Abstract
In order to assess the functional roles of heat stress-induced class B-heat shock factors in Arabidopsis, we investigated T-DNA knockout mutants of AtHsfB1 and AtHsfB2b. Micorarray analysis of double knockout hsfB1/hsfB2b plants revealed as strong an up-regulation of the basal mRNA-levels of the defensin genes Pdf1.2a/b in mutant plants. The Pdf expression was further enhanced by jasmonic acid treatment or infection with the necrotrophic fungus Alternaria brassicicola. The single mutant hsfB2b and the double mutant hsfB1/B2b were significantly improved in disease resistance after A. brassicicola infection. There was no indication for a direct interaction of Hsf with the promoter of Pdf1.2, which is devoid of perfect HSE consensus Hsf-binding sequences. However, changes in the formation of late HsfA2-dependent HSE binding were detected in hsfB1/B2b plants. This suggests that HsfB1/B2b may interact with class A-Hsf in regulating the shut-off of the heat shock response. The identification of Pdf genes as targets of Hsf-dependent negative regulation is the first evidence for an interconnection of Hsf in the regulation of biotic and abiotic responses.
Keywords: Abiotic/environmental stress, transcriptional control and transcription factors, transcriptome analysis, defense responses, disease resistance, Arabidopsis
INTRODUCTION
Environmental adaptation of plants depends on elaborate systems for stress sensing and signaling, common and stress-specific responses, and probably also on a hierarchical control of reactions. There is evidence that heat shock factors (Hsfs) play important roles in stress sensing and signaling of different environmental stresses and that different stresses, including also high temperature, induce reactive oxygen species (ROS) in plants (Dat et al., 1998). ROS, particularly H2O2, are important components in biotic and abiotic stress response and signaling mechanisms.
Hsfs recognize consensus binding motifs, so-called heat stress elements (HSE: 5′-nGAAnnTTCnnGAAn-3′ or 5′-nTTCnnGAAnnTTCn-3’) conserved in promoters of heat-inducible genes of all eukaryotes. The classical target genes are the heat shock protein (Hsp) genes, which are (including the HSE motifs in the promoter region) highly conserved in eukaryotes (Wu, 1995). Hsfs display a modular structure with a highly conserved N-terminal DNA-binding domain (DBD), which is characterized by a central helix-turn-helix motif, and an adjacent bipartite oligomerization domain (HR-A/B) composed of hydrophobic heptad repeats. Hsf trimerization via the formation of a triple stranded alpha-helical coiled coil is a prerequisite for high-affinity DNA binding and subsequently for transcriptional activation of heat shock genes. Other functional domains of Hsf include clusters of basic amino acid residues (NLS) essential for nuclear import, leucine-rich export sequences in the HR-C region (NES) and—less conserved—a C-terminal activator domain (CTAD) rich in aromatic, hydrophobic, and acidic amino acids, the so-called AHA motifs (Nover et al., 1996, Döring et al., 2000). Sequence comparisons indicate that the combination of a C-terminal activator motif (AHA) with the consensus sequence FWxx(F/L) (F/I/L) to an adjacent nuclear export signal (NES) represents a signature domain for many plant activator Hsfs (Koskull-Döring et al., 2007).
Based on structural characteristics and phylogenetic comparison, plant Hsfs are subdivided into three classes and several subgroups. Recent studies in Arabidopsis showed that different class A-Hsfs play important roles during early and late stages of stress response. Early Hsfs are considered to be constitutively expressed in the cell and may become activated immediately upon stress treatment. Late Hsf functions are considered for those Hsfs whose expression is significantly enhanced upon stress treatment, which suggests that these Hsfs require the action of early Hsfs for their own expression (Wunderlich et al., 2007).
Most research on Hsfs was focused on the functional characterization of class A-Hsfs of Arabidopsis, which comprises 15 different genes/proteins, some of which are known to function as transcriptional activators of stress target genes. Recent evidence obtained from the identification of class A-Hsf-knockout mutants and microarray expression profiling indicates that some early, like HsfA1a and HsfA1b (Busch et al., 2005), and the late HsfA2 share an overlapping set of target genes.
Much less is known about the function of class B-Hsfs, which differ from class A by a structural variation within the oligomerization domain and the lack of an AHA-motif that is required for transcriptional activation function of class A-Hsfs (Koskull-Döring et al., 2007). There are only five different class B-Hsf genes present in the Arabidopsis genome, two of which are considered to have early functions (B3 and B4), three are considered to act as late Hsfs (HsfB1, HsfB2a, HsfB2b) because their mRNA expression is significantly increased upon heat stress and there is evidence that this heat-induced expression requires the combined action of the early class A-HsfA1a and -A1b (Lohmann et al., 2004; Busch et al., 2005).
In contrast to Hsfs of class A, classes B- and C-Hsfs have no evident function as transcription activators on their own (Kotak et al., 2004; Czarnecka-Verna et al., 2000, 2004). There is evidence that tomato HsfB1 interacts with HsfA1a and may also be involved in CREB-dependent expression of housekeeping genes during recovery (Bharti et al., 2004). Interestingly, the orthologous Arabidopsis HsfB1 did not show such properties in the same assay systems (Bharti et al., 2004). In Arabidopsis, HsfB1 is the most strongly heat-induced class B-Hsf (Busch et al., 2005). Surprisingly, transgenic overexpression of Hsf4 (synonymous for HsfB1) had no effect on the expression of heat shock proteins (Hsp) or the development of thermotolerance (Prändl et al., 1998). Since class B-Hsfs have the capacity to bind to similar or the same sites in the heat shock gene promoters as class A-Hsfs, it was proposed that they may act as repressors of target gene expression (Czarnecka-Verna et al., 2000, 2004).
In the present study, we investigated T-DNA knockout mutants of HsfB1 and HsfB2b for understanding their functional roles during the later phases of heat shock response. HsfB1/B2b are the only two out of three class B-Hsfs, which are clear targets of class Hsf-dependent enhancement of expression and for which viable knockout mutants were available. Microarray analysis for the identification of putative target genes revealed that, in hsfB1/hsfB2b double knockout plants, the major targets are Pdf1.2 genes, which are involved in immunity against infection by necrotrophic microorganisms. The identification of Pdf genes as targets of Hsf-dependent negative regulation is the first evidence for an interconnection of Hsf in the regulation of biotic and abiotic responses.
RESULTS
Isolation and Characterization of hsfB1 and hsfB2b Knockout Mutants
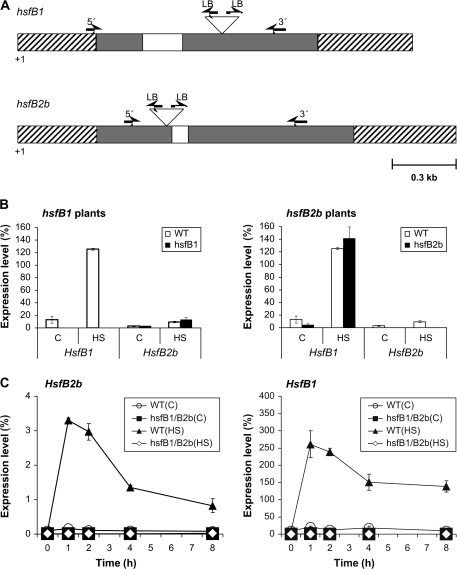
In order to analyze the function of the heat shock-induced Hsf genes, AtHsfB1 and AtHsfB2b, a loss-of-function strategy was employed. The T-DNA insertion lines Salk_012292 (insertion in HsfB1) and Salk_047291 (insertion in HsfB2b) were obtained from the stock collection of mutants available at the Signal Salk institute (http://signal.salk.edu/cgi-bin/tdnaexpress) and characterized. Homozygous mutants were identified via PCR screening using gene-specific and T-DNA-specific primers. The positions of the T-DNA insertions were confirmed and precisely mapped by DNA sequencing. In HsfB1, the insertion maps to the second exon 367 nucleotides upstream of the stop codon, in HsfB2b to a position 277 nucleotides downstream from the first exon border (Figure 1A). The homozygous mutants hsfB1–/– and hsfB2b–/– are designated as hsfB1 and hsfB2b, respectively.
Figure 1.
mRNA Quantification of HsfB1 and HsfB2b in Single and Double Mutants.
