Abstract
Many stress responsive genes have been reported with an effect on improving stress resistance in model plants under greenhouse conditions. Towards identification of genes for drought resistance breeding, seven well documented genes (CBF3, SOS2, NCED2, NPK1, LOS5, ZAT10, and NHX1) in stress resistance were selected in this study and transformed into rice cultivar Zhonghua 11 under the control of constitutive promoter Actin1 and stress-inducible promoter of a rice HVA22 homolog, and transgenic rice were tested for drought resistance under field conditions. A total of 1598 independent transgenic T0 plants were generated. The percentages of single copy and expression of the transgenes were 36.7% and 57.6%, respectively. For each gene construct, 30 T1 families with expression of transgene were selected for drought resistance testing at the reproductive stage in field, and 10 of them were tested in PVC pipes with a defined stress protocol at the same stage. Relative yield and relative spikelet fertility were used as two major criteria to evaluate drought resistance performance because significantly decreased yield was observed in the T1 generation. Transgenic families of eight constructs (HVA22P:CBF3, HVA22P:NPK1, Actin1:LOS5, HVA22P:LOS5, Actin1:ZAT10, HVA22P:ZAT10, Actin1:NHX1, and HVA22P:NHX1) showed significantly higher RY than wild-type (WT) under both drought stress field and PVC tube conditions. Transgenic families of 9 constructs (HVA22P:SOS2 and CBF3, LOS5, ZAT10, and NHX1 by both promoters) showed significantly higher relative spikelet fertility than WT in the field or PVC pipes. In the field drought resistance testing of T2 families derived from the T1 families with relatively lower yield decrease, transgenic families of seven constructs (HVA22P:CBF3, Actin1:NPK1, HVA22P:NPK1, Actin1:LOS5, HVA22P:LOS5, Actin1:ZAT10, and HVA22P:ZAT10) showed significantly higher yield per plant than WT, and families of nine constructs (Actin1:CBF3, HVA22P:CBF3, HVA22P:SOS2, HVA22P:NPK1, Actin1:LOS5, HVA22P:LOS5, Actin1:ZAT10, HVA22P:ZAT10, and Actin1:NHX1) had higher spikelet fertility than WT. In general, LOS5 and ZAT10 showed relatively better effect than the other five genes in improving drought resistance of transgenic rice under field conditions. The results and experience obtained from this study could be a useful reference for drought resistance engineering in rice.
Keywords: Oryza sativa, abiotic stress, grain yield, transformation
INTRODUCTION
Water deficit, more commonly referred to as ‘drought’, causes major economic losses in crop production throughout the world. For example, global losses of the two major cereal crops, rice and maize, to drought are estimated to be more than $5 billion annually (Data from Food Security, the Rockefeller Foundation). Drought has been, and continues to be, the single most devastating factor that is menacing food production and food security, especially in areas with inadequate agricultural water resources. Consequently, with the global shortage of water, reducing water consumption in crop production has now been generally recognized as an essential strategy for sustainable agriculture. In China, for example, the estimated annual loss of national economy from water shortage alone reaches more than $25 billion (Deng, 1999). It has been estimated that rice production consumes about half of the total water consumption of the country. However, drought stress is still the single most important constraint in rice production (Lin and Shen, 1996), mostly due to variation in the rainfall patterns from one year to another, and also uneven distribution of rainfall in the rice growing season. This is especially true in the marginal mountain areas that are far away from river basins. Thus, there is an urgent need for reducing water consumption especially in rice production, by increasing the tolerance of this crop to reduced water supply.
In recent years, the idea of developing drought-tolerant crops has been well recognized as the most promising and effective strategy for food security against drought and water shortage. However, drought tolerance is a complex trait that involves numerous aspects of developmental, physiological, biochemical, and molecular adjustments. These include, for example, changes in root growth, guard cell regulation, osmotic adjustment, alterations in photosynthesis, and synthesis of protective proteins and antioxidants. The regulatory pathways leading to these adjustments are poorly understood and remain a focal point of research (Zhu, 2002). Nevertheless, a number of genes have been demonstrated to be important for drought tolerance, and genetic engineering using some of these genes has shown promise in improving plant drought tolerance in laboratory tests.
Under drought stress, plants accumulate ABA, which is critical for stomatal closure leading to reduced transpirational water loss, and induces the expression of many genes with presumed protective roles (Zhu, 2002). Recently, all the genes encoding enzymes required for ABA biosynthesis have been cloned in Arabidopsis. Overexpression of two of these enzymes, NCED2 (Iuchi et al., 2001; Qin and Zeevaart, 2002; Thompson et al., 2000) and LOS5 (Xiong et al., 2001), led to increased ABA production, and reduced leaf transpiration under drought conditions, and consequently increased drought tolerance of the transgenic plants. Many reports have documented that engineered production of compatible osmolytes, such as trehalose and fructan, confers improved drought tolerance in plants (Holmstrom et al., 1996; Pilon-Smits et al., 1999). Like many other abiotic stresses, drought leads to the accumulation of reactive oxygen species, which causes damage to cellular structures, particularly membranes. Transgenic plants overexpressing enzymes that are involved in the detoxification of reactive oxygen species have improved tolerance to drought stress (McKersie et al., 1996). In addition, overexpression of certain individual stress protein or transcription factor regulating multiple stress proteins was shown to confer increased tolerance to drought as well as to salt and freezing stresses (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Xu et al., 1996). Drought and salt stress tolerance share many common mechanisms. However, unique to salt stress is the excess of toxic ions such as Na+ and Cl–. Overexpression of NHX1, a vacuolar Na+/H+ antiporter, has been reported to increase the capacity of plant cells to store Na+ in the vacuole, and to improve salt tolerance in transgenic plants (Apse et al., 1999). A regulatory pathway that coordinately controls the transport of Na+ across cellular membranes has been established (Zhu, 2002).
In rice, efforts have also been made in testing genes for drought tolerance. Expression of a fused bacteria gene TPS–TPP in rice significantly increased the level of trehalose, resulting in enhanced drought tolerance (Garg et al., 2002; Jang et al., 2003). Overexpression of a rice MAPK can significantly increase the tolerance of rice to drought, salinity, and cold (Xiong and Yang, 2003). Recently, transcription factor gene CBF3/DREB1 (Oh et al., 2005) and the rice DREB1 homologue (Ito et al., 2006) have been reported for their effectiveness on improving stress tolerance in transgenic rice. Although these results were from greenhouse tests, it suggests that genetic engineering is a promising strategy to explore a wide range of genes that are potential for drought tolerance improvement in economically important crops. Hu et al. (2006) reported that overexpression of a stress-responsive transcription factor in rice resulted in significantly improved drought resistance under the field conditions, further supporting the possibility of developing drought resistance rice by transgenic approach.
