Abstract
To study cold signaling, we screened for Arabidopsis mutants with altered cold-induced transcription of a firefly luciferase reporter gene driven by the CBF3 promoter (CBF3-LUC). One mutant, chy1-10, displayed reduced cold-induction of CBF3-LUC luminescence. RNA gel blot analysis revealed that expression of endogenous CBFs also was reduced in the chy1 mutant. chy1-10 mutant plants are more sensitive to freezing treatment than wild-type after cold acclimation. Both the wild-type and chy1 mutant plants are sensitive to darkness-induced starvation at warm temperatures, although chy1 plants are slightly more sensitive. This dark-sensitivity is suppressed by cold temperature in the wild-type but not in chy1. Constitutive CBF3 expression partially rescues the sensitivity of chy1-10 plants to dark treatment in the cold. The chy1 mutant accumulates higher levels of reactive oxygen species, and application of hydrogen peroxide can reduce cold-induction of CBF3-LUC in wild-type. Map-based cloning of the gene defective in the mutant revealed a nonsense mutation in CHY1, which encodes a peroxisomal β-hydroxyisobutyryl (HIBYL)–CoA hydrolase needed for valine catabolism and fatty acid β-oxidation. Our results suggest a role for peroxisomal metabolism in cold stress signaling, and plant tolerance to cold stress and darkness-induced starvation.
Keywords: Cold stress, signal transduction, gene regulation, CHY1, β-hydroxyisobutyryl–CoA hydrolase
INTRODUCTION
Throughout growth and development, plants are subjected to various environmental challenges, including interactions with microorganisms and changes in temperature, light intensity, and soil water potential. Unlike animals, plants cannot move but are forced to adapt to these biotic and abiotic stresses by metabolic modifications. For instance, plant acclimation to cold conditions involves complicated metabolic changes, including alterations in lipid composition, accumulation of compatible osmolytes such as sugars and proline, and changes in the expression of hundreds of genes (Thomashow, 1999). These changes underlie mechanisms used by plants from temperate regions to increase freezing tolerance following pre-exposure to low, non-freezing temperatures (0–10°C), namely cold acclimation (Levitt, 1980) or acquired freezing tolerance (Kaplan et al., 2004).
Low temperatures induce expression of diverse plant genes, known as COR (cold regulated), KIN (cold induced), LTI (low temperature induced), or RD (responsive to dehydration) genes (Medina et al., 1999; Nordin et al., 1993; Welin at al., 1995). Three transcription factors known as CBFs (CRT/DRE binding factor) or DREBs (DRE binding protein) can bind to DRE/CRT (dehydration-responsive element/C-repeat) cis-elements in promoters and activate transcription of the COR/KIN/LTI/RD genes (Stockinger et al., 1997; Yamaguchi-Shinozaki and Shinozaki, 1994). The CBF genes are transiently induced by low temperature, and this induction precedes that of COR/KIN/LTI/RD genes. Ectopic overexpression of CBF1 or CBF3 in Arabidopsis results in constitutive expression of downstream cold-inducible genes, even at warm temperatures, elevated levels of proline and soluble sugars (Gilmour et al., 2000), and increased freezing tolerance (Gilmour et al., 2000; Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Liu et al., 1998). These studies demonstrate a critical role for the CBF regulon in cold acclimation, although it is not the only regulon contributing to plant cold responses (Xin and Browse, 1998; Zhu et al., 2005). Studies using a cbf2 loss-of-function mutant suggest that CBF2 negatively regulates the expression of CBF1 and CBF3 in the cold (Novillo et al., 2004).
Cold stress causes dramatic changes to plant metabolism, as a result not only of general reductions in enzyme activities and reaction rates in the cold, but also active reconfiguration of the metabolome under the control of cold signaling (Cook et al., 2004; Kaplan et al., 2004; Levitt, 1980). The CBF regulon has a major role in the active reconfiguration of metabolite profiles (Cook et al., 2004). Whether and how metabolic status affects cold signaling and cold regulation of gene expression are not understood.
Forward genetic screening has been used to identify the upstream regulators of the CBF regulon. ICE1 is a positive regulator of CBFs and encodes a MYC-like bHLH transcriptional activator (Chinnusamy et al., 2003). The ice1 mutation blocks CBF3 expression, decreases expression of many downstream genes, and significantly reduces chilling and freezing tolerance (Chinnusamy et al., 2003). Conversely, the Arabidopsis HOS1 protein is a negative regulator of CBF genes. CBFs and their downstream COR genes show enhanced cold induction in hos1 mutant plants (Ishitani et al., 1998). HOS1 encodes a RING-finger ubiquitin E3 ligase that targets ICE1, a positive regulator of CBFs for ubiquitination and proteosomal degradation (Lee et al., 2001; Dong et al., 2006).
In this report, we present evidence supporting the importance of cellular metabolism in cold regulation and plant cold tolerance. In a screen for Arabidopsis mutants with deregulated expression of a luciferase reporter gene driven by the CBF3 promoter (CBF3-LUC), we isolated a mutant, chy1-10, with reduced CBF3-LUC and endogenous CBF3 expression in response to cold. chy1 mutant plants are less tolerant to freezing stress compared to wild-type after cold acclimation. In addition, chy1 is more sensitive than the wild-type to cold and dark treatment. Constitutive CBF3 expression partially rescues the cold- and dark-sensitive phenotype of chy1-10. The defective gene was isolated by map-based cloning and encodes a peroxisomal β-hydroxyisobutyryl (HIBYL)–CoA hydrolase needed for fatty acid β-oxidation and valine catabolism. Our findings reveal a requirement for peroxisome function in the cold-induction of CBF3, for plant freezing tolerance, and survival following dark treatment, particularly in the cold. We suggest that metabolism is not passively regulated by cold stress, but that cellular metabolic status plays an important role in gene regulation in the cold. Peroxisomes are an important source of reactive oxygen species (ROS) (Corpas et al., 2001) and ROS can modulate the concentration of cytosolic free calcium (Price et al., 1994), which is known to be a second messenger that can activate cold-responsive gene expression (Catala et al., 2003; Knight et al., 1996; Monroy and Dhindsa, 1995). We provide evidence that metabolic status might be connected to cold regulation through ROS.
RESULTS
Isolation of chy1-10, a Mutant Defective in Cold Signaling
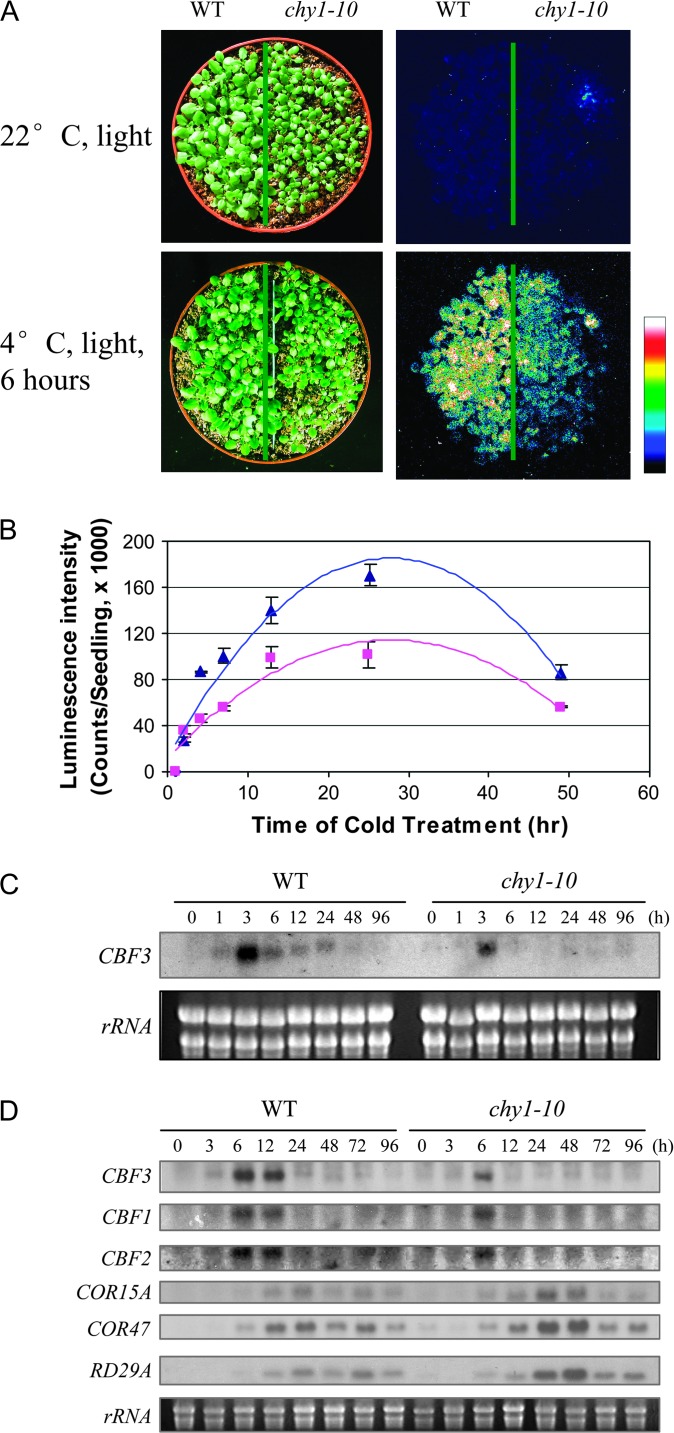
Transgenic Arabidopsis plants (ecotype Columbia) containing CBF3-LUC emit bioluminescence in response to low temperatures (Chinnusamy et al., 2003). CBF3-LUC plants (herein referred to as wild-type) were mutagenized with EMS and mutants with altered luminescence patterns in response to cold treatment were selected by luciferase imaging. We cloned the defective gene in one mutant, which revealed that the mutant is allelic (see below) to chy1 (Zolman et al., 2001a). We therefore designated it chy1-10. Figure 1A shows luminescence images of chy1-10 and wild-type seedlings with or without cold treatment (4°C) in the light. After 6 h of cold treatment, chy1-10 plants showed only ∼50% of the luminescence found in wild-type plants (Figure 1A and 1B).