(A) T-DNA insertion sites in hsfB1 and hsfB2b lines as determined by sequencing PCR products obtained from genomic DNA of the tagged lines using either gene-specific 5’ or 3’ primers in combination with T-DNA left border (LB) primers. Triangles indicate the T-DNA elements. Exons are depicted as grey boxes, introns as open boxes, non-translated regions as hatched boxes. The position and direction of primers for Hsf genes or T-DNA sequences are marked by horizontal arrows. T-DNA elements and primers are not drawn to scale.
Poly (A)+-RNA was isolated from leaves and analyzed by qRT–PCR for the mRNA levels of HsfB1 and HsfB1b of wild-type (WT) and single mutant (hsfB1 and hsfB2b) plants that had been subjected to heat stress (37°C, 1 h) or control temperature (22°C, 1 h) (B), or double knockout (hsfB1/B2b) plants that had been subjected to heat stress (37°C) or control temperature (22°C) for 0–8 h (C).
PCR levels were normalized with respect to Actin2 mRNA (= 100%). Data points show means (n = 2) and range (B), and means (n = 3) and S.D. (C), respectively.
The expression of the respective Hsf in hsfB1 and hsfB2b plants was investigated at the transcript level using qRT–PCR both at control temperature and after 1 h of heat stress in comparison to wild-type (WT) plants. It was shown that, at 22°C, HsfB1 and HsfB2b are expressed at low levels (15% for HsfB1 and 4% for HsfB2b compared to actin2 = 100%) in WT plants (Figure 1B). After heat stress, the transcript levels of HsfB1 were increased in WT and hsfB2b plants (approximately 12- and 30-fold) but no HsfB1 transcripts were detected in hsfB1 plants with or without HS. HsfB2b mRNA levels were up-regulated in WT and hsfB1 plants by factors of three to five-fold after HS but HsfB2b transcripts were not detected in hsfB2b plants with or without HS. These results indicate that the T-DNA insertions caused a complete knockout of the expression of the respective Hsf genes in hsfB1 and hsfB2b single mutant plants.
However, there was no obvious phenotypic effect on morphology or thermotolerance associated with either of the single hsfB1 or hsfB2b mutant lines. Therefore, we isolated a double knockout mutant hsfB1–/–/hsfB2b–/– (abbreviated hsfB1/B2b). Following a crossing of the homozygous single knockout lines, we isolated a double knockout mutant homozygous for both Hsf mutations, in subsequent generations by PCR screening. The transcript levels of HsfB1 and HsfB2b were determined in a time course experiment from 0 to 8 h heat stress in the hsfB1/B2b double mutant compared to WT plants using qRT–PCR. As previously shown by Lohmann et al. (2004), HsfB1 and HsfB2b mRNAs are expressed at low levels (20 and0.2%, respectively, compared to actin2) at 22°C in WT. The expression levels are significantly increased after heat stress (Figure 1C): for HsfB1 to a high level (from 20 to 275% of actin2), for HsfB2b to a lower level (from 0.2 to 3.5% of actin2) after 1–2 h heat stress. No mRNAs were detected for either of the two Hsfs in the double knockout mutant hsfB1/B2b, neither at control temperature nor after heat stress. This indicates that HsfB1/B2b expression is completely eliminated in hsfB1/B2b plants. Despite this clear knockout effect at the level of gene expression, there was no obvious phenotypic difference with respect to growth, fertility and thermotolerance between hsfB1/B2b, single knockout, or WT plants.
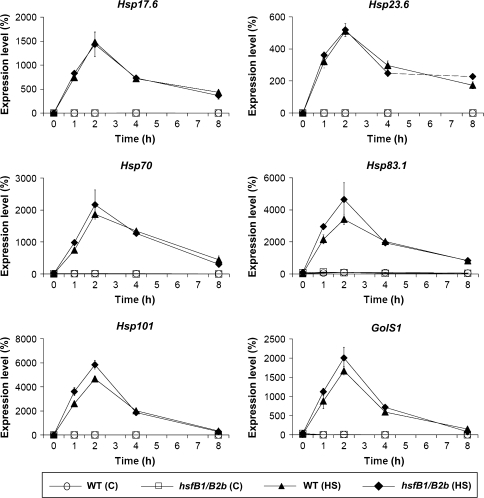
In order to test whether the combined functions of HsfB1 and HsfB2b are generally required for late effects in heat shock gene expression, we determined the mRNA expression profiles of a number of known Hsf target genes (Hsp17.6, Hsp23.6, Hsp70, Hsp83.1, Hsp101, and GolS) in a time course experiment from 0 to 8 h heat stress (Figure 2). Interestingly, the mRNA profiles, which differ individually from each other, are almost identical for WT and hsfB1/B2b double knockout plants. Hence, the deficiency of HsfB1 and HsfB2b has little if any effect on the expression profiles of typical Hsf-A1a/1b target genes (Lohmann et al., 2004; Panikulangara et al., 2004; Busch et al., 2005).
Figure 2.
mRNA Expression Profiles of Heat Shock Factor Target Genes.
Levels of mRNAs of different Hsp and GolS1 in wild-type (WT) and hsfB1/B2b double mutant plants during heat stress (HS). Poly (A)+-RNA was isolated from leaves that had been heat-stressed at 37°C for the indicated times, converted into cDNA and subjected to real-time PCR. Relative amounts were calculated and normalized with respect to Actin2 mRNA (= 100%). Data points show means and range (n = 3).
Expression Profiling: Identification of Putative HsfB1/HsfB2b Target Genes
In order to understand the functions of HsfB1 and HsfB2b, we tried to identify genes whose expression is affected in hsfB1/B2b plants, at either 22°C or after heat stress. Considering the heat-induced expressions of HsfB1 and HsfB2b, it was implicated that the effect should become more obvious after longer times of heat stress. As suggested by Figure 2, we applied heat stress at 37°C for 2 h to detect differences in gene expression between WT and hsfB1/B2b plants.
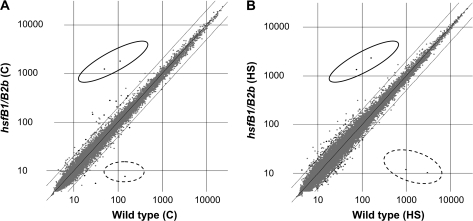
Whole genome Affymetrix microarray mRNA hybridizations were performed using RNA samples isolated from leaves that had been subjected to either 2 h at 22°C (control) or heat stress temperature. By conducting a Welch's t-test (confidence 95%, Multiple Testing Correction Benjamini and Hochberg) on the normalized hybridization data of the two control mRNA samples (not subjected to heat stress), it was examined whether HsfB1 and HsfB2b exert an influence on basal gene expression. The datasets of genes expressed at 22°C in WT versus hsfB1/B2b plants were compared. Only 16 genes appeared to be differentially expressed by a factor of more than two (Figure 3; black spots, left-hand panel), two of them, Pdf1.2a and Pdf1.2b, were outstanding with respect to the extraordinary differences in expression levels, with a more than 15-fold increase in hsfB1/B2b plants (Table 1). The expression of Pdf genes is known to be induced in response to necrotrophic pathogens and the defensins exhibit protective functions. Only four of the differentially regulated genes are down-regulated in hsfB1/B2b plants by a factor of more than two. Among the down-regulated genes, the strongest effect was on HsfB1 (18.44-fold), which is consistent with the knockout mutation in this gene. These data indicate that, at 22°C, the expression levels of >99.93% of all genes represented on the chip are identical in WT and hsfB1/B2b plants.
Figure 3.
Comparison of Expression Characteristics Wild-Type versus hsfB1/B2b Double Mutant Plants.
Expression levels (relative units) of all genes (gray spots) detected by microarray hybridization; black spots indicate genes differentially expressed by the following criteria: more than two-fold difference, confidence 95%, multiple testing correction Benjamini and Hochberg. The data points for Pdf1.2a and Pdf1.2b are marked with a full circle; dashed circles mark the expression levels genes (HsfB1 and HsfB2b) significantly down-regulated mutant plants.
(A) Control condition (room temperature: 22°C) comparison of wild-type (WT) and hsfB1/B2b double mutant.
(B) Heat shock (37°C) comparison of WT and hsfB1/B2b double mutant.
Table 1.