Even though many stress resistance genes have been identified in non-crop species such as Arabidopsis, comparison of the effect of these genes on improving stress resistance in a given crop has seldom been reported. Toward a long-term goal of developing drought-resistant irrigated rice with good adaptation to drought-prone areas, we tested several functionally characterized genes in this study for their effects on improving drought tolerance in rice by developing transgenic rice lines overexpressing these genes and testing for drought resistance under field conditions.
RESULTS
Rice Transformation and Identification of Transgenic Plants
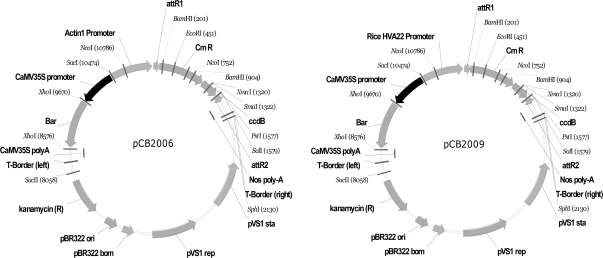
To harbor target genes, two backbone vectors containing a constitutive promoter (from rice Actin1 gene) and an inducible promoter (from a rice homologous gene of HVA22), respectively, were constructed (Figure 1). Then, seven genes (CBF3, SOS2, NCED2, NPK1, LOS5, ZAT10, and NHX1) reported to confer stress tolerance in model plants were constructed into these two vectors and the resultant gene constructs were introduced into Agrobacterium strain EHA105 by electroporation and used for transforming rice Zhonghua 11 (Oryza sativa L. ssp. japonica). A total of 1598 independent T0 transformants were generated for the 14 constructs and seeds were available for 1130 T1 families (Table 1).
Figure 1.
The Sketch Structure of pCB2006 and pCB2009 Vectors Used for Rice Transformation.
Vectors pCB2006 and pCB2009 contained the constitutive Actin1 promoter and the stress-inducible HVA22P promoter from rice, respectively.
Table 1.
Rice Transformation and Molecular Identification of Candidate Genes.
| Gene | Promoter | Total T0 plants | T1 families with seeds availablea | Transgenic copy |
Overexpression (%)b | ||
| Single copy (%)b | 2–3 copies (%)b | ||||||
| CBF3 | Actin1 | 130 | 87 | 34.5 | 46.4 | 62.3 | |
| HVA22P | 142 | 112 | 33.6 | 44.2 | 55.5 | ||
| SOS2 | Actin1 | 78 | 72 | 32.2 | 43.6 | 68.5 | |
| HVA22P | 82 | 81 | 34.6 | 41.3 | 51.4 | ||
| NCED2 | Actin1 | 96 | 84 | 36.8 | 47.4 | 66.1 | |
| HVA22P | 78 | 74 | 34.1 | 45.6 | 49.8 | ||
| NPK1 | Actin1 | 100 | 63 | 41.2 | 48.2 | 59.3 | |
| HVA22P | 95 | 60 | 38.4 | 45.7 | 43.2 | ||
| LOS5 | Actin1 | 181 | 111 | 36.3 | 46.5 | 68.2 | |
| HVA22P | 102 | 88 | 34.8 | 47.5 | 55.4 | ||
| ZAT10 | Actin1 | 92 | 63 | 36.6 | 44.4 | 62.6 | |
| HVA22P | 196 | 66 | 41.2 | 48.6 | 50.8 | ||
| NHX1 | Actin1 | 127 | 94 | 40.5 | 48.1 | 69.6 | |
| HVA22P | 99 | 75 | 39.5 | 45.2 | 43.4 | ||
| Total | 1598 | 1130 | Avg. | 36.7 | 45.9 | 57.6 | |
Seeds were harvested only from the T0 plants without obvious morphological change compared to the wild-type plants.
More than 50 independent T0 plants for each construct were used for detection of copy number and expression level.
Transgenic plants were identified by PCR using primers specific to the Bar gene. Among more than 900 independent plants randomly selected from transformants of 14 constructs, more than 95% of them are PCR-positive. Southern blotting was performed to detect the copy number of transgene by using the Bar gene fragment (amplified by the Bar-specific primers) as a probe. The percentage of single copy transgene (more than 700 PCR-positive independent plants have been tested) was 36.7%, and the percentage of two to three copies was 45.9% (Table 1 and Supplemental Figure 1). The expression level of the transgenes in the mixed leaf tissues from each T1 family derived from the T0 transgenic plants was checked by RNA gel blot analysis using the sequences of transgenes as probes (Supplemental Figure 1). Among more than 700 independent families tested, the percentage of overexpression was about 57.6%. The percentage of overexpression from constitutive expression vector pCB2006 was much higher than that from vector pCB2009 for each of the transgenes (Table 1).
Pre-Screening of Drought Resistance in Drought-Prone Field
Considering the huge number of transgenic plants generated and the limited space of drought testing shelter, drought resistance pre-screening was performed in drought-prone field for T1 families from the T0 plants with available seeds. A total of 1130 T1 families for 14 constructs were used for pre-screening in the drought-prone field (Table 2).
Table 2.
Sensitivity of T1 Transgenic Families to Drought Stress Evaluated by Leaf Drying Score.
| Gene | Promoter | T1 families screened | Families with LDS < 1 | Families with LDS at 1–3 | % of relatively resistant families |
| CBF3 | Actin1 | 87 | 8 | 55 | 72.4 |
| HVA22P | 112 | 13 | 62 | 67.0 | |
| SOS2 | Actin1 | 72 | 6 | 27 | 45.8 |
| HVA22P | 81 | 4 | 34 | 46.9 | |
| NCED2 | Actin1 | 84 | 2 | 28 | 35.7 |
| HVA22P | 74 | 3 | 34 | 50.0 | |
| NPK1 | Actin1 | 63 | 0 | 33 | 52.4 |
| HVA22P | 60 | 0 | 31 | 51.7 | |
| LOS5 | Actin1 | 111 | 7 | 64 | 64.0 |
| HVA22P | 88 | 6 | 52 | 65.9 | |
| ZAT10 | Actin1 | 63 | 7 | 31 | 60.3 |
| HVA22P | 66 | 6 | 33 | 59.1 | |
| NHX1 | Actin1 | 94 | 0 | 32 | 34.0 |
| HVA22P | 75 | 0 | 35 | 46.7 | |
| Total | 1130 | 62 | 551 |
LDS: Leaf drying score based on visual evaluation of the dried area of top leaves at 1 week after flowering. 0: no dried leaves; 1: fewer than 5%; 2: 5–10%; 3: 10–15%; 4: 15–20%; 5: >20%. Scoring was performed at noon when the LDS of neighboring control plants was more than 4. The score for each family is the average performance of the 16 plants in the middle of each plot.