Figure 1.
The chy1-10 Mutation Alters Cold Regulated Gene Expression.
(A) Luminescence images of wild-type (left) and chy1-10 (right) plants taken with or without cold treatment at 4°C in the light for 6 h. The color scale at the right shows the luminescence intensity from dark blue (lowest) to white (highest).
(B) Time course of luminescence intensity in wild-type (triangles) and chy1-10 (squares) plants during cold treatment in the light. Shown are the mean values ± standard deviation (n = 20).
(C) Northern blot analysis showing reduced induction of CBF3 in 2-week-old WT and chy1-10 mutant plants following cold treatment in the light for the indicated time (h). A photo of the ethidium bromide-stained rRNA is included as a loading control.
(D) RNA was prepared from WT and chy1-10 seedlings treated at 4°C in the dark for the indicated times. Gene probes used for RNA gel blot hybridization are indicated at left.
To determine if the chy1-10 mutation affects expression of endogenous CBF3, we extracted total RNA from mutant and wild-type seedlings treated with low temperature in the light. Consistent with the CBF3-LUC imaging result, cold induction of CBF3 was reduced in chy1-10 plants (Figure 1C). Wild-type plants showed CBF3 induction after 1 h of cold treatment; expression peaked at 3 h. In contrast, CBF3 induction in chy1-10 was observed only after 3 h, and the expression level was substantially decreased. Reduced CBF3 induction also was observed following cold and dark treatment of chy1-10 (Figure 1D). In addition, levels of CBF1 and CBF2 mRNAs were somewhat lower in the chy1-10 mutant than in wild-type (Figure 1D). We also examined induction of several CBF target genes following cold and dark treatment. Surprisingly, transcript levels of COR15A, COR47A, and RD29A in the chy1-10 mutant were not lower than in wild-type (Figure 1D), supporting the hypothesis that the CBFs are not the sole transcriptional regulators of these target genes (Zhu et al., 2005).
The chy1-10 mutant plants were backcrossed to CBF3-LUC wild-type, and the resulting F1 seedlings showed wild-type CBF3-LUC luminescence after 6 h of cold treatment. The F2 population from the self-fertilized F1 plant segregated in ∼3:1 ratio of wild-type to mutant (data not shown), indicating that the mutant defect is caused by a recessive mutation in a single nuclear gene. All subsequent characterization was performed using chy1-10 mutant plants backcrossed to wild-type four times to remove potential unlinked mutations.
chy1-10 Is Defective in Acquired Freezing Tolerance
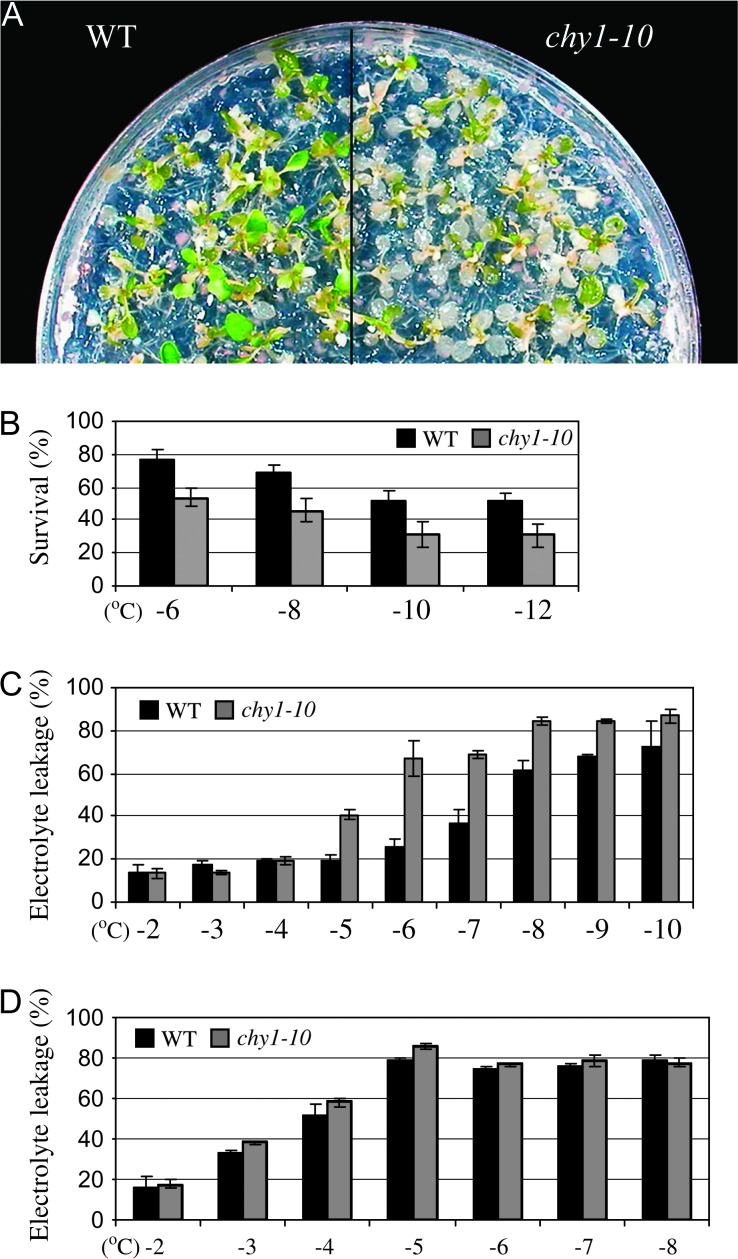
Because the chy1-10 mutant shows reduced cold induction of the CBF3 gene, which is important for freezing tolerance, we examined the effect of cold acclimation on whole plant freezing tolerance and leaf electrolyte leakage. Ten-day-old chy1-10 and wild-type seedlings grown on agar plates were cold acclimated at 4°C in the light for 4 d, subjected to freezing treatment (see Methods), then returned to normal growth conditions for 2 d and examined. The chy1-10 mutant was less freezing-tolerant than wild-type at temperatures below –5°C (Figure 2A and 2B). For example, only ∼46% of chy1-10 plants survived freezing at –8°C, whereas ∼69% of wild-type plants survived (Figure 2B). To assay the effect of cold acclimation on leaf electrolyte leakage, wild-type and chy1-10 plants were treated at 4°C in the light for 4 d, then the leaves were excised and subjected to freezing conditions followed by electrolyte leakage assays. chy1-10 leaves showed higher levels of electrolyte leakage than wild-type at most freezing temperatures (Figure 2C). In contrast, without cold acclimation, wild-type and chy1-10 leaves showed similar increases in electrolyte leakage (Figure 2D). These results show that the mutant plants are defective in acquired freezing tolerance.
Figure 2.
Freezing Sensitivity of chy1-10 Mutant Plants.
(A) Decreased survival of chy1-10 mutant plants 2 d after freezing treatment at –8°C following a 4-d cold acclimation.
(B) Quantification of wild-type and chy1-10 survival 2 d after freezing treatment following a 4-d cold acclimation. Data shown are mean values with standard deviation (n = 8).
(C) Comparison of leaf electrolyte leakage after freezing treatments at the indicted temperatures (see Methods) following a 4-d cold acclimation at 4°C in the light. Data shown are mean values with standard errors (n = 10).