List of Up-Regulated (A) and Down-Regulated (B) Genes in hsfB1/B2b Double Mutant Compared to WT at Control Temperature.
| (A) | |||||||
| Affymetrix identifier | WT (C) | WT (HS) | hsfB1/B2b (HS) | hsfB1/B2b (C) | 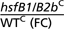 |
AGI | Annotation |
| 249052_at | 48.86 | 73.46 | 1,346.00 | 1,242.00 | 25.43 | At5g44420 | Plant defensin protein, putative (PDF1.2a) |
| 257365_x_at | 110.70 | 156.80 | 2,341.00 | 1,802.00 | 16.28 | At2g26020 | Plant defensin protein, putative (PDF1.2b) |
| 259169_at | 26.62 | 27.75 | 116.00 | 145.70 | 5.47 | At3g03520 | Phosphoesterase family protein |
| 245228_at | 89.70 | 48.74 | 118.20 | 271.00 | 3.02 | At3g29810 | Phytochelatin synthetase-like protein |
| 266841_at | 64.77 | 17,191.00 | 17,090.00 | 280.30 | 3.00 | At2g26150 | Putative heat shock transcription factor, AtHsfA2 |
| 263374_at | 48.03 | 7,591.00 | 7,769.00 | 141.00 | 2.94 | At2g20560 | DNAJ heat shock family protein |
| 256518_at | 18.43 | 1,518.00 | 1,415.00 | 44.14 | 2.40 | At1g66080 | Expressed protein |
| 260025_at | 93.37 | 8,128.00 | 8,786.00 | 207.30 | 2.22 | At1g30070 | SGS domain-containing protein |
| 248258_at | 164.30 | 759.50 | 990.50 | 352.00 | 2.14 | At5g53400 | Nuclear movement family protein |
| 249867_at | 12.72 | 59.00 | 98.56 | 25.98 | 2.04 | At5g23020 | 2-isopropylmalate synthase-like protein |
| 247771_at | 153.00 | 527.90 | 606.70 | 311.70 | 2.04 | At5g58590 | Ran binding protein 1 homolog-like |
| 262656_at | 112.20 | 2,449.00 | 2,263.00 | 226.80 | 2.02 | At1g14200 | Zinc finger (C3HC4-type RING finger) family protein |
| (B) | |||||||
| Affymetrix identifier | WT (C) | WT (HS) | hsfB1/B2b (HS) | hsfB1/B2b (C) | 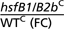 |
AGI | Annotation |
| 246214_at | 141.60 | 2,986.00 | 10.25 | 7.68 | 18.44 | At4g36990 | Heat shock transcription factor Hsf4 |
| 261662_at | 30.64 | 6.71 | 6.59 | 5.50 | 5.58 | At1g18350 | MAP kinase kinase 5, putative |
| 245449_at | 179.90 | 176.20 | 126.30 | 76.94 | 2.34 | At4g16870 | Retrotransposon-like protein |
| 247213_at | 17.51 | 13.60 | 13.80 | 7.92 | 2.21 | At5g64900 | Expressed protein |
FC, fold change; AGI, Arabidopsis genome initiative gene model (www.arabidopsis.org); HS, heat stress (37°C); C, control (22°C).
In a second comparative analysis, a t-test was applied for a dataset comprising the more than two-fold differences in RNA levels of heat-stressed WT versus heat-stressed hsfB1/B2b plants (Figure 3; right-hand panel). The scattering of the expression levels (gray spots) indicates the robustness of the analysis. The black spots mark the expression levels of 31 genes, which were differentially expressed; 15 genes showed significantly higher, and 16 genes lower levels in WT compared to hsfB1/B2b plants (Table 2). Thus, only a relatively small fraction (0.93%) of 3333 heat stress-affected genes (data not shown) can be attributed to the functions of HsfB1/B2B in WT. Interestingly, Pdf1.2a and Pdf1.2b appeared to be also up-regulated (fold changes of 14–18) in hsfB1/B2b plants; both are expressed at approximately the same high levels after heat stress as compared to control temperature. Among the genes down-regulated in hsfB1/B2b plants are, as expected, HsfB1 (fold-change 291) and HsfB2b (fold-change 82) mRNAs. This clearly proves the Hsf-gene knock character of the mutant line. All other differentially expressed genes, up- or down-regulated in hsfB1/B2b plants, show only relatively little differences in expression (fold change 2–3) compared to WT.
Table 2.
List of Down-Regulated (A) and Up-Regulated (B) Genes in hsfB1/B2b Double Mutant Compared to WT After Heat Stress.
| (A) | |||||||
| Affymetrix identifier | WT (C) | WT (HS) | hsfB1/B2b (HS) | hsfB1/B2b (C) | 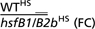 |
AGI | Annotation |
| 246214_at | 141.6 | 2,986 | 10.25 | 7.68 | 291.2 | At4g36990 | Heat shock transcription factor Hsf-B1 |
| 254878_at | 16.5 | 980.1 | 11.9 | 8.95 | 82.37 | At4g11660 | Heat shock transcription factor-like protein, Hsf-B2b |
| 256400_at | 9.51 | 40.66 | 15.35 | 8.256 | 2.649 | At3g06140 | Zinc finger (C3HC4-type RING finger) family protein |
| 253916_at | 46.93 | 33.14 | 13.63 | 51.46 | 2.431 | At4g27240 | Zinc finger (C2H2 type) family protein |
| 247141_at | 45.29 | 28.95 | 12.47 | 51.44 | 2.321 | At5g65560 | Pentatricopeptide (PPR) repeat-containing protein |
| 252268_at | 10.41 | 24.49 | 10.71 | 12.5 | 2.288 | At3g49650 | Kinesin motor protein-related |
| 246314_at | 69.22 | 69.06 | 31.97 | 69.19 | 2.16 | At3g56850 | ABA-responsive element binding protein 3 |
| 254489_at | 6.139 | 14.96 | 6.938 | 5.966 | 2.156 | At4g20800 | FAD-binding domain-containing protein |
| 261381_at | 14.19 | 28.85 | 13.53 | 11.06 | 2.132 | At1g05460 | RNA helicase SDE3 (SDE3) |
| 263507_s_at | 6.432 | 14.5 | 6.849 | 5.708 | 2.118 | At2g07684 | Hypothetical protein |
| 246200_at | 153.6 | 100.7 | 48.71 | 136.1 | 2.067 | At4g37240 | Expressed protein |
| 263795_at | 15.31 | 33.61 | 16.41 | 14.21 | 2.048 | At2g24610 | Cyclic nucleotide-regulated ion channel, putative |
| 264689_at | 68.18 | 30.83 | 15.11 | 65.42 | 2.04 | At1g09900 | Pentatricopeptide (PPR) repeat-containing protein |
| 260198_at | 6.09 | 12.08 | 5.945 | 6.849 | 2.032 | At1g67635 | Hypothetical protein |
| 247922_at | 64.07 | 23.14 | 11.4 | 64.12 | 2.03 | At5g57500 | Expressed protein |
| (B) | |||||||
| Affymetrix identifier | WT (C) | WT (HS) | hsfB1/B2b (HS) | hsfB1/B2b (C) | 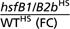 |
AGI | Annotation |
| 249052_at | 48.86 | 73.46 | 1,346 | 1,242 | 18.33 | At5g44420 | Plant defensin protein, putative (PDF1.2a) |
| 257365_x_at | 110.7 | 156.8 | 2,341 | 1,802 | 14.93 | At2g26020 | Plant defensin protein, putative (PDF1.2b) |
| 247529_at | 515.7 | 15.3 | 45.02 | 545.7 | 2.943 | At5g61520 | Monosaccharide transporter STP3 |
| 247533_at | 70.64 | 4.264 | 11.7 | 57.39 | 2.743 | At5g61570 | Protein kinase family protein |
| 248450_at | 27.4 | 17.55 | 45.88 | 25.66 | 2.614 | At5g51290 | Ceramide kinase-related |
| 254300_at | 47.41 | 41.1 | 102.7 | 57.52 | 2.498 | At4g22780 | Translation factor EF-1 alpha-like protein |
| 259502_at | 1,908 | 61.25 | 150.4 | 2,613 | 2.455 | At1g15670 | Kelch repeat-containing F-box family protein |
| 250379_at | 5.032 | 6.455 | 14.69 | 4.682 | 2.276 | At5g11590 | Transcription factor TINY |
| 256940_at | 15.97 | 24.15 | 54.48 | 22.56 | 2.256 | At3g30720 | Expressed protein |
| 246405_at | 1,008 | 19.82 | 44.24 | 865.6 | 2.232 | At1g57630 | Disease resistance protein (TIR class), putative |
| 257129_at | 9.129 | 16.92 | 35.97 | 9.101 | 2.125 | At3g20100 | Cytochrome p450 family |
| 257876_at | 10.42 | 11.45 | 24.13 | 14.04 | 2.106 | At3g17130 | Invertase/pectin methylesterase inhibitor family protein |
| 256595_x_at | 32.25 | 30.15 | 61.96 | 38.15 | 2.055 | At3g28530 | Expressed protein |
| 251842_at | 8.311 | 9.711 | 19.74 | 8.103 | 2.033 | At3g54580 | Proline-rich extensin-like family protein |
| 251756_at | 23.5 | 13.95 | 28.26 | 28.71 | 2.026 | At3g55820 | Hypothetical protein |
FC, fold change; AGI, Arabidopsis genome initiative gene model (www.arabidopsis.org); HS, heat stress (37°C); C, control (22°C).