Among the 1130 families tested, 127 families showed visible morphological segregation, such as changes of plant height, flowering, leaf color or shape, panicle shape, etc. (data not shown), and these families were excluded from drought resistance scoring because these phenotypic changes were not likely a result of the transgenes, since most of the families with the expression of transgenes confirmed by Northern blotting showed normal phenotypes.
The drought stress in one replication developed unexpected slowly, probably because of the paddy soil and relatively low position of the field, and no significant stress was developed at the time of flowering. The evaluation was therefore based on another replication. The performance of drought resistance was evaluated mainly based on the degree of leaf drying score (LDS) at the end of flowering. Relative fertility was not used because an unexpected low temperature hit the flowering stage and the fertility was generally low (less than 50%) for all plants. In the pre-screening test, 62 families from expression constructs of five candidate genes (CBF3, SOS2, NCED2, LOS5, and ZAT10) exhibited very high resistance (LDS < 1) to drought stress compared to the wild-type (WT) plants (LDS ≥ 4); 551 families (1 ≤ LDS ≤ 3) from all the stress responsive genes showed fewer drying leaves than WT control (Table 2).
The percentage of relatively resistant families and the degree of resistance showed difference between genes and/or promoters. Generally, the transgenic plants of CBF3 and LOS5 had better drought resistance performance than other genes in the pre-screening. The effects of constitutive promoter and inducible promoter in improving drought resistance were not always consistent among these candidate genes. For example, the constitutive promoter had relatively better effect for CBF3, but the inducible promoter had relatively better effect for NCED2 and NHX1 (Table 2). The relatively resistant families from the pre-screening were selected for drought resistance in the field under the rain-off shelter and in PVC tubes for more accurate evaluation of drought resistance.
Drought Resistance Testing under Managed Drought-Stressed Conditions
Thirty transgenic families with expression of the transgene, one to three copies of T-DNA, and relative resistance with LDS from 0 (high resistance) to 3 (moderate resistance) obtained from the pre-screening were selected from each of the 14 gene constructs (seven candidate genes by two types of promoters) and tested for drought resistance under the managed drought-stressed conditions. Considering the segregation of transgenes in T1 or T2 generation, herbicide was applied at seedling stage to select positive transgenic plants for transplanting, since the constructs contain the Bar gene. Drought testing was conducted in two environments: refined field equipped with a movable rain-off shelter and PVC tubes placed in plastic tents.
In the field, yield-related traits (panicle number, grain number, spikelet fertility, 1000-grain-weight) and leaf drying score were measured after maturity. Leaf drying score was significantly negatively correlated with yield and yield-related traits, whereas yield and yield-related traits are positively correlated with each other (Table 3). Under the normal growth conditions, most of the transgenic families showed significantly lower yield per plant and spikelet fertility than the WT—a phenomenon referred to as yield decrease (Tables 4 and 5), which might be due to the negative effect of tissue culture, since the testing was conducted in early generation (T1). In fact, yield decrease seems to be very frequent in transgenic rice produced by tissue culture. For example, only 3–5% of Bt gene transgenic plants generated in our laboratory showed no yield decrease in early generations (Y. Lin, unpublished data). Therefore, drought resistance in this experiment was evaluated by using relative yield and relative spikelet fertility as major parameters, which were considered as the most indicative criteria for drought resistance (Yue et al., 2006).
Table 3.
Correlation Coefficient between Yield and Yield-Related Traits in Drought Testing in the Field.
| Grain yield per plant (GY) | Leaf drying score (LDS) | Panicle number (PN) | Grain number (GN) | Spikelet fertility (SF) | 1000-grain-weight (KGW) | |
| GY | 1.000 | |||||
| LDS | −0.307* | 1.000 | ||||
| PN | 0.7786* | −0.265* | 1.000 | |||
| GN | 0.989* | −0.309* | 0.773* | 1.000 | ||
| SF | 0.511* | −0.215* | 0.139* | 0.518* | 1.000 | |
| KGW | 0.453* | −0.197* | 0.267* | 0.347* | 0.225** | 1.000 |
Significant at P < 0.01 level.
Table 4.
Grain Yield of T1 Transgenic Families from Each Construct under Normal Growth and Two Drought Stress Conditions.
| Construct (promoter:gene) | Grain yield (g per plant)a |
Relative yieldb |
|||
| Normal growth | Drought stress in field | Drought stress in PVC | Drought stress field | Drought stress PVC | |
| Wild-type (ZH11) | 32.55 ± 1.98 | 7.64 ± 0.23 | 21.11 ± 0.92 | 0.23 | 0.65 |
| Actin1:CBF3 | 17.65 ± 0.89** | 3.74 ± 0.30* | 13.76 ± 0.62** | 0.21 | 0.78** |
| HVA22P:CBF3 | 14.59 ± 0.67** | 4.80 ± 0.25* | 12.42 ± 0.52** | 0.33** | 0.85** |
| Actin1:SOS2 | 17.70 ± 0.89** | 4.36 ± 0.33* | 12.32 ± 0.61** | 0.25 | 0.69 |
| HVA22P:SOS2 | 15.91 ± 0.81** | 7.30 ± 0.31 | 9.96 ± 0.55** | 0.46** | 0.59 |
| Actin1:NCED2 | 22.87 ± 0.79* | 4.89 ± 0.28* | 13.14 ± 0.59** | 0.21 | 0.57 |
| HVA22P:NCED2 | 22.20 ± 2.29* | 3.98 ± 0.28* | 13.96 ± 0.73** | 0.18 | 0.63 |
| Actin1:NPK1 | 18.50 ± 1.07** | 2.78 ± 0.19** | 14.73 ± 0.60** | 0.15* | 0.81** |
| HVA22P:NPK1 | 17.42 ± 1.10** | 5.15 ± 0.27 | 14.36 ± 0.69** | 0.30* | 0.83** |
| Actin1:LOS5 | 16.79 ± 0.66** | 7.81 ± 0.39 | 13.23 ± 0.61** | 0.47** | 0.79** |
| HVA22P:LOS5 | 14.14 ± 1.32** | 4.83 ± 0.34* | 11.10 ± 0.66** | 0.34** | 0.79** |
| Actin1:ZAT10 | 15.99 ± 0.79** | 6.62 ± 0.39 | 11.92 ± 0.54** | 0.41** | 0.75* |
| HVA22P:ZAT10 | 15.76 ± 0.85** | 6.08 ± 0.25 | 12.77 ± 0.51** | 0.39** | 0.81** |
| Actin1:NHX1 | 17.76 ± 0.90** | 6.75 ± 0.28 | 12.57 ± 0.56** | 0.38** | 0.71* |
| HVA22P:NHX1 | 13.05 ± 1.01** | 3.56 ± 0.25* | 9.97 ± 0.49** | 0.27* | 0.76* |
The value is mean ± SE (n = 30 and 10 families for treatments in the field and PVC, respectively), and 16 (in the field) or 10 (in PVC) plants for each family measured.