(D) Comparison of leaf electrolyte leakage after freezing treatments at the indicated temperatures without cold acclimation. Data shown are mean values with standard deviation (n = 10).
The chy1-10 Mutation Disrupts a Peroxisomal HIBYL–CoA Hydrolase
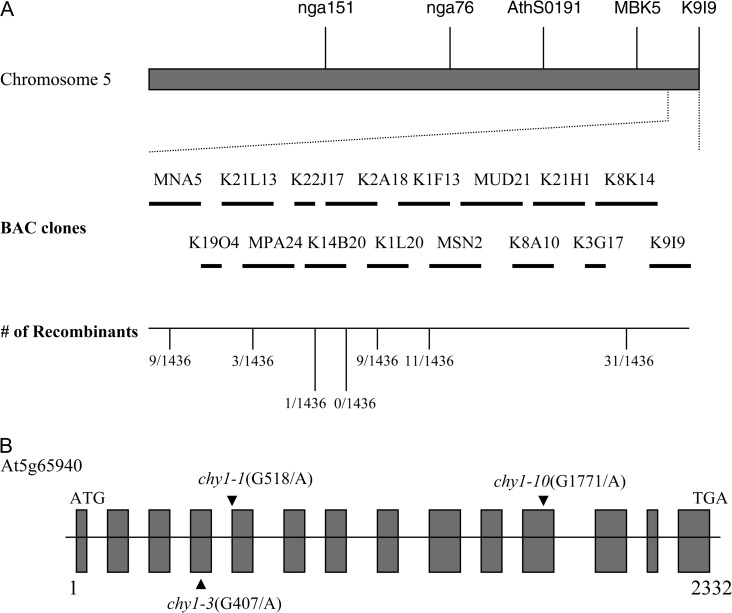
We used a map-based cloning strategy to identify the mutated gene in chy1-10 (see Methods). Initial mapping localized chy1-10 to the bottom of chromosome 5, between the markers MBK5 and K9I9 (Figure 3A). Subsequent mapping using new markers delimited chy1-10 to an approximately 50-kb region on two BAC clones, K14B20 and K2A18, between markers K14B20-56k and K2A18-24k. Candidate genes in this region were sequenced from chy1-10 mutant plants. This analysis revealed a single nucleotide substitution in At5g65940 (K14B20.11) in the mutant: a G-to-A change at position 34121 of the BAC clone, corresponding to position 1771 of the gene, creating a premature stop codon that would truncate the encoded enzyme (Figure 3B). No mutations were found in any other sequenced genes.
Figure 3.
Positional Cloning of chy1-10.
(A) Genetic mapping with PCR-based markers positioned chy1-10 on the BAC clone K14B20. The number of recombination events out of the total number of chromosomes examined is indicated.
(B) Sequence analysis of chy1-10 identified a single nucleotide change (G1771 to A1771) that creates a premature stop codon in At5g65940 (K14B20.11). Mutations previously found in chy1-1 and chy1-3 (Zolman et al., 2001a) also are indicated.
CHY1 encodes a peroxisomal β-hydroxyisobutyryl (HIBYL)–CoA hydrolase that hydrolyzes HIBYL–CoA to β-hydroxyisobutyrate and CoA during Val catabolism (Zolman et al., 2001a). The Arabidopsis CHY1 protein is 43% identical to a mammalian HIBYL-CoA hydrolase that functionally complements the Arabidopsis chy1 mutant when targeted to the peroxisome (Zolman et al, 2001a). Five chy1 mutant alleles had been previously described. chy1-1 has a G-to-A mutation at position 518 that alters the 3′-splice site of the fourth intron; chy1-2 has a 34-bp deletion from positions 1084–1118, which alters the 5′-splice site of the eighth intron; and chy1-3 has a G-to-A mutation at position 407, changing a conserved Gly to Ser (Zolman et al., 2001a) (Figure 3B). Two additional chy1 mutant alleles (dbr5-1 and dbr5-2) have G-to-A substitutions at the last base of introns 6 and 11, respectively, disrupting the corresponding 3′ splice sites (Lange et al., 2004).
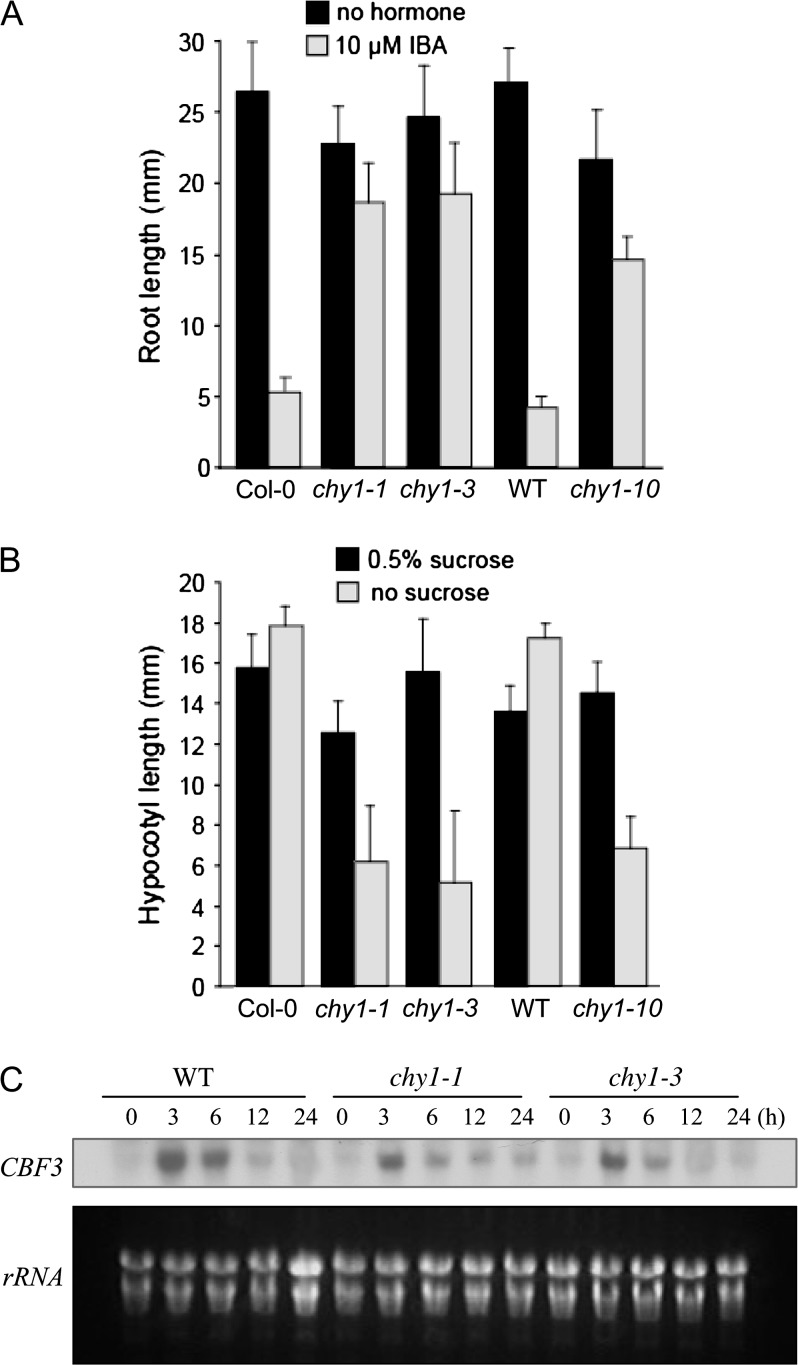
The original chy1 alleles were isolated as resistant to inhibition of root elongation and promotion of lateral root formation by the auxin precursor indole-3-butyric acid (IBA) (Zolman et al., 2001a) or the synthetic compound 2,4-dichlorophenoxybutyric acid (2,4-DB) (Lange et al., 2004). chy1 mutants also exhibit developmental phenotypes indicative of defects in peroxisomal fatty acid β-oxidation, including reduced hypocotyl elongation in the dark in the absence of exogenous sucrose (Lange et al., 2004; Zolman et al., 2001a) and slowed seed storage lipid catabolism during germination (Lange et al., 2004; Zolman et al., 2000). We found that chy1-10 displays the same IBA resistance and sucrose dependence as the original chy1 alleles (Figure 4A and 4B). We also compared CBF3 expression in the cold and light and observed reduced expression of CBF3 in the chy1-1 and chy1-3 mutant alleles (Figure 4C).
Figure 4.
Altered Phenotypic Responses and Impaired Induction of CBF3 in chy1 Mutant Plants.