In order to test the reliability of the microarray data, we re-examined the mRNA levels of Pdf1.2 and a number of other differentially expressed genes using qRT–PCR (Table 3). By and large, the results of qRT–PCR confirm the gene-chip hybridization data but, as expected, the PCR data of the strongly heat-induced genes such as AtHsfA2 and AtHsfB1 show a greater discrepancy in expression levels to the microarray data. Similar observations have been reported in the analysis of other hsf mutants (Busch et al., 2005).
Table 3.
Comparison of Gene Expression Levels Determined by Gene Chip Hybridization and qRT–PCR.
| Gene | WT (C) |
WT (HS) |
hsfB1/B2b (C) |
hsfB1/B2b (HS) |
||||
| RT–PCRa | GCb | RT–PCRa | GCb | RT–PCRa | GCb | RT–PCRa | GCb | |
| Pdf1.2a | 1.3 | 0.9 | 2.3 | 2.0 | 39.1 | 23.7 | 47.6 | 37.4 |
| Pdf1.2b | 1.4 | 2.1 | 2.1 | 4.2 | 36.2 | 34.4 | 61.8 | 65.1 |
| AtHsfA2 | 1.8 | 1.2 | 1965.4 | 459.5 | 7.6 | 5.3 | 1922.9 | 475.7 |
| DnaJ | 1.3 | 0.9 | 267.1 | 202.8 | 2.9 | 2.7 | 293.1 | 215.8 |
| Atmkk7 | 1.7 | 0.6 | 0.0 | 0.2 | 0.0 | 0.1 | 0.0 | 0.2 |
| AtHsfB1 | 12.9 | 2.7 | 238.6 | 79.6 | 0.5 | 0.1 | 0.4 | 0.3 |
| AtHsfB2b | 0.1 | 0.3 | 3.0 | 26.2 | 0.0 | 0.2 | 0.0 | 0.3 |
| Stp3 | 16.2 | 9.7 | 0.7 | 0.4 | 18.2 | 10.4 | 1.2 | 1.3 |
| F7H2.2 | 16.9 | 36.1 | 0.7 | 1.6 | 22.4 | 49.7 | 1.1 | 4.2 |
mRNA level determined quantitative PCR. Relative amounts were calculated and normalized with respect to Act2 mRNA (= 100%). Data represents mean ± range (n = 2).
Expression estimates based on the gene chip (GC) analysis; data represent normalized and averaged intensity values of the replicates relative to the signal of Act2 mRNA hybridization (= 100%).
Analysis of Pdf1.2 Gene Promoter Region and Hsf–HSE Binding
Pdf1.2 genes are up-regulated in hsfB1 and hsfB1/B2b plants and therefore we inspected the promoter region of Pdf1.2a and Pdf1.2b. Both genes are highly conserved in coding and in promoter regions. Using the internet tool http://arabidopsis.med.ohio-state.edu/AtcisDB/, several other putative binding motifs in the promoter sequence of Pdf1.2a and Pdf1.2b were identified. Both promoters share a number of common motifs, such as DPBF1 and -2, RAV1-A, BoxII, GATA box, GCC-box, and Ibox. The GCC-box mediates jasmonic acid-induced activation of Pdf1.2 expression in Arabidopsis (Brown et al., 2003).
However, there was no perfect match to the three-boxed HSE consensus sequences nGAAn/nTTCn/nGAAn, the preferred binding site for Hsfs. One variant HSE, representing only two boxes (nGAAn/nTTCn), could be detected in Pdf1.2b at position –629 to –619 upstream of the transcription start site, while this was absent in Pdf1.2a. However, another imperfect HSE (nGAAn/nATCn) that represents only one complete box consensus sequence was present in both promoters, at positions –594/–584 of Pdf1.2a and at positions –678/–668 in Pdf1.2b. Since both genes and promoters appear highly homologous and co-regulated, we concentrated further investigations only on the Pdf1.2a promoter.
Using EMSA, we were unable to identify any difference in the formation of binding-specific complexes in the Pdf1.2a promoter between WT and hsf-mutant plants, neither without nor after heat stress (not shown). This result suggests that no Hsf can directly bind to the promoter upstream region.
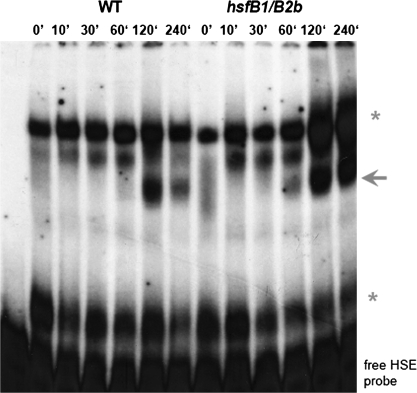
In a second approach, we assayed for any changes in late Hsf binding capacity in WT and hsfB1/B2b plants by using synthetic HSEs, which contain the perfect three-box consensus sequence nGAAn/nTTCn/nGAAn (Lohmann et al., 2004) as a probe. In a time course experiment covering up to 4 h heat stress prior to protein extraction, there was a reproducible difference in the EMSA profiles between WT and hsfB1/B2b plants (Figure 4). The hsfB1/B2b plants show an earlier onset (60 min), a prolonged appearance (240 min), and a stronger intensity of a signal that represents the major late Hsf–HSE binding complex. In WT, the same signal appears with a maximum after 2 h heat stress and is already diminished after 240 min.
Figure 4.
The Formation of Late Hsf-HSE Binding Complexes in Wild-Type and hsfB1/hsfB2b Plants.
Leaves of Arabidopsis wild-type (WT) (Col-0) and double Hsf mutant (hsfB1/hsfB2b) plants were incubated at 37°C for different periods of time (0, 10, 30, 60, 120, and 240 min). Total protein was extracted from each sample and subjected to EMSA using radioactive-labeled synthetic HSE consensus sequence as a probe. The first lane contains only the free probe without protein. The major late Hsf–HSE binding complex (Lohmann et al., 2004) is marked by an arrowhead; the unspecific constitutive binding complexes (Lohmann et al., 2004) are marked by asterisks.