The values are the ratios of measurement under stress to that under normal growth.
** and * indicate significant difference (by LSD test) between transgenic families from each construct and the wild-type at the probability level of P = 0.01 and P = 0.05, respectively. Significantly higher values in transgenic rice are highlighted in bold.
Table 5.
Spikelet Fertility of T1 Transgenic Families from Each Construct under Normal Growth and Two Drought Stress Conditions.
| Construct | Spikelet fertility (%) |
Relative spikelet fertility |
|||
| Normal growth | Drought stress in field | Drought stress in PVC | Drought stress field | Drought stress PVC | |
| Wild-type (ZH11) | 76.7 ± 1.1 | 34.4 ± 1.0 | 58.7 ± 3.7 | 0.45 | 0.77 |
| Actin1:CBF3 | 62.5 ± 2.8** | 21.2 ± 1.0* | 53.6 ± 3.3 | 0.34 | 0.86* |
| HVA22P:CBF3 | 61.5 ± 3.0** | 15.7 ± 1.2** | 43.8 ± 3.0** | 0.26* | 0.89** |
| Actin1:SOS2 | 62.0 ± 1.4** | 30.1 ± 1.4 | 50.6 ± 3.3 | 0.49 | 0.82 |
| HVA22P:SOS2 | 62.3 ± 1.1** | 48.4 ± 1.2** | 46.3 ± 3.2* | 0.78** | 0.74 |
| Actin1:NCED2 | 61.7 ± 1.3** | 13.5 ± 1.1** | 45.5 ± 4.0* | 0.22* | 0.73 |
| HVA22P:NCED2 | 60.0 ± 1.6** | 21.2 ± 1.0* | 47.6 ± 3.1* | 0.35 | 0.79 |
| Actin1:NPK1 | 64.4 ± 2.4* | 13.6 ± 0.7** | 59.7 ± 3.2 | 0.21* | 0.82 |
| HVA22P:NPK1 | 64.7 ± 1.1* | 26.4 ± 1.2 | 55.7 ± 3.8 | 0.41 | 0.72 |
| Actin1:LOS5 | 64.3 ± 1.5** | 26.1 ± 1.0 | 53.6 ± 3.5 | 0.41 | 0.83* |
| HVA22P:LOS5 | 68.3 ± 1.9* | 26.3 ± 1.5 | 56.1 ± 4.3 | 0.39 | 0.83* |
| Actin1:ZAT10 | 70.4 ± 2.7 | 40.8 ± 1.6* | 60.4 ± 3.2 | 0.58* | 0.86* |
| HVA22P:ZAT10 | 61.9 ± 2.8* | 27.0 ± 1.1 | 60.2 ± 3.7 | 0.44 | 0.97** |
| Actin1:NHX1 | 68.7 ± 1.8* | 23.0 ± 0.9** | 57.3 ± 2.9 | 0.33 | 0.93* |
| HVA22P:NHX1 | 69.5 ± 1.7 | 19.9 ± 1.2** | 50.2 ± 3.9* | 0.29* | 0.86* |
The value is mean ± SE (n = 30 and 10 families for treatments in field and PVC, respectively). For each family, 16 and 10 plants were measured for treatments in the field and PVC, respectively.
** and * indicate significant difference (by LSD test) between transgenic families from each construct and the wild-type at the probability level of P = 0.01 and P = 0.05, respectively. Significantly higher values in transgenic rice are highlighted in bold.
Transgenic families from nine constructs (involving all the other six genes, excluding NCED2) showed significantly higher relative yield than WT in the drought testing in the field, whereas transgenic plants for 10 constructs (involving all the other five genes, excluding NCED2 and SOS2) showed significantly higher relative yield than WT under the drought stress in the PVC pipes (Table 4). Transgenic families of eight constructs (HVA22P:CBF3, HVA22P:NPK1, Actin1:LOS5, HVA22P:LOS5, Actin1:ZAT10, HVA22P:ZAT10, Actin1:NHX1, and HVA22P:NHX1) involving five genes (CBF3, NPK1, LOS5, ZAT10, and NHX1) showed significantly higher relative yield than WT under both drought stress field and PVC tube conditions. Among seven stress responsive genes used in this study, transgenic families of three genes (LOS5, ZAT10, and NHX1) under the control of both stress-inducible promoter (Actin1) and constitutive promoter (HVA22P) showed significantly higher relative yield than WT in two drought-stressed conditions.
When relative spikelet fertility was used as an evaluation parameter of drought resistance (Table 5), only the transgenic families of two gene constructs (HVA22P:SOS2 and Actin1:ZAT10) showed significantly higher relative spikelet fertility than WT in the drought testing under field conditions. However, transgenic plants of four genes (CBF3, LOS5, ZAT10, and NHX1) under the control of both promoters showed significantly higher relative spikelet fertility than WT in PVC pipes.