(A) Root elongation on IBA. Roots of 8-day-old seedlings grown without hormone (black bars) or on 10 μM IBA (gray bars) were measured as described (Zolman et al., 2000). chy1-1 and chy1-3 are in the Col-0 background; chy1-10 is in the WT background. Error bars indicate the standard deviation of the means (n > 12).
(B) Hypocotyl elongation in the dark in the absence of sucrose. Hypocotyl lengths of 5-day-old seedlings grown in the dark on medium supplemented with 0.5% sucrose (black bars) compared to sucrose-free medium (gray bars). Error bars indicate the standard deviation of the means (n > 12).
(C) Northern blot analysis shows impaired induction of CBF3 in 2-week-old chy1-1 and chy1-3 mutant plants following cold treatment in the light for the indicated times. A photo of the ethidium bromide-stained rRNA is included as a loading control.
To confirm that the HIBYL–CoA hydrolase defect causes the reduced cold-induction of CBF3-LUC in chy1-10, a 35S-CHY1 overexpression construct (Zolman et al., 2001a) was introduced into chy1-10 plants via Agrobacterium-mediated transformation. T2 progeny of 50 Basta-resistant transformants were subjected to luminescence imaging and found to have wild-type luminescence patterns in the cold (Supplemental Figure 1). This result showed that the HIBYL–CoA hydrolase gene rescued the luminescence phenotype of chy1-10 mutant plants. In addition, we crossed chy1-1 and chy1-3 with chy1-10 and analyzed the resulting F1 progeny by luminescence imaging for expression of the CBF3-LUC transgene. As expected, CBF3-LUC expression in both chy1-10/chy1-1 and chy1-10/chy1-3 F1 plants was significantly reduced in response to cold treatment compared to wild-type (data not shown).
Together, these results indicate that chy1-10 is allelic to chy1-1 and chy1-3, and that all of the examined alleles disrupt HIBYL–CoA hydrolase activity similarly.
Sensitivity of chy1 to Darkness-Induced Starvation
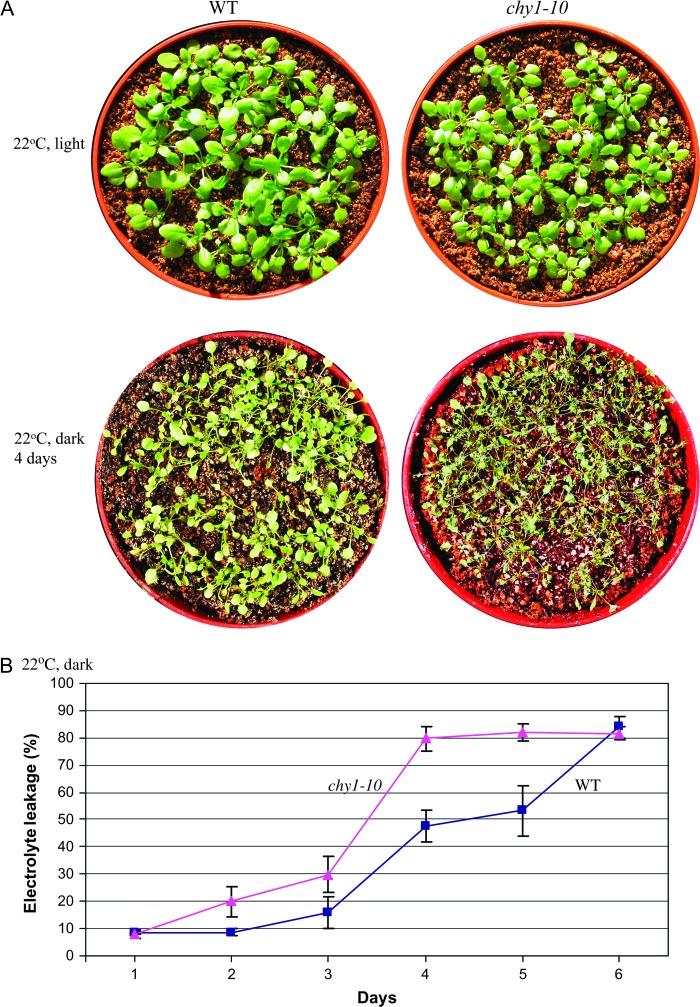
As mutations in CHY1 disrupt peroxisomal fatty acid β-oxidation required for plant growth during early seedling development in the absence of exogenous sugar (Lange et al., 2004; Zolman et al., 2001a), we examined the sensitivity of chy1-10 plants to darkness-induced starvation. Leaf senescence and seedling injury occurred rapidly in the dark at room temperature in both wild-type and chy1-10 plants. Interestingly, the mutant appeared more sensitive than wild-type to the dark treatment (Figure 5A). Measurement of leaf injury using electrolyte leakage confirmed the rapid damage suffered by both wild-type and the chy1 mutant during dark treatment at room temperature and the increased sensitivity of chy1-10 to these conditions (Figure 5B).
Figure 5.
Sensitivity of Wild-Type and chy1-10 Mutant Plants to Darkness-Induced Starvation at Room Temperature.
(A) Fourteen-day-old wild-type and chy1-10 mutant plants grown at 22°C were shifted to the dark at room temperature for 4 d. Plants were examined 2 d after return to normal growth conditions.
(B) Ion leakage was assayed using 14-day-old soil-grown plants treated in the dark at room temperature for the indicated number of days. Data shown are mean values with standard deviation (n = 10).
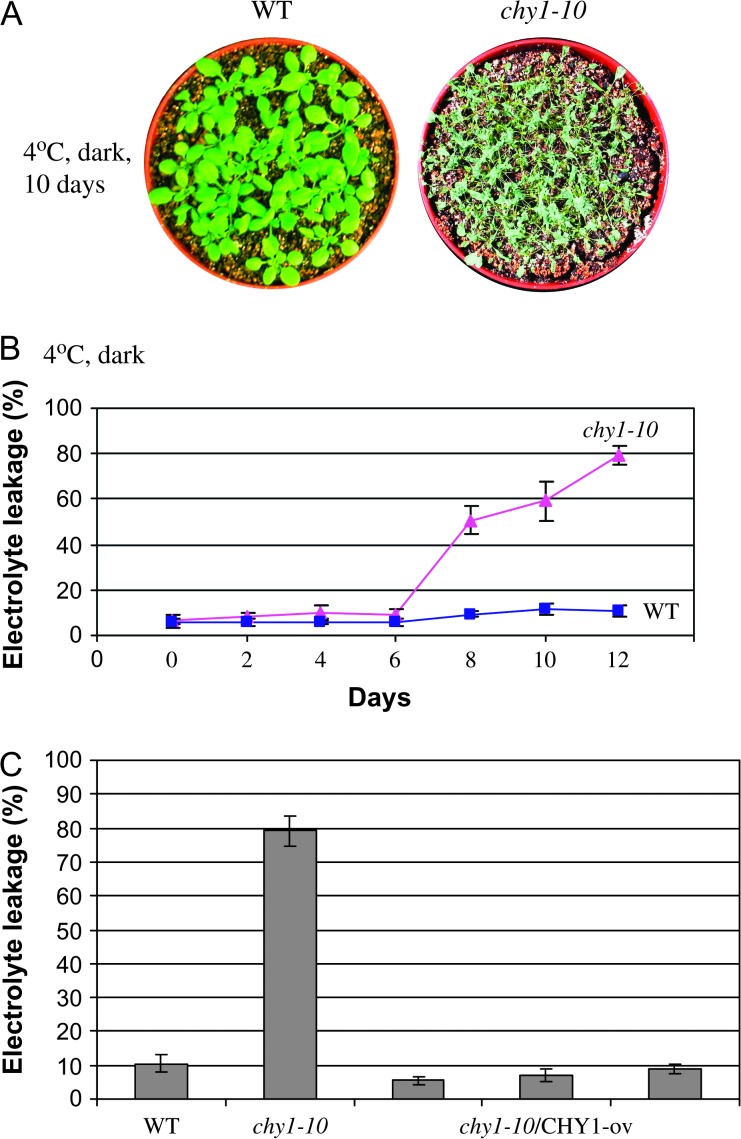
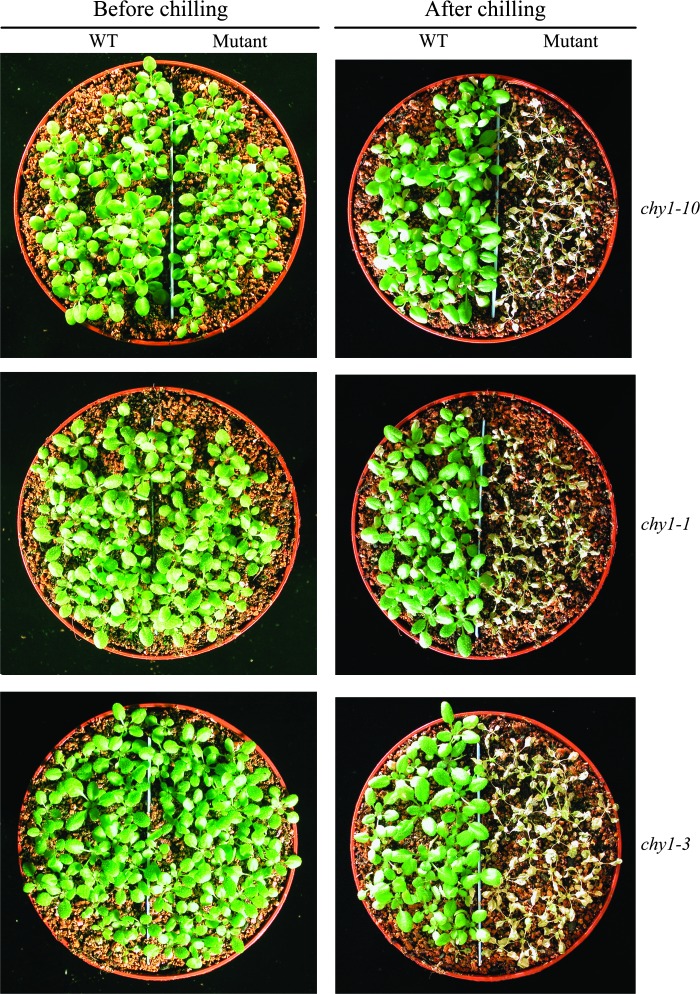
Due to the impaired cold-regulated CBF3 expression in the mutant (Figure 1C and 4C), we analyzed the performance of the chy1-10 mutant in the cold and dark. The damage caused by dark (Figure 5) was prevented by cold temperature in the wild-type but not in the mutant (Figure 6A). After treatment at 4°C in the dark for 10 d, chy1-10 mutant plants were severely damaged and died, whereas the same treatment did not significantly damage wild-type plants (Figure 6A). We examined the chy1-1 and chy1-3 mutant alleles under the cold and dark treatment, and observed the same phenotype as in chy1-10 (Figure 7).