Effects of Pathogen Infection on Pdf Expression and Pathogen Resistance in WT and Hsf-Knockout Plants
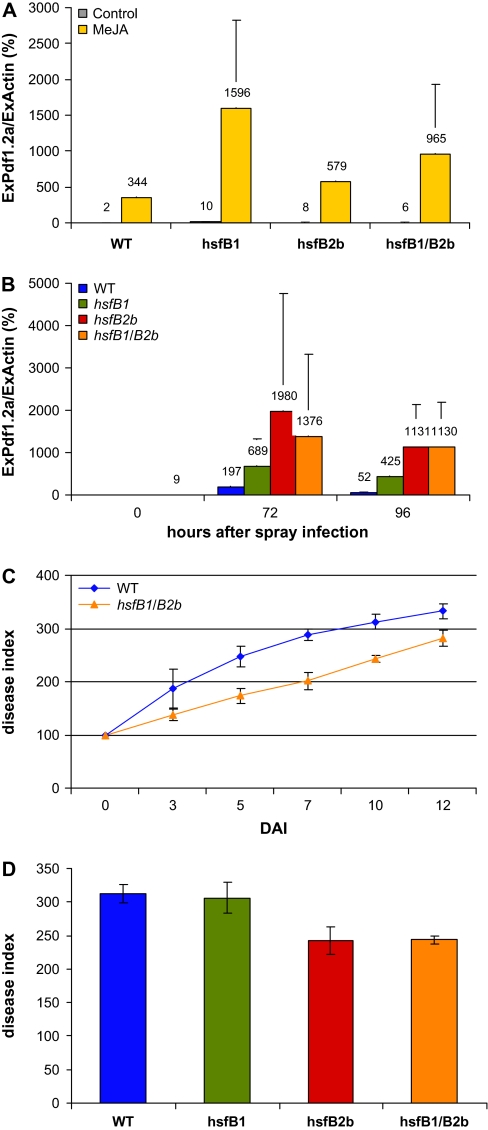
There is ample evidence that Pdf1.2a and Pdf1.2b are induced during pathogen attack or jasmonic acid application (Penninckx et al., 1998). We tested whether Pdf genes are still responsive to jasmonic acid induction in double mutants (hsfB1/B2b) and single Hsf-mutant (hsfB1 or hsfB2b) plants. Leaves of 5-week-old plants were sprayed with 100 μM MeJ and samples were collected after 6 h treatment for mRNA analysis. The results show (Figure 5A) that (1) Pdf1.2a mRNA is induced by MeJ to higher levels in hsf mutant plants, and (2) the induced levels in the mutants exceed the induced levels of WT plants.
Figure 5.
Effects MeJ and A. brassicicola Infection on Pdf1.2a Expression and Pathogen Resistance in Arabidopsis Wild-Type, Single, and Double Knockout Hsf Mutants.
Expression of Pdf1.2a was quantified after 6 h treatment with MeJ (A) or after 72 and 96 h post infection with A. brassicicola (B). Data were normalized with respect to Actin 2 expression (= 100%), points show means and S.D. (n = 3).
(C) Leaves of 6-week-old Arabidopsis wild-type (WT) (Col-0), and hsfB1/B2b plants were drop-inoculated with spores of A. brassicicola. The disease index was calculated 3, 5, 7, 10, and 12 d after inoculation (DAI). Data points show means and S.D. (n = 3–5).
(D) The disease index calculated on day 10 post inoculation of WT (Col-0), double knockout hsfB1/B2b, and single knockout mutants hsfB1, hsfB2b plants. Columns represent means and S.D. (n = 3–5). Significant differences are indicated by asterisks (α = 5).
We examined whether the infection with the necrotrophic fungus Alternaria brassicicola leads to enhanced induction of Pdf1.2a expression and an improved pathogen tolerance in Hsf-knockout plants. Leaves of 6-week-old plants were inoculated with A. brassicicola in a spray assay for the analysis of Pdf1.2a mRNA levels using qRT–PCR (Figure 5B) and in a spot assay (two 5-μL drops of 5 × 105 conidial spores suspended in water per leaf) for the determination of the disease index (Figure 5C). The results show that Pdf1.2a expression is significantly induced at 72 h post inoculation. This timing in Arabidopsis WT is in good agreement with published data (Schenk et al., 2003) showing a significant increase in mRNA levels 72 h after infection. The mRNA levels appeared to be induced to higher levels in the Hsf knockout plants compared to WT.
The disease resistance of WT and mutant lines was monitored on days 3, 5, 7, 10, and 12 after infection with A. brassicicola (Figure 5C). Infected leaves of double knockout plants (hsfB1/hsfB2b) provided significantly higher levels of tolerance (lower disease index) against A. brassicicola infection compared to WT plants. These differences in pathogen resistance can be seen from the beginning (3 days after infection (DAI)) throughout the whole experiment (12 DAI). Comparing the disease index of all four lines on day 10 after infection, there is a significant difference between hsfB2b and hsfB1/hafB2b plants, which were more resistant, compared to WT and single knockout hsfB1 plants (Figure 5D).
DISCUSSION
Considering the complexity of the Hsf gene family, the isolation of knockout mutants for individual Hsf genes is crucial for the determination of their functional role and biological importance in plants. Although single and double mutations in HsfB1 and HsfB2b lead to a complete loss of mRNA accumulation, there was no detectable effect on the heat stress response. This finding contrasts the functions of class A-Hsfs in which double knockout mutations in hsfA1a/hsfA1b caused a strong attenuation of the induction of a number of target genes including Hsp17.6, Hsp70, Hsp83.1, and Hsp101 (Lohmann et al., 2004). In our present study, the class B-Hsf mutations hsfB1/B2B had no effect on the expression of these HsfA1a/HsfA1b target genes or any other known heat shock genes in Arabidopsis. Hence, HsfB1 and HsfB2b are not directly involved in the regulation of the onset of the heat shock response. This is reminiscent to tomato, in which, when the expression of LbHsfB1 was compromised in a transgenic approach, plants showed no obvious effect on the heat shock response (Mishra et al., 2002).
Plant Defensin Genes Are De-Repressed in the hsfB1/B2b Mutant Plants
In order to understand the biological roles of HsfB1/B2b, we performed microarray expression analysis to identify possible target genes and pathways affected in hsfB1/B2b mutant plants. The total numbers of differentially expressed genes (control versus heat stress) in WT (3333 = 15%) and hsfB1/B2b plants (2875 = 13%) are relatively high but only 44 genes were identified as targets of the HsfB1/B2b-dependent transcriptional regulation. Comparable expression profiling results have been previously obtained by Busch et al. (2005) in a different genetic background of Arabidopsis (ecotype Wassilewskija), where a total of 112 class A-Hsf target genes has been identified.
Out of the 44 putative HsfB1/B2B-dependent target genes, 15 were differentially regulated (12 up and three down-regulated) at control temperature, 29 (15 up- and 14 down-regulated) upon heat stress in mutant plants compared to WT. The major effect was the enhanced expression (16 to 25-fold) of Pdf1.2a and Pdf1.2b, both at heat stress and control temperature conditions. Pdf1.2a and Pdf1.2b encode small polypeptides that protect plants against pathogen attack (Penninckx et al., 1996, 1998; Broekaert et al., 1995; Thomma et al., 2002).
The almost selective up-regulation of Pdf1.2a and Pdf1.2b does not exclude the importance of other differentially expressed genes with a fold change of two to three. Besides Pdf1.2a and Pdf1.2b, three other genes de-repressed in hsfB1/B2b plants are involved in plant defense: (1) Stp3, a green leaf-specific, low-affinity monosaccharide transporter (Buttner et al., 2000), (2) a kelch repeat-containing F-box family protein, which has been shown to be induced after pathogen attack (www.genevestigator.ethz.ch/), and (3) a TIR class disease resistance gene, which is highly induced by pathogens (www.genevestigator.ethz.ch/). These genes are down-regulated after heat stress in WT but heat stress has only little effect on the repression of these genes in hsfB1/B2b plants. Another gene, encoding a 2-isopropylmalate synthase-like protein (also known as methylthioalkymalate synthase-like protein), which might be involved in resistance to insect herbivores (Kroymann et al., 2003), shows an enhanced mRNA level (approximately two-fold) in hsfB1/B2b plants at control temperature.
Interestingly, among the other genes up-regulated in hsfB1/B2b plants is the class-A transcription factor AtHsfA2 (approximately three-fold up-regulated basal level), which is the strongest heat-inducible class A-Hsf of Arabidopsis (Busch et al., 2005). It is proposed that AtHsfA2 forms a heteromeric complex with AtHsfA1a and AtHsfA1b to regulate the expression of target genes during early and late stages of the heat shock response (Schramm et al., 2006). AtHsfA2 appears to act dominantly as a late, and AtHsfA1a and AtHsfA1b as early Hsfs that share a number of common target genes including Apx2, Hsp26.5, Hsp25.3, Hsp22.0, Hsp18.1, Hsp70, Hsp101, and DNAJ (Wunderlich et al., 2007).