Drought Resistance Re-Testing of T2 Transgenic Families
The above drought resistance testing of T1 families was compared on the basis of construct-wise. Considering the relative high frequency of yield decrease in transgenic rice, especially in T1 generation (Table 4), T2 seeds were harvested from the T1 families without obvious yield decrease in the previous experiment and selected for further drought resistance testing. Generally, more families without yield decrease were identified from the inducible constructs than from the constitutive constructs; the selected families from each gene construct for drought resistance re-testing are listed in Table 6. Obviously, T2 transgenic families from each construct had no significant difference in yield/spikelet fertility with the WT under normal irrigation growth conditions, suggesting that the yield decrease of selected transgenic families was very low in T2 generation and could be neglected in this experiment. Therefore, absolute value of yield per plant or spikelet fertility was used for assessing drought resistance of each construct and the WT. Under the severe drought stress field conditions, seven constructs (HVA22P:CBF3, Actin1:NPK1, HVA22P:NPK1, Actin1:LOS5, HVA22P:LOS5, Actin1:ZAT10, and HVA22P:ZAT10) showed significantly enhanced drought resistance compared to the WT measured by yield per plant, and nine constructs (Actin1:CBF3, HVA22P:CBF3, HVA22P:SOS2, HVA22P:NPK1, Actin1:LOS5, HVA22P:LOS5, Actin1:ZAT10, HVA22P:ZAT10, and Actin1:NHX1) measured by spikelet fertility (Table 6). Overall, transgenic families from constructs HVA22P:CBF3, HVA22P:NPK1, Actin1:LOS5, HVA22P:LOS5, Actin1:ZAT10, and HVA22P:ZAT10 exhibited significantly higher yield per plant and spikelet fertility than the WT, while transgenic families from four constructs (Actin1:SOS2, Actin1:NCED2, HVA22P:NCED2, and HVA22P:NHX1) showed no effect on drought resistance using both yield per plant and spikelet fertility as criteria, and four constructs (Actin1:CBF3, HVA22P:SOS2, Actin1:NPK1, and Actin1:NHX1) exhibited drought resistance only at the level of either yield per plant or spikelet fertility.
Table 6.
Yield and Spikelet Fertility of T2 Transgenic Families from Each Construct under Normal Growth and Stress Conditions in the Drought Resistance Re-Testing.
| Construct | No. of tested familiesa | Yield per plant (g)b |
Spikelet fertility (%)b |
||
| Normal growth | Drought stress | Normal growth | Drought stress | ||
| Wild-type (ZH11) | 20 | 29.5 ± 3.5 | 5.3 ± 0.5 | 72.4 ± 5.6 | 10.2 ± 1.2 |
| Actin1:CBF3 | 8 | 27.2 ± 0.5 | 5.5 ± 0.6 | 70.8 ± 8.5 | 12.3 ± 2.2* |
| HVA22P:CBF3 | 8 | 27.3 ± 2.7 | 5.9 ± 0.6* | 72.6 ± 3.4 | 14.5 ± 2.4** |
| Actin1:SOS2 | 4 | 27.1 ± 2.1 | 4.8 ± 0.7 | 70.4 ± 3.9 | 10.1 ± 2.0 |
| HVA22P:SOS2 | 6 | 27.3 ± 3.6 | 5.2 ± 0.4 | 68.6 ± 7.3 | 12.3 ± 2.4* |
| Actin1:NCED2 | 4 | 27.7 ± 3.2 | 5.4 ± 0.5 | 68.2 ± 6.1 | 10.5 ± 2.6 |
| HVA22P:NCED2 | 8 | 27.8 ± 4.3 | 4.8 ± 0.8 | 68.4 ± 8.3 | 11.3 ± 3.6 |
| Actin1:NPK1 | 10 | 29.3 ± 1.8 | 5.9 ± 1.1* | 66.5 ± 6.2 | 10.3 ± 2.3 |
| HVA22P:NPK1 | 11 | 28.4 ± 3.1 | 6.8 ± 0.8** | 69.0 ± 7.3 | 12.5 ± 2.3* |
| Actin1:LOS5 | 7 | 28.5 ± 2.1 | 6.4 ± 0.7** | 74.2 ± 3.4 | 15.6 ± 2.5** |
| HVA22P:LOS5 | 8 | 27.3 ± 2.3 | 6.7 ± 1.2** | 67.7 ± 7.9 | 15.3 ± 2.1** |
| Actin1:ZAT10 | 8 | 27.8 ± 3.4 | 6.2 ± 0.8** | 73.2 ± 1.8 | 14.4 ± 2.1** |
| HVA22P:ZAT10 | 10 | 28.4 ± 1.5 | 7.2 ± 1.2** | 66.9 ± 6.5 | 16.6 ± 3.2** |
| Actin1:NHX1 | 5 | 28.5 ± 3.1 | 5.6 ± 0.5 | 71.2 ± 2.5 | 12.1 ± 2.2* |
| HVA22P:NHX1 | 5 | 28.4 ± 2.3 | 5.4 ± 0.6 | 69.4 ± 6.7 | 11.3 ± 2.8 |
Families were selected from 30 families for each construct and had no significant difference with the wild-type for grain yield per plant under unstressed field conditions (P = 0.05).
The value is the mean of tested families without yield decrease for each gene construct in terms of yield per plant and spikelet fertility. The value of the wild-type is the mean of 20 plots distributed evenly in the field.
** and * indicate significant difference (by LSD test) between transgenic families from each construct and the wild-type at the probability level of P = 0.01 and P = 0.05, respectively. Significantly higher values in transgenic rice are highlighted in bold.
In summary, transgenic families from candidate genes LOS5, ZAT10, CBF3, and NPK1 exhibited significantly enhanced drought resistance measured by yield per plant and/or spikelet fertility under the control of the constitutive (Actin1) and stress-inducible (HVA22P) promoters. Transgenic families of SOS5 and NHX1 showed significantly improved spikelet fertility only under the control of Actin1 or HVA22P promoter. Strangely, no drought resistance phenotype was detected for NCED2 gene controlled by either promoters measured by either yield or spikelet fertility (Table 6). Though the number of families without yield decrease was rather limited for each construct in this experiment, families with significantly improved drought resistance can still be identified in drought stress field conditions, suggesting that overexpression of these genes can improve drought resistance of rice.
DISCUSSION
Improving the Drought Resistance of Rice by Means of the Genetic Manipulation of Stress Responsive Genes in Field Conditions
To date, there have been many reports of the development of transgenic plants with improved drought resistance by manipulation of the expression of stress-related genes in laboratory or greenhouse conditions (Dubouzet et al., 2003; Garg et al., 2002; Holmstrom et al., 1996; Park et al., 2005; Shen et al., 1997; Xiong and Yang, 2003; Xu et al., 1996). However, there are very few studies in which drought resistance of transgenic plants has been tested in the field (Hu et al., 2006; Wang et al., 2005). The results obtained under laboratory or greenhouse conditions may be partially consistent with those obtained in the field, but must be further confirmed in drought-stressed field environments (Shao et al., 2005).