Figure 6.
Chilling Sensitivity of chy1-10 Mutant Plants in the Dark at 4°C.
(A) Fourteen-day-old wild-type and chy1-10 mutant plants grown at 22°C were shifted to 4°C and grown in the dark for 10 d.
(B) Ion leakage was assayed using wild-type and chy1-10 leaves from 14-day-old plants grown in soil at 22°C and then shifted to 4°C in the dark for the indicated number of days. Data shown are mean values with standard deviation (n = 10).
(C) Leaf ion leakage in 14-day-old WT, chy1-10, and three transgenic lines of chy1-10 transformed with 35S-CHY1 (chy1-10/CHY1-ov) after cold treatment at 4°C for 10 d in the dark. Data shown are mean values with standard deviation (n > 10).
Figure 7.
chy1-1 and chy1-3 Mutant Plants Are Chilling Sensitive in the Dark.
Fourteen-day-old chy1-1 and chy1-3 mutant plants grown at 22°C were shifted to 4°C and grown for 10 d in the dark.
To examine the extent of chy1-10 leaf injury, leaves excised from soil-grown plants treated at 4°C in the dark for different time periods were assayed for electrolyte leakage (Ishitani et al., 1998). Wild-type leaves showed little increase in electrolyte leakage during a 12-d cold treatment in darkness (Figure 6B). In contrast, chy1-10 leaves showed a dramatic increase in electrolyte leakage beginning after 8 d of cold treatment in the dark. After a 12-d dark treatment, approximately 80% of cellular electrolytes in chy1-10 leaves were lost (Figure 6B).
We also exposed 50 transgenic lines of chy1-10 plants overexpressing CHY1 (see above) to 4°C in the dark for 10 d. All survived similarly to the wild-type (data not shown) and had only minor electrolyte leakage (Figure 6C), indicating that the HIBYL–CoA hydrolase gene rescued the chy1-10 mutant phenotypes. These results show that cold treatment can prevent darkness-induced injury in wild-type but not in chy1 mutant plants, consistent with an important role of CHY1 in not only dark, but also cold responses.
CBF3 Overexpression in chy1-10 Partially Rescues the Mutant Defects in Cold and Dark Responses
The chy1-10 mutation substantially decreases CBF3 expression (Figure 1). We hypothesized that reduced CBF3 expression may be partly responsible for some of the mutant phenotypes, and that enhancing CBF3 expression in chy1-10 mutant plants may suppress the defects. To test this hypothesis, CBF3 was expressed behind a strong constitutive promoter (Gong et al., 2002) in chy1-10 mutant plants. Thirty-five independent transgenic lines were obtained from Agrobacterium-mediated transformation and 10 were selected for detailed characterization.
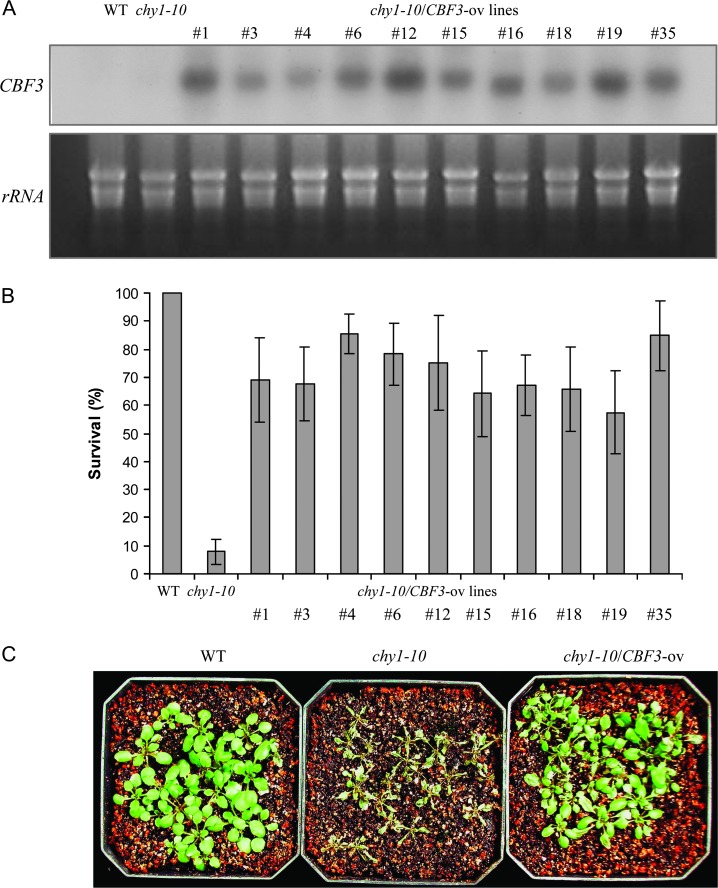
RNA blot analysis demonstrated CBF3 overexpression in all of the transgenic lines under normal growth conditions, when endogenous CBF3 mRNA is not detected (Figure 8A). Interestingly, constitutive expression of CBF3 significantly increased the survival of chy1-10 plants in the cold and dark (Figure 8B). A representative homozygous T3 population after cold treatment in the dark for 10 d is shown in Figure 8C. These results are consistent with the possibility that the mutant survival phenotypes can be attributed at least in part to altered CBF3 expression, and the CBF regulon has an important function in chilling tolerance in the dark.
Figure 8.
Ectopic Expression of CBF3 Suppresses the Chilling-Sensitivity of chy1-10 Mutant Plants.
(A) Northern blot of CBF3 expression in WT, chy1-10, and chy1-10 lines transformed with a CBF3 overexpression construct grown without cold treatment. A photo of the ethidium bromide stained rRNA is included as a loading control.
(B) chy1-10 mutant plants overexpressing CBF3 (CBF3-ov) were chilling resistant. Comparison of survival after cold treatment at 4°C for 10 d in the dark. Data shown are mean values from three experiments with standard deviation (n = 15).
C) Fourteen-day-old WT, chy1-10, and chy1-10/CBF3-ov (#35) were transferred to 4°C for 10 d in the dark.