HsfB2b Is a Negative Regulator of Defensin Gene Expression and Pathogen Resistance
There is ample evidence for the involvement of jasmonic acid in the regulation of Pdf1.2 expression and the protective role of defensins in plant pathogen resistance (Brown et al., 2003; Nandi et al., 2005; Thomma et al., 1998). Jasmonic acid application induces the expression of Pdf1.2 genes. The Arabidopsis triple mutant, fad3-2 fad7-2 fad8, which is unable to accumulate jasmonic acid, is susceptible to infection by Pythium spp. (Vijayan et al., 1998). Moreover, the mutants in jasmonate signaling pathway such as coi1 and jar1 showed enhanced susceptibility to fungal pathogens, such as A. brassicicola, Botrytis cinerea, etc. (Penninckx et al., 1996; Thomma et al., 1998) and suppressed Pdf1.2 expression (Penninckx et al., 1998).
MeJ treatment led to further enhanced Pdf1.2 mRNA levels in hsfB1, hsfB2b, and in double knockout plants by a factor of approximately 100. There is a clear tendency that the mutant lines accumulate much higher mRNA levels compared to WT. This indicates that, in Hsf-mutant plants, the de-repression is not only restricted to the low basal levels of Pdf1.2a expression.
The effects of MeJ on Pdf1.2a expression are largely reproduced in experiments using biotic stress infection with A. brassicicola for induction. Pdf1.2a and Pdf1.2b are induced to very high levels when plants are infected with A. brassicicola compared to other pathogens (genevestigator data).
The results of our patho-assays are in good agreement with the hypothesis that defensin expression correlates with pathogen resistance. The single mutant hsfB2b and the double mutant hsfB1/B2b showed significantly improved resistance levels (lower disease index) compared to WT and, surprisingly, to hsfB1 plants. The discrepancy between high levels of Pdf1.2 mRNA and only WT-level of disease resistance suggests that other molecular processes of the pathogen response are negatively affected by HsfB2b and thus result in improved pathogen resistance in plants unable to express HsfB2b. HsfB2b acts as a negative regulator of Pdf1.2 expression and pathogen resistance. The effect of hsfB2b is reminiscent to another recessive mutation, the overexpressor of cationic peroxidase 3 (Ocp3), which confers a constitutive expression of Pdf1.2 that results in an enhanced resistance of ocp3 plants to B. cinerea and P. cucumerina (Coego et al., 2005). The protective role of defensin gene expression against fungal infections in plants has been further demonstrated for transgenic rice (Kanzaki et al., 2002) or potato (Khan et al., 2006) plants overexpressing the wasabi defensin gene.
HsfB1 and HsfB2b seem to exert similar effects on Pdf expression, suggesting that they cooperate in repressing the basal and pathogen-induced levels. The involvement of HsfB1 in Pdf1.2 expression and pathogen response is strongly supported by expression profiling data (www.genevestigator.ethz.ch/at/). Many pathogens (P. syringae, P. infestans, B. cinerea, A. brassicicola) and a number of signaling mutants like Cpr5 (Bowling et al., 1997; Clarke et al., 2001), and nahG (Delaney et al., 1994) affect the expression of both Pdf genes.
Functional Roles of HsfB1 and HsfB2b
The regulation of Pdf1.2 expression through jasmonate signaling seems to require a region located at position –255 to –277 bp upstream of the Pdf1.2a transcription start site. This region includes the GCC-box, which is a common motif in the promoters of various genes encoding defense-related proteins, and has been identified as the specific binding site for members of the ERF subfamily of AP2/ERF transcription factors (Buttner and Singh, 1997; Ohme-Takagi and Shinshi, 1995; Zhou et al., 1997). It is possible that general transcription factors have general binding capability to different cis-regulatory elements and may activate gene expression through interactions with ERFs. Co-regulation of gene expression by the GCC-box and other promoter elements has been observed. For example, the GCC-box is required but not sufficient for high-level induction of the tobacco osmotin gene by ethylene (Raghothama et al., 1997). A G-box is also linked to jasmonate responsiveness in defense-associated genes in various plant species (Kim et al., 1992; Mason et al., 1993).
Perfect HSEs (three-box sequences), the regulatory elements for Hsf-dependent gene expression, are present neither in this region of the Pdf1.2a promoter nor in the introns or the other non-translated regions of the gene. An imperfect two-box variant HSE is unable to bind Hsf, as indicated by our negative results in detecting any changes in protein binding to promoter segments (not shown). This suggests that the negative effect of HsfB1/B2b on Pdf expression in WT must be indirect, possibly through interaction with other proteins in the chromatin or in the cell. According to the Arabidopsis Small RNA Project Database (http://asrp.cgrb.oregonstate.edu/; Gustafson et al., 2005), there are no microRNAs in Arabidopsis thaliana that target the transcripts of the Pdf1.2.
Using perfect synthetic HSE sequences as a probe, we were able to detect a positive effect on the formation of the late Hsf–HSE complex, which appears earlier, persists longer, and is more intense in hsfB1/B2b plants compared to WT. The formation of this complex requires HsfA2, the major heat stress- and high light-induced class A-Hsf in Arabidopsis (Wunderlich et al., 2007). A higher binding capacity of the HsfA2-dependent late complex in hsfB1/B2b plants suggests that HsfB1/B2b may interact with class A-Hsf in regulating the shut-off of the heat shock response. The absence of other HsfB1/B2b in knockout plants was not associated with the loss of HSE-binding capacity. This further supports the hypothesis that these Hsfs are not directly involved in binding to the HSE in vivo. It is conceivable that these class B-Hsf form heteromeric complexes with class A-Hsf with an increasing probability during the heat shock response. There is evidence that the tomato LpHsfB1 (ortholog of AtHsfB1) can interact with the class A-Hsf LpHsfA1a but also with other general factors like HAC1/CBP and thus specifying a co-activator function (Bharti et al., 2004). In a similar way, the Arabidopsis HsfB1 may interact with other Hsfs (possibly with HsfA2) and/or other general transcription factors or chromatin components for exerting a negative (repressive) role on target gene expression. Further studies identifying target genes during later stages of the heat shock response or during recovery and interaction partners of class B-Hsf will shed more light on the functions and molecular mechanism of Hsf-dependent repression of genes.
Biological Role of Limiting Pdf Expression
The identification of Pdf genes as targets of Hsf-dependent negative regulation is the first evidence for an interconnection of Hsf in the regulation of biotic and abiotic responses. The question arises as to why plant defensin expression is down-regulated by heat-inducible Hsfs. Regarding plant defense, a simple explanation may be that the pathogens are not capable of efficient infection at high temperature and thus plants do not require the expression of defensins for protection. This is consistent with our findings that A. brassicicola does not grow under heat stress temperature conditions, that a heat stress early after infection causes a delay in the development of disease symptoms, and that jasmonate induction of Pdf1.2 expression is blocked by heat treatment (not shown). Moreover, higher levels of Pdf1.2 expression may be detrimental to plant growth and development, which might become effective only on a long-term evolutionary scale. In eukaryotic cells, it has been demonstrated that beta-defensins have little effect on the epithelial cells at any concentration. In contrast, alpha-defensin promotes proliferation of the epithelial cells at low concentration but has a cytotoxic effect at high concentration and may have adverse effects on the host (Nishimura et al., 2004). In a similar way, Arabidopsis plants carrying the iop1 mutation show much higher induced expression levels of Pdf1.2, but, during their entire lifespan, the iop1 mutants stayed significantly smaller (Penninckx et al., 2003).
METHODS
Plant Material, Growth Condition, and Heat Treatment
Arabidopsis thaliana (Columbia-0) was used in experiments. Col-0 T-DNA insertion lines were obtained from the Signal Salk institute (http://signal.salk.edu/tdnaprimers.2.html).
Seeds were spread on soil or MS media and kept in the dark at 4–8°C for 2 d to achieve a uniform germination. Ten DAG (days after germination), seedlings were singled out in individual pots and grown in soil with a light/dark cycle of 16/8 h at 22°C, 60% relative humidity and a light intensity of 5000 lumen m−2. All experiments were carried out with 5-week-old plants unless specified.
For heat treatment (HS), fully expanded leaves were collected and pooled, incubated at 37°C (for 1, 2, 4, or 8 h, depending on the experiment performed) in pre-warmed SIB (1 mM potassium phosphate, pH 6, 1% (W/V) sucrose), then placed in a shaking water bath with 40 oscillations per minute. As a control (C), leaf pools were incubated under the same experimental conditions but at room temperature (22°C). After heat treatment, excess SIB was rinsed off with sterile water; leaf tissue was blotted dry with filter paper and immediately frozen in liquid nitrogen. Samples were stored at –70°C for future use.