In this study, T1 transgenic families were firstly pre-screened for drought resistance in terms of leaf drying score (LDS), and some morphological traits were also investigated in the field (Table 2). Visual phenotypes of drought resistance and morphologic traits from the drought resistance pre-screening were beneficial for excluding the families with abnormal morphology and reducing the workload of field screening. These transgenic families with rather low LDS were selected for drought resistance testing in the field and PVC tubes conditions. Relative yield and relative spikelet fertility were used as two major criteria to evaluate drought resistance performance considering the severe yield decrease in T1 generation. The results (Tables 4 and 5) showed that constitutive and/or inducible overexpression of stress responsive genes (CBF3, LOS5, NCED2, NHX1, SOS2, ZAT10, and NPK1) conferred a different extent of drought resistance, reflected by the relative yield and relative spikelet fertility. For major crops (e.g. rice, wheat, and maize), yield is the ultimate goal for crop production and yield performance in the drought-stressed field is the most important criterion for assessing drought resistance (Turner, 1997). Thus, T2 families derived from the T1 families without obvious yield decrease in the above experiment were selected for further drought resistance retesting in field conditions using yield per plant and spikelet fertility as criteria. The results from drought resistance re-testing showed that constitutive and/or inducible overexpression of all the candidate genes except NCED2 can contribute to the improvement of drought resistance at the level of yield per plant and/or spikelet fertility. The number of transgenic families used in drought resistance experiments was perhaps limited for NCED2 (four families for Actin1:NCED2, eight families for HVA22P:NCED2 construct), which might be the reason why the promising transgenic families with significant drought resistance have not been identified from this gene. Although the overexpression of a single stress-responsive gene can contribute to the improvement of drought resistance to some extent, the level of drought resistance had a big gap towards practical agriculture production. Gene stacking by overexpressing a series of stress-responsive genes was undoubtedly a promising strategy for comprehensively improving the resistance of drought and other abiotic stresses (Chen et al., 2005). Different stress responsive genes may be clustered by means of crossing some transgenic families with promising drought resistance conferred by different candidate genes in this study.
Yield Decrease Associated with Agrobacterium-Mediated Transformation in Rice
Although the T1 families selected from drought pre-screening exhibited the same phenotypes as WT for the majority of morphological traits (e.g. plant height, plant structure, number of tillers, and flowering time), most of these transgenic families had significantly lower grain yield than WT under normal irrigation conditions (Tables 4 and 5). This yield decrease may be due to several effects associated with the Agrobacterium-mediated transformation of rice. First, the transgenic plants were derived from tissue culture that may have potential detrimental effects, particularly in the early generation, on growth and productivity (Chen et al., 2005; Kasuga et al., 1999; Liu et al., 1998; Stam et al., 1997). Second, since Agrobacterium-mediated transformation generates random insertions of T-DAN into the recipient genome (Hiei et al., 1994; Wu et al., 2003) and yield is associated with many genes (Yoon et al., 2006), it is possible that some genes related to yield were disrupted. Third, introduction of the transgene may lead to genetic or physiological incompatibility (Holmberg and Bulow, 1998; Meyer, 2000; Romero et al., 1997; Stempak et al., 2005).
In this study, a large number of transgenic families were pre-screened for drought resistance. About 100 transgenic families with improved drought resistance were selected from more than 1500 independent transformants in the re-testing of drought resistance. In order to obtain transgenic families with better drought resistance, more transgenic families should be screened through many cycles of drought stress testing. We believe that such a bipartite (stress and non-stress) in-field screening protocol, with minor modification, can be successfully applied to the field testing of other transgenic crops.
Conclusions
Taking all the results together, most of the exogenous candidate genes showed certain effects in improving drought resistance of rice at the level of yield per plant and/or spikelet. These transgenic families with significantly improved drought resistance, normal phenotype, single copy and stable overexpression of transgene may be selected with priority for producing rice cultivars for drought resistance in the next phase. We have also learned that, for the purpose of producing drought-resistant rice, it is very important to screen a relatively large population of independent transgenic families for yield under both normal and stressed conditions.
METHODS
Construction of Expression Binary Vectors
To facilitate the construction of a fairly large number of candidate genes in a short time, we designed and constructed a versatile base vector pCB2003 featuring efficient replacement of promoters and high-throughput insertion of candidate genes. The base vector pCB2003 was derived from pCAMBIA3301, which was based on the pZP200 binary vector featuring a plasmid high yield due to the high copy number replication origin of pBR322, relatively small size and capability of triparental mating. In pCAMBIA3301, a CaMV 35S promoter-driving Bar cassette is placed next to the left T-DNA border within the T-DNA, while a GUS reporter cassette is placed next to the T-DNA right border. To create the base vector pCB2003, a multiple cloning site composed of SacI–DraIII (a)–AscI–AvrII–SwaI–DraIII (b) (GGG in DraIII (a) and TTT in DraIII (b)) was first inserted upstream of the 35S promoter, then the whole GUS reporter cassette was replaced by a Gateway Conversion fragment that contains the chloramphenicol-resistance gene for counter selection and the ccdB gene for negative selection. This base vector pCB2003 is 9983 bp in size and features the glufosinate ammonium herbicide selection in both monocot and dicot plants, a versatile multiple cloning site for directional insertion of promoters, and the high throughput insertion of candidate genes using Gateway Technology.
Based on the vector pCB2003, several expression binary vectors were derived, including pCB2006 and pCB2009 for constitutive and stress-inducible expression of candidate genes, respectively (Lei et al., 2007). The rice Actin1 promoter was amplified by PCR based on the published sequence of the Actin1 promoter (McElroy et al., 1990) inserted into pCB2003 between two DraIII sites, which resulted in expression vector pCB2006 (Figure 1). To construct a drought-inducible vector, we first isolated from rice a drought-inducible promoter of the HVA22 homologous gene (LOC_Os08g36440) that encodes a late embryogenesis protein and shows above 80% similarity of amino acid sequence to the barley HVA22 gene (Shen et al., 2001). This promoter, named HVA22P, was then inserted into pCB2003 using DraIII sites, resulting in an expression construct pCB2009 for driving candidate genes (Figure 1).
The cDNA templates of candidate genes (CBF3, SOS2, NCED2, NPK1, LOS5, ZAT10, and NHX1) were provided by the original inventors. PCR products of candidate genes were obtained by amplification of the full-length cDNAs of the genes with BP-reaction-adapted primers and cloned into Donor vector pDNOR207. Then, the candidate genes were introduced into expression vectors pCB2006 and pCB2009 by LR recombination reaction following the standard protocol of Gateway recombination cloning Technology.
Rice Transformation
All expression vectors were introduced into japonica rice Zhonghua 11 (drought-sensitive) by Agrobacterium-mediated transformation (Hiei et al., 1994; Lin and Zhang, 2005). The embryonic calli from Zhonghua 11 seeds were cultured for 3 d at 28°C with the Agrobacterium strain EHA105 that carried the cDNA constructs and then transferred to the selection medium containing 200 μg mL−1 PPT (phosphinothricin) and 200 μg mL−1 carbenicillin. After two to three cycles (2 weeks per cycle) of selection, resistant calli were transferred to the pre-regeneration medium containing 100 μg mL−1 PPT. After 7 d, the resistant calli were transferred to the regeneration medium without PPT to regenerate plantlets.