Accumulation of Reactive Oxygen Species in chy1-10
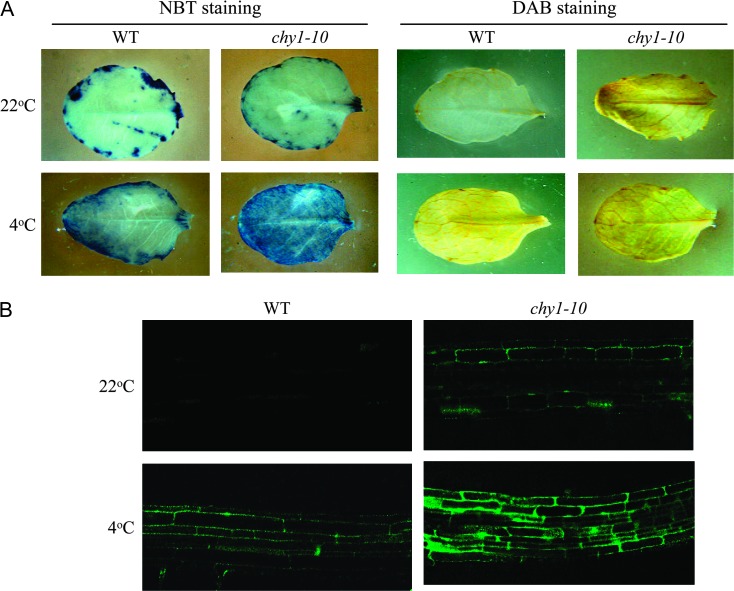
The peroxisome is an important source of reactive oxygen species (ROS) (Corpas et al., 2001). ROS can modulate calcium signaling in plants (Price et al., 1994), and calcium is a second messenger that can mediate cold regulation of gene expression (Catala et al., 2003; Knight et al., 1996; Monroy and Dhindsa, 1995). We assayed wild-type and chy1-10 plants for ROS levels using nitroblue tetrazolium (NBT) staining for superoxide and 3,3′-diaminobenzidine (DAB) staining for hydrogen peroxide (Lee et al., 2002a). Without cold treatment, wild-type leaves showed minimal NBT or DAB staining, indicating low superoxide and hydrogen peroxide levels (Figure 9A). After a 2-d cold treatment in the dark, substantial superoxide staining was detected in both wild-type and chy1-10, but the level in the mutant appeared higher than in the wild-type (Figure 9A). Interestingly, DAB staining was elevated in chy1-10 leaves even without cold and dark treatment, suggesting a high basal level of hydrogen peroxide in the mutant (Figure 9A). After the cold and dark treatment, high levels of hydrogen peroxide were detected in both the wild-type and mutant (Figure 9A).
Figure 9.
Detection of Reactive Oxygen Species in WT and chy1-10 Mutant Plants.
(A) Wild-type and chy1-10 leaves stained with NBT (dark blue) for superoxide or DAB (brown) for hydrogen peroxide after incubation at 22°C or following cold treatment at 4°C for 2 d.
(B) Detection of reactive oxygen species in WT and chy1-10 roots using CM-H2DCF diacetate, acetyl ester fluorescence imaging of seedlings grown at 22°C or following cold treatment for 2 d.
We also examined the level of reactive oxygen species in root tissues of chy1-10 mutants using CM-H2DCF diacetate acetyl ester fluorescence imaging (Shin and Schachtman, 2004). Consistent with the DAB staining, this assay showed higher basal levels of ROS in chy1-10 than in the wild-type (Figure 9B). After the cold and dark treatment, the levels of reactive oxygen species in chy1-10 again were substantially higher than those in the wild-type. These results suggest that disruption of the HIBYL–CoA hydrolase causes accumulation of reactive oxygen species.
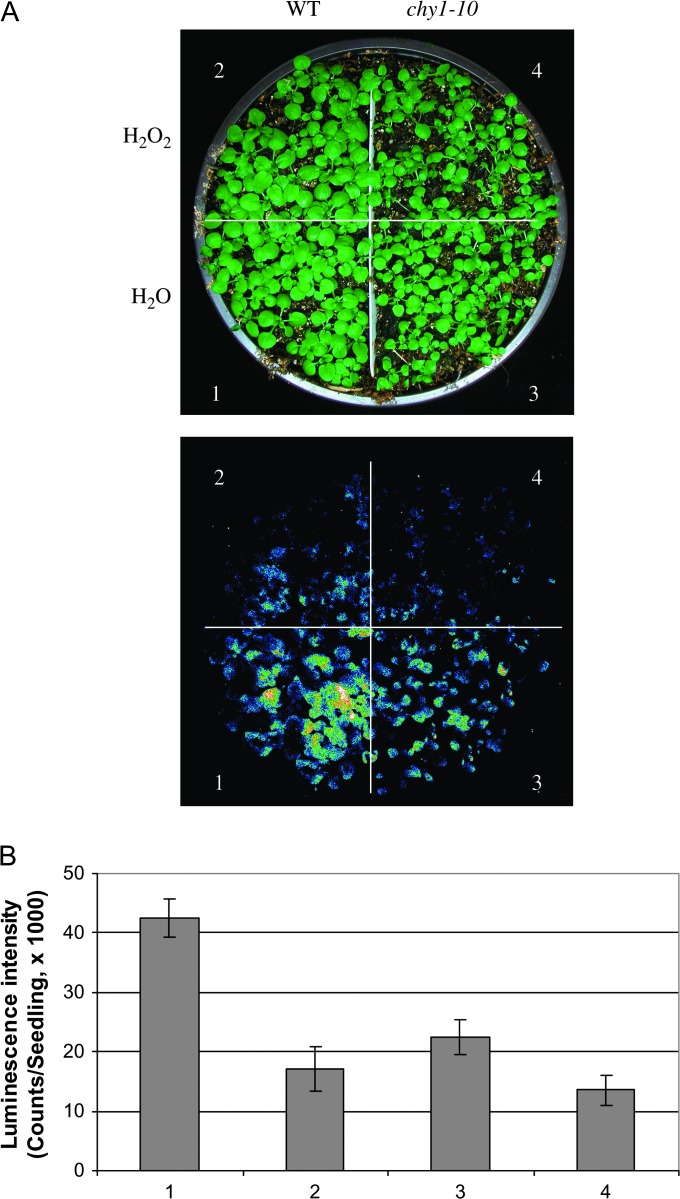
To begin to investigate whether ROS may influence cold responsive gene expression, we tested the effect of exogenous application of H2O2 on CBF3-LUC expression. Soil-grown CBF3-LUC seedlings were sprayed with 5 mM H2O2 or an H2O control, held at 22°C for 5 h, transferred to 0°C for 12 h, and then imaged for luciferase expression. We found that exogenous application of hydrogen peroxide significantly reduced cold-regulated expression of CBF3-LUC both in the wild-type and in the chy1-10 mutant (Figure 10A and 10B).
Figure 10.
Effect of Hydrogen Peroxide on Cold-Regulated Expression of CBF3-LUC.
(A) Luminescence images of CBF3-LUC plants. CBF3-LUC seedlings of the WT or chy1-10 genotype were sprayed with H2O (lower panel) or 5 mM H2O2 (upper panel). Luminescence images of the seedling were taken following cold treatment at 0°C for 12 h.
(B) Quantification of luminescence intensity of the CBF3-LUC seedlings treated with or without 5 mM hydrogen peroxide. Data shown are mean values with standard deviation (n = 10).
DISCUSSION
In this study, a forward genetics screen led to the unexpected finding that disruption of a peroxisomal metabolic enzyme impairs cold regulated expression of CBF3, which encodes a key transcription factor that controls plant freezing tolerance (Stockinger et al., 1997; Yamaguchi-Shinozaki and Shinozaki, 1994). The chy1 mutation impairs the ability of the mutant plants to tolerate freezing stress following cold acclimation and to survive dark treatment, especially in the cold. Although mutations in CHY1 previously had been shown to confer developmental defects on seedlings grown in the absence of exogenous sucrose and decreased catabolism of seed storage lipids during germination (Lange et al., 2004; Zolman et al., 2000, 2001a), the importance of CHY1 for plant responses to cold stress and starvation in the dark was not known. Our results suggest a role for peroxisomal metabolism in cold-responsive gene expression and plant responses to cold and dark stresses.
Several lines of evidence support a role for CHY1 in cold signaling and cold tolerance. First, chy1 mutant plants show reduced expression of CBF3-LUC and endogenous CBFs in response to cold treatment (Figure 1). Second, the mutant plants are defective in cold acclimation, as revealed by freezing tolerance assays (Figure 2). Third, cold treatment can prevent darkness-induced injury in the wild-type but not in chy1 mutant plants (Figure 6). Fourth, ectopic expression of CBF3 can partially rescue the chy1 mutant sensitivity to the cold and dark (Figure 8), although the partial rescue of mutant defects might be a general effect of CBF3 overexpression. CHY1 also has a role in plant survival during darkness-induced starvation, as the chy1 mutant has reduced survival and greater electrolyte leakage in darkness even at normal growth temperatures, although both wild-type and mutant plants are more sensitive to dark treatment at room temperature than in the cold (Figure 5).
