. doi: 10.1105/tpc.006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo SA, Tax FE. Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 2006;9:460–469. doi: 10.1016/j.pbi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Morris ER, Walker JC. Receptor-like protein kinases: the keys to response. Curr. Opin. Plant Biol. 2003;6:339–342. doi: 10.1016/s1369-5266(03)00055-4. [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Nishimura R, et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002;420:426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Common patterns of evolution of gene expression level and protein sequence in Drosophila. Mol. Biol. Evol. 2004;21:1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
- Oakley TH, Gu Z, Abouheif E, Patel NH, Li WH. Comparative methods for the analysis of gene-expression evolution: an example using yeast functional genomic data. Mol. Biol. Evol. 2005;22:40–50. doi: 10.1093/molbev/msh257. [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K. Leucine-rich repeat receptor-like kinase 1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell. 2005;17:1105–1119. doi: 10.1105/tpc.104.027474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr. Opin. Plant Biol. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Pires-daSilva A, Sommer RJ. The evolution of signalling pathways in animal development. Nat. Rev. Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- Rabbani MA, et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Russinova E, et al. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, Carvalho-Niebel F, Duc G, Frugoli J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 2005;58:809–822. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]
- Searle IR, et al. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl Acad. Sci. U S A. 2001a;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE. 2001b;113:RE22. doi: 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- Shiu SH, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004;131:1491–1501. doi: 10.1242/dev.01028. [DOI] [PubMed] [Google Scholar]
- Shpak ED, Lakeman MB, Torii KU. Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell. 2003;15:1095–1110. doi: 10.1105/tpc.010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- Sivaguru M, et al. Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 2003;132:2256–2266. doi: 10.1104/pp.103.022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Stein JC, Howlett B, Boyes DC, Nasrallah ME, Nasrallah JB. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl Acad. Sci. U S A. 1991;88:8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Stracke S, et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- Strain E, Muse SV. Positively selected sites in the Arabidopsis receptor-like kinase gene family. J. Mol. Evol. 2005;61:325–332. doi: 10.1007/s00239-004-0308-0. [DOI] [PubMed] [Google Scholar]
- Suzaki T, et al. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development. 2004;131:5649–5657. doi: 10.1242/dev.01441. [DOI] [PubMed] [Google Scholar]
- Takahashi S, et al. Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Mol. Biol. 2004;56:29–55. doi: 10.1007/s11103-004-2200-0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, et al. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan MJ, Pollard KS. A new algorithm for hybrid hierarchical clustering with visualization and the bootstrap. J. Statist. Plann. Inference. 2003;117:275–303. [Google Scholar]
- Verica JA, Chae L, Tong H, Ingmire P, He ZH. Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis. Plant Physiol. 2003;133:1732–1746. doi: 10.1104/pp.103.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Decoupled evolution of coding region and mRNA expression patterns after gene duplication: implications for the neutralist-selectionist debate. Proc. Natl Acad. Sci. U S A. 2000;97:6579–6584. doi: 10.1073/pnas.110147097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, Kohorn BD. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 2001;13:303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC, Zhang R. Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of Brassica. Nature. 1990;345:743–746. doi: 10.1038/345743a0. [DOI] [PubMed] [Google Scholar]
- Walker MG, Volkmuth W, Sprinzak E, Hodgson D, Klingler T. Prediction of gene function by genome-scale expression analysis: prostate cancer-associated genes. Genome Res. 1999;9:1198–1203. doi: 10.1101/gr.9.12.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell. 2005;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Parniske M. Regulation of plant symbiosis receptor kinase through serine and threonine phosphorylation. J. Biol. Chem. 2005;280:9203–9209. doi: 10.1074/jbc.M411665200. [DOI] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. [Google Scholar]
- Zhang XS, Choi JH, Heinz J, Chetty CS. Domain-specific positive selection contributes to the evolution of Arabidopsis leucine-rich repeat receptor-like kinase (LRR RLK) genes. J. Mol. Evol. 2006;63:612–621. doi: 10.1007/s00239-005-0187-z. [DOI] [PubMed] [Google Scholar]
- Zhao DZ, Wang FG, Speal B, Ma H. The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 2002;16:2021–2031. doi: 10.1101/gad.997902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Wang H, Walker JC, Li J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004;40:399–409. doi: 10.1111/j.1365-313X.2004.02214.x. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Wang X. Large-scale profiling of the Arabidopsis transcriptome. Plant Physiol. 2000;124:1472–1476. doi: 10.1104/pp.124.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, et al. Towards elucidating global gene expression in developing Arabidopsis: parallel analysis of 8300 genes. Plant Physiol. Biochem. 2001;39:221–242. [Google Scholar]
- Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.