PCR, Southern, and RNA Gel Blot Analysis
Genomic DNA was extracted by using the CTAB method (Zhang et al., 1992). The Bar gene-specific primers (5′-AACCCACGTCATGCCAGTT-3′ and 5′-TCGTCAACCACTACATCGAGA-3′) were used to identify positive transgenic plants. PCR reaction was conducted in a volume of 20 μL containing 100 ng genomic DNA, 2 mM MgCl2, 0.2 mM of each dNTP, 1 PCR buffer, 0.2 μM of each primer, and 1 unit rTaq Polymerase (TaKaRa, Dalian, China). The PCR reaction was performed at 94°C for 5 min, then with 30 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 1 min, and finally at 72°C for 5 min.
Copy number of transgene was determined by Southern blot analysis using the Bar gene fragment as a probe (amplified by the two Bar-specific primers). Three micrograms of genomic DNA from each sample was digested with EcoRI, fractionated on 0.7% agarose gel, and blotted onto nylon membranes that were hybridized with a 32P-dCTP-labeled Bar-specific probe using standard protocols (Sambrook et al., 1989).
Total RNA samples analyzed in this study were isolated from leaf tissues using TRIzol reagent (Life Technologies, Rockville, MD). Mixed leaf tissues from normal growing (for constitutive pCB2006 constructs) or drought-stressed (leaf rolled, for pCB2009 construct) transgenic T1 families were used for the expression identification of seven stress-responsive genes. Fifteen micrograms of total RNA from each sample were separated on 1.2% agarose gel containing formaldehyde and then transferred onto a nylon membrane, and hybridized with the respective 32P-labeled candidate gene-specific fragment. Hybridization and washing conditions were based on standard protocols (Sambrook et al., 1989).
Drought Resistance Testing of Transgenic Rice
Drought stress experiments were made up of the pre-screening, testing, and re-testing of drought resistance. The drought stress pre-screening was performed in Hainan in the winter season of 2005. Twenty plants for each family were grown in two rows with planting density similar to real agricultural fields. Two replications were placed in two separate fields. Considering the potential variation of soil at different locations of the field, WT control (Zhonghua 11) plants were inserted after every 10 transgenic families for comparison. The irrigation was stripped 1 month after transplanting, corresponding to the initiation of panicle development.
The drought resistance testing was performed in the summer of 2005. Thirty T1 families with one to three copies of T-DNA (checked at T0 generation), expression of the transgene and relatively resistant to drought stress obtained from pre-screening were selected from each of the 14 gene constructs (seven candidate genes by two types of promoters) and tested for drought resistance. Herbicide was applied at the seedling stage to select positive transgenic plants for transplanting. Drought testing was conducted in two different environments: the isolated field equipped with a movable rain-off shelter and PVC tubes (1 m in length and 0.2 m in diameter) placed under plastic tents (length × width × height: 26 × 6 × 3.6 m) with foldable roofs. In the field conditions, 20 plants of each family were planted in a plot and a plot of the WT rice was inserted every 10 transgenic plots. Drought stress was applied by stripping watering at booting stage (∼2 weeks before flowering) in the field and the stress was ceased when the WT plants became completely leaf-rolled (the rolled leaf could not re-expand after irrigation). Such severe drought stress corresponded to a water content of 15–18% (vol./vol.) in the soil determined by Time Domain Reflectometry meters placed in the field. The same set of materials were planted in the paddy field without drought treatment. The other drought testing was conducted in PVC tubes. Twenty transgenic plants of each transgenic family were planted in PVC tubes (one plant per tube). Only 10 families of the 30 selected families used in the field for each construct were tested in PVC tubes because of the limited capacity of the facility. For each family, 20 plants were divided into two groups for drought stress and normal growth treatments, respectively. The WT plants were inserted after every 10 transgenic plants for comparison. Drought stress was initiated at the panicle development stage (∼2 weeks before flowering) by discharging water through a hole near the bottom of the pipes. Each plant was stressed to the same degree at which leaves of main tillers were completely rolled (observed at 6:00 PM), then irrigated thoroughly overnight and immediately subjected to another round of stress until complete leaf-rolling. After two rounds of drought stress, plants were irrigated to allow recovery at the flowering and seed maturation stages. The details of drought treatment and trait measurement followed our previous study (Xiao et al., 2007; Yue et al., 2006).
In the drought resistance re-testing experiment of 2006, T2 families derived from T1 families without obvious yield decrease in the above experiment were selected for further drought resistance re-testing in the field. Experiment design and drought stress treatment were the same as the above field experiment of drought stress, except that the planting density of the WT was different from the above experiment: a plot of WT plants were inserted every five transgenic plots.
Data Collection and Statistical Analysis
In the drought pre-screening experiment, leaf drying score was used as the major criterion to evaluate the drought resistance performance of T1 transgenic families; the score for each family is the average performance of the 16 plants in the middle of each plot. In the drought resistance testing, relative yield (the ratio of yield per plant in stress treatment to that in normal growth) and relative spikelet fertility (the ratio of spikelet fertility in stress treatment to that in normal growth) were used as the major criteria to evaluate the drought resistance performance of transgenic plants, and some yield-related traits (leaf drying score, panicle number, grain number, 1000-grain-weight) were also measured. In the drought resistance re-testing experiment, yield per plant and spikelet fertility were directly used as the major criteria to evaluate the drought resistance of transgenic families. For each T1 or T2 family in the field, yield per plant, spikelet fertility, and yield-related traits of 16 plants from each plot (excluding four plants, two on each side of the plot) were measured, and the mean value of the 16 plants in each plot was used for statistical analysis. For drought resistance testing in the PVC pipes, yield and spikelet fertility of all the plants were individually measured, and the yield and spikelet fertility values of each plant under drought and normal growth conditions were used for statistical analysis.
The data on grain yield per plant, spikelet fertility, relative yield, and relative spikelet fertility were analyzed by one-way analysis of variance (ANOVA). The subsequent multiple comparisons among the means of transgenic families or lines and WT were examined based on the least significant difference (LSD) test. Correlation coefficient between yield and yield-related traits (leaf drying score, panicle number, grain number, spikelet fertility, 1000-grain-weight) in the drought field was analyzed by a linear correlation model. All statistical analysis was performed using the SPSS package (Version 12.0).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This research was supported mainly by the Rockefeller Foundation, and partially by the National Program on Basic Research and Development, the National Special Key Project on Functional Genomics of Rice, and the National Natural Science Foundation of China.