Interestingly, although chy1-10 plants are impaired in cold induction of CBF3-LUC in the light (Figure 1) as well as in the dark (data not shown), chy1-10 plants are chilling sensitive in the dark but not in the light (data not shown). Light also alleviates the cold sensitivity of los4-1 mutant plants, which were similarly identified as having decreased CBF expression (Gong et al., 2002). los4-1 mutant plants, which are defective in an RNA helicase, are severely damaged after cold treatment for 2 weeks in the dark but not in the light. Although photosynthesis is inefficient at low temperatures (Stitt and Hurry, 2002), light may activate certain cold-tolerance mechanisms, including changes in gene expression. For example, light-induced proline and sucrose accumulation may help protect cells from cold injury (Gong et al., 2002). Indeed, applying a 3% sucrose solution to leaves of soil-grown chy1-10 plants or incubating chy1 seedlings on medium supplemented with 1% sucrose rescued the mutant plants from darkness damage at either 4 or 22°C (data not shown). The protective effects of applied sucrose are consistent with either osmotic protection from chilling damage or alleviation of carbon starvation in the dark.
Peroxisomes are respiratory microbodies bound by a single membrane. Peroxisomal matrix proteins are encoded in the nucleus, synthesized on free ribosomes, and post-translationally imported into the organelle. In the early 1960s, when peroxisomes were first characterized from mammalian tissues, their main function was thought to be removal of toxic hydrogen peroxide. Peroxisomes are now known to be involved in a range of important cellular functions in eukaryotic cells. In plants, peroxisomes are important for fatty acid β-oxidation, the glyoxylate cycle, and photorespiration (reviewed by Olsen, 1998). As a result, peroxisome-defective mutants have seedling establishment defects attributed to decreased seed storage oil utilization during germination (Hayashi et al., 1998; Zolman et al., 2000). Analysis of peroxisome-defective mutants also has revealed that the naturally occurring auxin IBA is converted to the active auxin IAA in the peroxisome, and that this process is important for lateral root formation in developing seedlings (Zolman et al., 2000, 2001b, 2005; Zolman and Bartel, 2004). Moreover, biosynthesis of the defense hormone jasmonic acid requires three rounds of peroxisomal β-oxidation (reviewed by Creelman and Mullet, 1997). Peroxisomal processes also include metabolism of the branched-chain amino acids (Zolman et al., 2001a), propionate, and isobutyrate (Lucas et al., 2007). In addition, a peroxisome biogenesis protein can influence photomorphogenesis (Hu et al., 2002). The dominant ted3 mutation in a peroxisomal protein (PEX2) suppresses the pleiotropic de-etiolated mutant phenotypes of det1 and cop1. Although the precise mechanism is unclear, peroxisomes may release signaling molecules that affect the expression of nuclear genes controlling photomorphogenesis (Hu et al., 2002).
The original chy1 mutant alleles were isolated based on resistance to the inhibitory effects of exogenous IBA on root elongation (Zolman et al., 2001a). These mutants exhibit developmental defects in the absence of exogenous sucrose (Zolman et al., 2001a) and catabolize seed storage lipids poorly during germination (Zolman et al., 2000), indicating defects in peroxisomal fatty acid β-oxidation. Two additional chy1 alleles (dbr5-1 and dbr5-2) were isolated in a screen for 2,4-DB resistant mutants (Lange et al., 2004). CHY1 encodes HIBYL–CoA hydrolase, which is involved in Val catabolism in humans; a human HIBYL–CoA hydrolase cDNA modified to contain a peroxisomal targeting signal can functionally complement the chy1 mutant (Zolman et al., 2001a). As expected, chy1 mutants show reduced levels of Val catabolism (Lange et al., 2004). chy1 mutants also show enhanced sensitivity to exogenous isobutyrate and propionate (Lucas et al., 2007).
The CHY1-encoded HIBYL–CoA hydrolase is involved in catabolism of Val and isobutyrate. During this process, methylacrylyl–CoA is converted to HIBYL–CoA in a reversible reaction. A peculiarity in these pathways is that CHY1 hydrolyzes the CoA thioester, which is later reformed. Disruption of this hydrolysis is hypothesized to cause HIBYL–CoA accumulation and shift the equilibrium from HIBYL–CoA back to methylacrylyl–CoA. Methylacrylyl–CoA is toxic, reacting rapidly with nucleophiles such as cysteine, cysteamine, and glutathione; this conjugation may covalently inactivate enzymes with active site nucleophiles (Brown et al., 1982; Dearfield et al., 1991). Indeed, activity of the β-oxidation enzyme 3-ketoacyl–CoA thiolase is significantly decreased in chy1 mutants (Lange et al., 2004); the IBA-response phenotype and sucrose dependence of chy1 mutants is likely an indirect effect of methylacrylyl–CoA accumulation leading to thiolase inactivation. In addition, methylacrylyl–CoA may disrupt cellular membranes in chy1 mutants, perhaps causing accumulation of reactive oxygen species, electrolyte leakage and impairing cold-induced gene expression. Alternatively, methylacrylyl–CoA may be sequestered in the peroxisome and the ensuing damage may be limited to this organelle. Peroxisomal defects including reduced β-oxidation may disrupt a peroxisome-derived signal that affects cold-induced nuclear gene expression, in a manner analogous to the effect of the ted3 mutation on nuclear gene expression (Hu et al., 2002). It also is possible that potential alterations in auxin response or homeostasis in the chy1 mutant may contribute to the impaired cold stress tolerance of the mutant.
In this work, we demonstrate a connection between peroxisomal metabolism, cold-regulated CBF3 expression, and cold tolerance. chy1 mutants have reduced CBF expression levels. However, interestingly, the levels of three cold-regulated transcripts that are activated by CBFs, including COR15A, COR47A, and RD29A, were not lower in chy1-10 than wild-type. Mutations that either decrease (Lee et al., 2002a, 2002b) or increase (Zhu et al., 2005) RD29A expression in the cold without changing the expression of CBFs have been reported. These observations, together with our findings reported here, suggest that the CBF genes are not the sole transcriptional regulators for RD29A and several other COR/RD genes. The increased expression of RD29A and other COR/RD genes in chy1 despite reduced CBF levels implies that the chy1 mutation enhances a CBF-independent pathway for the activation of the COR/RD genes. The effect of decreased induction of CBFs on cold tolerance in the chy1 mutant plants may be mediated by CBF target genes other than the COR/RD genes examined here. Alternatively, the peroxisomal defect in chyl may affect CBF-independent pathways for cold tolerance. Further examination of chy1 and other peroxisome-defective mutants may clarify the connection between reduced CBF levels, downstream gene expression, and changes in cold and freezing responses.
Our study has uncovered a new role for plant peroxisomes in responses to cold and dark treatment. Low temperatures have large impacts on cellular metabolism (Levitt, 1980). We suggest that integration of metabolic status (e.g. peroxisomal activities) affects plant adaptive responses to cold treatment. Our results suggest the possibility that metabolic status might be connected to cold regulation through ROS. The elucidation of the precise molecular mechanisms underlying the effect of peroxisomes on cold-regulated gene expression will require further investigation.
METHODS
Plant Materials and Mutant Isolation
Construction of CBF3-LUC and generation of transgenic Arabidopsis plants was previously described (Chinnusamy et al., 2003). Arabidopsis thaliana ecotype Columbia (with the glabrous1 mutation) containing CBF3-LUC (hereafter referred to as wild-type) was mutagenized using EMS (ethyl methanesulfonate). M2 seedlings were screened for mutants defective in cold-regulated CBF3-LUC expression by luminescence imaging as described (Chinnusamy et al., 2003). Mutants were backcrossed to wild-type four times to eliminate unlinked mutations.
Gene Expression Analysis
Two-week-old wild-type and mutant plants grown in soil were treated at low temperature (0–4°C) in the light (20 μmol m−2 s−1) or dark. Total RNA was extracted from whole seedlings (above-ground parts) and RNA analysis was performed using gene-specific probes as described (Ishitani et al., 1998).
Electrolyte Leakage and Freezing Tolerance Assays
For chilling tolerance, plants grown in soil in a growth chamber with 16 h light at 22°C, 8 h dark at 18°C, and 75% relative humidity for 2 weeks were transferred to 4°C in dark or constant light. Plants were photographed 24 h after transfer from 4°C to room temperature.
The freezing tolerance assay was carried out as described (Chinnusamy et al., 2003). Ten-day-old seedlings of wild-type and chy1-10 grown on 0.7% agar media were cold acclimated at 4°C in the light for 4 d. These plates were placed on ice in darkness in a freezing chamber set to –1°C for 16 h. Ice chips were sprinkled on the plants before the chamber was programmed to cool at –1°C per hour. Petri dishes of plants were removed after being frozen at the desired temperature for 2 h, thawed at 4°C overnight, and then transferred to a growth chamber at 22°C. Survival of the seedlings was scored visually after 2 d.