RNA Isolation, Labeling, and Microarray Hybridization
Frozen leaf tissue (–80°), which had been collected from 45 individual plants per biological replicate, was crushed by using mortar and pistil and RNA was extracted with the Plant RNeasy Mini Kit (Qiagen, Hilden, Germany). A total of 5 μg RNA was used as starting material for double-stranded cDNA synthesis using the Superscript Choice System (Invitrogen, Karlsruhe, Germany) and an oligo dT-T7 primer (Genset, Paris, France). Biotinylated cRNA was synthesized from cDNA template using BioArray High Yield Transcript Labeling Kit (Enzo, Farmingdale, NY, USA). RNeasy columns (Qiagen) were employed to clean the biotinylated cRNA. The fragmentation of cRNA was performed according to GeneChip protocol (Affymetrix, Santa Clara, CA, USA).
The fragmented cRNAs were hybridized to Arabidopsis ATH 1-12151 (Affymetrix Gene-Chips), which represents 22 810 probe sets. GeneChip arrays were hybridized according to the manufacturer's protocol (Affymetrix). Hybridized GeneChips were scanned with the GeneChip Scanner 3000 (Affymetrix). Samples were quality-controlled by examining of 3’ to 5’ intensity ratios of control genes.
Analysis of Expression Data
Data were imported in Genespring 7.2 (Silicon Genetics, Redwood City, CA, USA) and retransformed in linear values. Data for every condition were available in two replicates of each experiment. Raw data were quantile-normalized and expression estimates were calculated by gcRMA (Wu et al., 2004) implemented in R. Statistical analysis of data was carried out on gene sets after eliminating all genes that showed less then two-fold change between conditions, by Welch's t-test to compare the different conditions and to find differentially expressed genes (selected P-value 0.05); the Benjamini and Hochberg False Discovery Rate Multiple Testing Correction was used to reduce the detection of false positives. The data are deposited in a MIAME compliant fashion in the ArrayExpress database (www.ebi.ac.uk/microarray/) under accession E-MEXP-1725.
Isolation of mRNA and Preparation of cDNA for Quantitative Real-Time PCR
This method has been used for the verification of knockout mutants and the expression profiles of individual Hsf target genes. Poly (A)+-RNA was isolated from 60–80 mg frozen leaf material using the chemagic mRNA Direct kit (Chemagen) and the amounts were quantified using the dye RiboGreen. cDNA was synthesized from 100 ng of mRNA using the iScript™ cDNA synthesis kit (Biorad). The amount of cDNA was quantified using PicoGreen. Quantitative RT–PCR was performed in triplicates using undiluted (1 ng) and 1/8 and 1/64 diluted cDNA as template. Real-time PCR was performed in a 50-μl reaction volume. The primers shown in Table 4 were used.
Table 4.
Primers Used for Real-Time PCR.
| Target gene | Primer sequence |
| Actin2 | 5′-AAGCTGGGGTTTTATGAATGG-3’ |
| (At3g18780) | 5′-TTGTCACACACAAGTGCATCAT-3’ |
| Hsp17.6 | 5′-GGTGAGTGGCAAAAGACAGA-3’ |
| (At1g59860) | 5′-AAACTTCCCCATCCTCCTCT-3’ |
| Hsp23.6 | 5′-CGATGAGATTAAGGCGGAGA-3’ |
| (At4g25200) | 5′-TCGACGTTTTTAGTTGATCTCG-3’ |
| Hsp70 | 5′-AGGAGCTCGAGTCTCTTTGC-3’ |
| (At2g32120) | 5′-AGGTGTGTCGTCATCCATTC-3’ |
| Hsp83.1 | 5′-GCTGCTAGGATTCACAGGATG-3’ |
| (At5g52640) | 5′-TCCTCCATCTTGCTCTCTTCA-3’ |
| Hsp101 | 5′-TAACGGGCCAAAGAGAAGTG-3’ |
| (At1g74310) | 5′-CACACGTTGGAGGTCAAGACT-3’ |
| AtHsfB1 | 5′-GGACCGGGATGAAAAGAATTA-3’ |
| (At36990) | 5′-CACGCTGGTTTGAACAGTCTT-3’ |
| AtHsfB2b | 5′-TGGAGGAGAATAACTCCGGTAA-3’ |
| (At11660) | 5′-ATGCAATGGGGATTCAGTAACA-3’ |
| GolS1 | 5′-AGCTTAGCCACAATATAATCATCG-3’ |
| (At1g56600) | 5′-ATCCTCCAAAACCCATAAAAATTA-3’ |
| Pdf1.2a | 5′-CCAAGTGGGACATGGTCAG-3’ |
| (At5g44420) | 5′-ACTTGTGTGCTGGGAAGACA-3’ |
| Pdf1.2b | 5′-GGTACTTGGTCAGGAGTTTGC-3’ |
| (At2g26020) | 5′-ACTTGTGAGCTGGGAAGACA-3’ |
Isolation of mRNA and Preparation of cDNA for Quantitative Real-Time PCR
This method was used for determining the Pdf1.2-mRNA expression after MeJ treatment or Alternaria spray assay. Poly (A)+-RNA was isolated from 50–70 mg frozen leaf material using the chemagic mRNA Direct kit (Chemagen). A total volume of 15 μl was used for cDNA synthesis using the iScript™cDNA kit (Biorad). Quantitative RT–PCR was performed with three dilutions (1, 1/8, and 1/64) of cDNA and in duplicate samples. As a control, undiluted mRNA was used. The reaction volume was 30 μl. mRNA expression level of Pdf1.2a was analyzed with respect to Actin2. The following primers were used:
Actin2-F3 (5′-AAGCTGGGGTTTTATGAATGG-3’),
Actin2-R3 (5′-TTGTCACACACAAGTGCATCAT-3’),
Pdf1.2a_for (5′-TGCTTTCGACGCACCGGC-3’),
Pdf1.2a_rev (5′-TGTAAAATACACACGATTTAGCACC-3’)
Pathogen Treatment: Spot Assay
Plants were grown in soil with a light/dark cycle of 8/16 h at 22°C, 100% relative humidity and a light intensity of 5000 lumen m−2. Pathogen (Alternaria brassicicola) treatment was carried out by apoplectic puncturing 6-week-old plants. Care was taken to choose leaves with approximately equal size and having the same developmental stage in WT as well as in mutants.
Two leaves per plant were chosen for fungal inoculation. Plants were inoculated at a concentration of 5 × 105 conidial spores per milliliter. Each leaf was inoculated with two 5-μL drops of a suspension in water. Inoculated plants were incubated at 100% RH and 22°C for control conditions and 35°C for heat stress treatment, respectively. The spread of fungal colonization on each leaf was analyzed 3, 5, 7, 10, and 12 DAI. As a control, one plant per line was treated with water.
Detection of Pathogen Colonization on Inoculated Plants and Disease Index
Pathogen-inoculated leaves were bonitated according to the disease symptoms shown in Table 5.
Table 5.
Pathogen Inoculated Leaves Bonitated According to Disease Symptoms.
| Score | Symptoms |
| 1 | No symptom |
| 2 | Little brownish at point of inoculation |
| 3 | Dark-brown spot at point of inoculation |
| 4 | Necrotic spots spreading around the point of inoculation |
| 5 | Maceration |
| 6 | Sporulation |
| 7 | Leaf detachment |
The disease index (DI) for each DAI was calculated from the results of bonitation by the following formula:
Methyl Jasmonate Treatment
Five-week-old plants were sprayed with methyl jasmonate (MeJ) solution (100 μM). Mock inoculations were only sprayed with water.
Combined heat stress and MeJ treatment was carried out as follows: plants were first sprayed with MeJ and then kept in a growth chamber (set at 37°C, light/dark cycle of 16/8 h, relative humidity 60%, light intensity 5000 lumen m−2). As a control, water-sprayed plants were kept in a growth chamber at 22°C (light/dark cycle of 16/8 h, relative humidity 60%, light intensity 5000 lumen m−2).
Pathogen Treatment: Spray Assay
Plants were grown in soil with a light/dark cycle of 8/16 h at 22°C, 100% relative humidity and light intensity 5000 lumen m−2. For pathogen treatment (Alternaria brassicicola), 6-week-old plants were sprayed with a concentration of 5 × 105 conidial spores per milliliter. Leaf material was collected immediately after spray infection and after 72 and 96 h post inoculation, respectively.