Acknowledgments
We thank Dr Abraham Blum for suggestions on drought resistance testing protocol of rice in the field. No conflict of interest declared.
References
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- Chen H, Tang W, Xu C, Li X, Lin Y, Zhang Q. Transgenic indica rice plants harboring a synthetic cry2A*? gene of Bacillus thuringiensis exhibit enhanced resistance against lepidopteran rice pests. Theor. Appl. Genet. 2005;111:1330–1337. doi: 10.1007/s00122-005-0062-8. [DOI] [PubMed] [Google Scholar]
- Deng N. Formulating the outline of a national program of agricultural science and enhancing the revolution of agricultural science and technology development in China. Review China Agri. Sci. Tech. 1999;1:3–8. [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl Acad. Sci. U S A. 2002;99:15898–15903. doi: 10.1073/pnas.252637799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Holmberg N, Bulow N. Improving stress tolerance in plants by gene transfer. Trends Plant Sci. 1998;3:61–66. [Google Scholar]
- Holmstrom K, Mantyla E, Welin B, Mandal A, Palva E. Drought tolerance in tobacco. Nature. 1996;379:683–684. [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl Acad. Sci. U S A. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Jang IC, et al. Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol. 2003;131:516–524. doi: 10.1104/pp.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Lei ZY, Zhao P, Cao MJ, Cui R, Chen X, Xiong LZ, Zhang QF, Oliver DJ, Xiang CB. High-throughput binary vectors for plant gene function analysis. J. Integr. Plant Biol. 2007;49:556–567. [Google Scholar]
- Lin JY, Shen M. Rice production constraints in China. In: Evenson RE, Herdt RW, Hossain M, editors. Rice Research in Asia, Progress and Priorities. Wallingford, UK: CAB International, and Los Baños, Philippines: IRRI; 1996. pp. 161–178. [Google Scholar]
- Lin YJ, Zhang Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 2005;23:540–547. doi: 10.1007/s00299-004-0843-6. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell. 1990;2:163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKersie BD, Bowley SR, Harjanto E, Leprince O. Water-deficit tolerance and field performance of transgenic alfalfa overexpressing superoxide dismutase. Plant Physiol. 1996;111:1177–1181. doi: 10.1104/pp.111.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P. Transcriptional transgene silencing and chromatin components. Plant Mol. Biol. 2000;43:221–234. doi: 10.1023/a:1006483428789. [DOI] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005;138:341–351. doi: 10.1104/pp.104.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Li J, Pittman JK, Berkowitz GA, Yang H, Undurraga S, Morris J, Hirschi KD, Gaxiola RA. Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc. Natl Acad. Sci. U S A. 2005;102:18830–18835. doi: 10.1073/pnas.0509512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Terry N, Sears T, van Dun K. Enhanced drought resistance in fructan-producing sugar beet. Plant Physiol. Biochem. 1999;37:313–317. [Google Scholar]
- Qin X, Zeevaart JA. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002;128:544–551. doi: 10.1104/pp.010663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero C, Bellés J, Vayá J, Serrano R, Culiánez-Macià F. Expression of the yeast trehalose-6-phosphate synthasegene in transgenic tobacco plants: pleitropic phenotypes include drought tolerance. Planta. 1997;201:293–297. doi: 10.1007/s004250050069. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shao HB, Liang ZS, Shao MA. LEA proteins in higher plants: structure, function, gene expression and regulation. Colloids Surf. B Biointerfaces. 2005;45:131–135. doi: 10.1016/j.colsurfb.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Shen B, Jensen RG, Bohnert HJ. Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to chloroplasts. Plant Physiol. 1997;113:1177–1183. doi: 10.1104/pp.113.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Chen CN, Brands A, Pan SM, Ho TH. The stress- and abscisic acid-induced barley gene HVA22: developmental regulation and homologues in diverse organisms. Plant Mol. Biol. 2001;45:327–340. doi: 10.1023/a:1006460231978. [DOI] [PubMed] [Google Scholar]
- Stam M, Mol JN, Kooter JM. The silence of genes in transgenic plants. Ann. Bot. 1997;79:3–12. [Google Scholar]
- Stempak JM, Sohn KJ, Chiang EP, Shane B, Kim YI. Cell and stage of transformation-specific effects of folate deficiency on methionine cycle intermediates and DNA methylation in an in vitro model. Carcinogenesis. 2005;26:981–990. doi: 10.1093/carcin/bgi037. [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB. Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol. Biol. 2000;42:833–845. doi: 10.1023/a:1006448428401. [DOI] [PubMed] [Google Scholar]
- Turner NC. Further progress in crop water relations. Adv. Agron. 1997;58:293–339. [Google Scholar]
- Wang Y, et al. Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J. 2005;43:413–424. doi: 10.1111/j.1365-313X.2005.02463.x. [DOI] [PubMed] [Google Scholar]
- Wu C, Li X, Yuan W, Chen G, Kilian A, Li J, Xu C, Zhou DX, Wang S, Zhang Q. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003;35:418–427. doi: 10.1046/j.1365-313x.2003.01808.x. [DOI] [PubMed] [Google Scholar]
- Xiao BZ, Huang YM, Tang N, Xiong LZ. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 2007;115:35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- Xiong L, Yang Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell. 2003;15:745–759. doi: 10.1105/tpc.008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–2083. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho T, Wu R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon DB, Kang KH, Kim HJ, Ju HG, Kwon SJ, Suh JP, Jeong OY, Ahn SN. Mapping quantitative trait loci for yield components and morphological traits in an advanced backcross population between Oryza grandiglumis and the O. sativa japonica cultivar Hwaseongbyeo. Theor. Appl. Genet. 2006;112:1052–1062. doi: 10.1007/s00122-006-0207-4. [DOI] [PubMed] [Google Scholar]
- Yue B, Xue W, Xiong L, Yu X, Luo L, Cui K, Jin D, Xing Y, Zhang Q. Genetic basis of drought resistance at reproductive stage in rice: separation of drought tolerance from drought avoidance. Genetics. 2006;172:1213–1228. doi: 10.1534/genetics.105.045062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Saghai Maroof MA, Lu TY, Shen BZ. Genetic diversity and differentiation of indica and japonica rice detected by RFLP analysis. Theor. Appl. Genet. 1992;83:495–499. doi: 10.1007/BF00226539. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]