The electrolyte leakage test was performed to compare membrane integrity and chilling sensitivity. Two-week-old plants grown in soil were treated at 4 or 22°C in the dark. Several rosette leaves from treated and untreated plants were detached and transferred to tubes with 10 mL deionized water. The conductivity of the solution was measured after shaking overnight at room temperature. After measurement, the samples were autoclaved for 30 min. After shaking at room temperature for two additional hours, the conductivity of the solution was measured again. The percent electrolyte leakage was calculated as the percentage conductivity before versus after autoclaving (Lee et al., 2002b). The leaf electrolyte leakage assay after freezing treatments was carried out as described previously (Ishitani et al., 1998).
Map-Based Cloning of chy1-10
For recombination mapping, the chy1-10 mutant in the Arabidopsis Columbia ecotype was crossed with wild-type plants of the Ler ecotype. F1 plants from the cross were self-fertilized, and the resulting F2 seeds were collected. A total of 718 homozygous chy1-10 mutants were selected from the segregating F2 population using the chilling-sensitive phenotype after cold treatment. Genomic DNA extracted from these seedlings was used for PCR-based mapping. Initial mapping linked the mutation to the markers MBK5 and K9I9 on the bottom of chromosome 5. New mapping markers on MNA5, MPA24, K14B20, K1L20, MSN2, and K8K14 BAC clones were developed based on insertion/deletions identified from the Cereon Arabidopsis polymorphism and Ler sequence collection (www.arabidopsis.org). Genomic DNA corresponding to candidate genes was PCR-amplified from mutant and wild-type plants and sequenced to identify the mutation.
Overexpression of CHY1 (HIBYL–CoA Hydrolase) and CBF3 cDNAs
For chy1-10 complementation, the CHY1 cDNA cloned behind the 35S promoter in a binary vector (Zolman et al., 2001a) was introduced into homozygous chy1-10 mutant plants by Agrobacterium tumefaciens strain GV3101 transformation. T2 transgenic lines resistant to Basta (glufosinate ammonium) were selected and analyzed. The cloning of CBF3 downstream of the strong constitutive super promoter was described (Gong et al., 2002). Agrobacterium strain GV3101 containing this construct was used to transform chy1-10 mutant plants. Hygromycin-resistant transgenic plants were selected and T2 progeny were examined.
Detection of Reactive Oxygen Species
For superoxide detection, leaves detached from 2-week-old plants were vacuum-infiltrated with 0.1 mg mL−1 nitroblue tetrazolium (NBT) in 25 mM Hepes buffer, pH 7.6. After 2 h incubation at room temperature in the dark, samples were transferred to 80% ethanol and treated at 70°C for 10 min. For hydrogen peroxide staining, leaves were vacuum-infiltrated with 0.1 mg mL−1 3,3′-diaminobenzidine (DAB) in 50 mM Tris-acetate buffer, pH 5.0. Samples were incubated for 24 h at room temperature in the dark before transferring to 80% ethanol. For chilling treatment, plants were placed at 4°C in the dark for 2 d before staining. Detection of the reactive oxygen species in seedling roots by CM-H2DCF diacetate, acetyl ester staining was as described (Shin and Schachtman, 2004). Fluorescence imaging was performed using confocal microscopy (Leica TCS SP2).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by NSF grants IBN-0212346, MCB-0241450 (J.-K.Z.), and IBN-0315596 (B.B.), USDA NRI grant 2003–00751 (J.-K. Z.), and the Robert A. Welch Foundation (C-1309 to B.B.).
Supplementary Material
Acknowledgments
We thank Ms. Weiping Tang and Ms. Xuhui Hong for excellent technical assistance. No conflict of interest declared.
References
- Brown GK, Hunt SM, Scholem R, Fowler K, Grimes A, Mercer JF, Truscott RM, Cotton RG, Rogers JG, Danks DM. Beta-hydroxyisobutyryl coenzyme A deacylase deficiency: a defect in valine metabolism associated with physical malformations. Pediatrics. 1982;70:532–538. [PubMed] [Google Scholar]
- Catala R, Santos E, Alonso JM, Ecker JR, Martinez-Zapater JM, Salinas J. Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell. 2003;15:2940–2951. doi: 10.1105/tpc.015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl Acad. Sci. U S A. 2004;101:15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Río LA. Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci. 2001;6:145–150. doi: 10.1016/s1360-1385(01)01898-2. [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:355–387. doi: 10.1146/annurev.arplant.48.1.355. [DOI] [PubMed] [Google Scholar]
- Dearfield KL, Harrington-Brock K, Doerr CL, Rabinowitz JR, Moore MM. Genotoxicity in mouse lymphoma cells of chemicals capable of Michael addition. Mutagenesis. 1991;6:519–525. doi: 10.1093/mutage/6.6.519. [DOI] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu J-K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl Acad. Sci. U S A. 2006;103:8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl Acad. Sci. U S A. 2002;99:11507–11512. doi: 10.1073/pnas.172399299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M. 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell. 1998;10:183–195. doi: 10.1105/tpc.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J. A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science. 2002;297:405–409. doi: 10.1126/science.1073633. [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Lee HJ, Stevenson B, Zhu JK. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell. 1998;10:1151–1161. doi: 10.1105/tpc.10.7.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004;136:4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell. 1996;8:489–503. doi: 10.1105/tpc.8.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange PR, Eastmond PJ, Madagan K, Graham IA. An Arabidopsis mutant disrupted in valine catabolism is also compromised in peroxisomal fatty acid β-oxidation. FEBS Lett. 2004;571:147–153. doi: 10.1016/j.febslet.2004.06.071. [DOI] [PubMed] [Google Scholar]
- Lee BH, Lee H, Xiong L, Zhu JK. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell. 2002a;14:1235–1251. doi: 10.1105/tpc.010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu JK. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bifunctional enolase. EMBO J. 2002b;21:2692–2702. doi: 10.1093/emboj/21.11.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt J. New York: Academic Press; 1980. Responses of Plants to Environmental Stress Vol. 1: Chilling, Freezing,and High Temperature Stress. (New York: Academic Press) [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas KA, Filley JR, Erb JM, Graybill ER, Hawes JW. Peroxisomal metabolism of propionic acid and isobutyric acid in plants. J. Biol. Chem. 2007;282:24980–24989. doi: 10.1074/jbc.M701028200. [DOI] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999;119:463–470. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy AF, Dhindsa RS. Low-temperature signal transduction: induction of cold acclimation-specific genes of alfalfa by calcium at 25 °C. Plant Cell. 1995;7:321–331. doi: 10.1105/tpc.7.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin K, Vahala T, Palva ET. Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 1993;21:641–653. doi: 10.1007/BF00014547. [DOI] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2004;101:3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen LJ. The surprising complexity of peroxisome biogenesis. Plant Mol. Biol. 1998;38:163–189. [PubMed] [Google Scholar]
- Price AH, Taylor A, Ripley SJ, Griffiths A, Trewavas AJ, Knight MR. Oxidative signals in tobacco increase cytosolic calcium. Plant Cell. 1994;6:1301–1310. doi: 10.1105/tpc.6.9.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Schachtman DP. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl Acad. Sci. U S A. 2004;101:8827–8832. doi: 10.1073/pnas.0401707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Hurry V. A plant for all seasons: alterations in photosynthetic carbon metabolism during cold acclimation in Arabidopsis. Curr. Opin. Plant Biol. 2002;5:199–206. doi: 10.1016/s1369-5266(02)00258-3. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl Acad. Sci. U S A. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Welin BV, Olson A, Palva ET. Structure and organization of two closely related low-temperature-induced dhn/lea/rab-like genes in Arabidopsis thaliana L. Heynh. Plant Mol. Biol. 1995;29:391–395. doi: 10.1007/BF00043662. [DOI] [PubMed] [Google Scholar]
- Xin Z, Browse J. Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl Acad. Sci. U S A. 1998;95:7799–7804. doi: 10.1073/pnas.95.13.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, et al. HOS10- encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl Acad. Sci. U S A. 2005;102:9966–9971. doi: 10.1073/pnas.0503960102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zolman BK, Bartel B. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc. Natl Acad. Sci. U S A. 2004;101:1786–1791. doi: 10.1073/pnas.0304368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17:3422–3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SPT, Bartel B. chy1, an Arabidopsis mutant with impaired β-oxidation, is defective in a peroxisomal β-hydroxyisobutyryl–CoA hydrolase. J. Biol. Chem. 2001a;276:31037–31046. doi: 10.1074/jbc.M104679200. [DOI] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 2001b;127:1266–1278. [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.