Isolation of Single and Double Hsf Mutants
Individual T-DNA insertion lines for AtHsfB1 and AtHsfB2b were obtained from the Salk Institute. Suitable primers for testing the T-DNA insertion (as described by the Salk website: http://signal.salk.edu/tdnaprimers.2.html) were generated and used in PCR reactions with genomic DNA as the template. Using 5’ and 3’ gene-specific primers, WT plants were identified and eliminated from segregating population. Mutants were identified by specific PCR fragments obtained using a T-DNA and a gene-specific primer.
To generate a double Hsf mutant, 8–9-week-old single homozygous mutant plants were selected for pollination. For ♀ (hsfB2b mutant plants), closed flower buds were selected. Flowers were opened with a pin set and anthers were removed. Flowers were then left opened for 2 d before pollination with pollen from hsfB1. After 2 d, healthy and non-pollinated stigmas were chosen for fertilization. For ♂ (hsfB1 mutant plants), fully mature anthers with ripened pollens were chosen.
The resulting F1 heterozygous population was selfed to get homozygous hsfB1/B2b double mutant plants. WT plants were identified using gene-specific primers for hsfB1 and hsfB2b. Accordingly, the double mutation was identified with T-DNA left border primer and the gene-specific primer (Table 6).
Table 6.
Double Mutations Identified with T-DNA Left Border Primer and the Gene-Specific Primers.
| AtHsfB1 (At36990) | 5′-AAAAGTTCGCCGGAGATGACG-3’ |
| 5′-GTCGCAACCTTCGCACTCACT-3’ | |
| AtHsfB2b (At11660) | 5′-CACAGAGGTCAATTCCGACGC-3’ |
| 5′-CTTCTTCCTCTGCAGCACCCA-3’ | |
| T-DNA (Lba1) | 5′-ATGGTTCACGTAGTGGGCCATC-3’ |
Electrophoretic Mobility Shift Assay (EMSA)
The Hsf binding to DNA has been tested essentially as described by Lohmann et al. (2004), and the same synthetic HSE sequences (5′-CCAGAAGCTTCCAGAAGCC) were used as a probe. Unspecific binding complexes are discriminated according to Lohmann et al. (2004) by the ability to be also formed with mutated HSE, while specific binding complexes are formed only with perfect consensus HSE sequences but not with mutated HSE.
FUNDING
The research was funded by grants of the Deutsche Forschungsgemeinschaft (SFB446, project A2).
Acknowledgments
We thank Dr Jasmin Doll and Markus Wunderlich (ZMBP-Allgemeine Genetik, Universität Tübingen) for advice and fruitful discussions. No conflict of interest declared.
References
- Bharti K, Koskull-Döring von P, Bharti S, Kumar P, Tintschl-Körbitzer A, Treuter E, Nover L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell. 2004;16:1521–1535. doi: 10.1105/tpc.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert WF, Terras FRG, Cammue BPA, Osborn RW. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995;108:1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003;132:1020–1032. doi: 10.1104/pp.102.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schöffl F. Identification of novel heat shock factor dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 2005;41:1–14. doi: 10.1111/j.1365-313X.2004.02272.x. [DOI] [PubMed] [Google Scholar]
- Buttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl Acad. Sci. U S A. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner M, Truernit E, Baier K, Scholz-Starke J, Sontheim M, Lauterbach C, Huss VAR, Sauer N. AtSTP3, a green leaf-specific, low affinity monosaccharide H +-symporter of Arabidopsis thaliana. Plant Cell Environ. 2000;23:175–184. [Google Scholar]
- Clarke JD, Aarts N, Feys BJ, Dong X, Parker JE. Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. 2001;26:409–420. doi: 10.1046/j.1365-313x.2001.2641041.x. [DOI] [PubMed] [Google Scholar]
- Coego A, Ramirez V, Gil MJ, Flors V, Mauch-Mani B, Vera P. An Arabidopsis homeodmain transcription factor, overexpressor of cationic peroxidase 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell. 2005;17:2123–2137. doi: 10.1105/tpc.105.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Pan S, Salem T, Gurley WB. Plant class B Hsfs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol. Biol. 2004;56:57–75. doi: 10.1007/s11103-004-2307-3. [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Yuan C-X, Scharf K-D, English G, Gurley WB. Plants contain a novel multi-member class of heat shock factors without transcriptional activator potential. Plant Mol. Biol. 2000;43:459–471. doi: 10.1023/a:1006448607740. [DOI] [PubMed] [Google Scholar]
- Dat JF, Foyer CH, Scott IM. Changes in salicylic acid and antioxidants during induction of thermotolerance in mustard seedlings. Plant Physiol. 1998;118:1455–1461. doi: 10.1104/pp.118.4.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, et al. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Döring P, Treuter E, Kistner C, Lyck R, Chen A, Nover L. The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell. 2000;12:265–278. [PMC free article] [PubMed] [Google Scholar]
- Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD. ASRP: the Arabidopsis small RNA project database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki H, Nirasawa S, Saitoh H, Ito M, Nishihara M, Terauchi R, Nakamura I. Overexpression of the wasabi defensin gene confers enhanced resistance to blast fungus (Magnaporthe grisea) in transgenic rice. Theor. Appl. Genet. 2002;105:809–814. doi: 10.1007/s00122-001-0817-9. [DOI] [PubMed] [Google Scholar]
- Khan SR, Nishihara M, Yamamura S, Nakamura I, Mii M. Transgenic potatoes expressing wasabi defensin peptide confer partial resistance to gray mold (Botrytis cinerea) Plant Biotech. 2006;23:179–183. [Google Scholar]
- Kim SR, Choi JL, Costa MA, An G. Identification of G-box sequence as an essential element for methyl jasmonate response of potato proteinase inhibitor II promoter. Plant Physiol. 1992;99:627–631. doi: 10.1104/pp.99.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskull-Döring P, Scharf k-D, Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007;12:452–457. doi: 10.1016/j.tplants.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Kotak S, Port M, Ganguli A, Bicker F, Koskull-Döring P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004;39:98–112. doi: 10.1111/j.1365-313X.2004.02111.x. [DOI] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc. Natl Acad. Sci. U S A. 2003;100:14587–14592. doi: 10.1073/pnas.1734046100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F. Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol. Gen. Genom. 2004;271:11–21. doi: 10.1007/s00438-003-0954-8. [DOI] [PubMed] [Google Scholar]
- Mason HS, DeWald DB, Mullet JE. Identification of a methyl jasmonate-responsive domain in the soybean vspB promoter. Plant Cell. 1993;5:241–251. doi: 10.1105/tpc.5.3.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Develop. 2002;16:1555–1567. doi: 10.1101/gad.228802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi A, Moeder W, Kachroo P, Klessig DF, Shah J. Arabidopsis ssi2-conferred susceptibility to Botrytis cinerea is dependent on EDS5 and PAD4. Molec. Plant Microbe Interact. 2005;18:363–370. doi: 10.1094/MPMI-18-0363. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Abiko Y, Kurashige Y, Takeshima M, Yamazaki M, Kusano K, Saitoh M, Nakashima K, Inoue T, Kaku T. Effect of defensin peptides on eukaryotic cells: primary epithelial cells, fibroblasts and squamous cell carcinoma cell lines. J. Dermatol. Sci. 2004;36:87–95. doi: 10.1016/j.jdermsci.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley WB. The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones. 1996;1:215–223. doi: 10.1379/1466-1268(1996)001<0215:thwcap>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikulangara TJ, Eggers-Schumacher G, Wunderlich M, Stransky H, Schöffl F. Galactinol synthase 1, a novel heat-inducible and Hsf-target gene responsible for heat-induced synthesis of Raffinose Family Oligosaccharides in Arabidopsis. Plant Physiol. 2004;136:3148–3158. doi: 10.1104/pp.104.042606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Schenk PM, van den Ackerveken G, Cammun PB, Thomma BP. The Arabidopsis mutant iop1 exhibits induced over-expression of the plant defensin gene PDF1.2 and enhanced pathogen resistance. Mol. Plant Pathol. 2003;4:479–486. doi: 10.1046/j.1364-3703.2003.00193.x. [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF. Pathogeninduced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Metraux JP, Broekaert WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prändl R, Hinderhofer K, Eggers-Schumacher G, Schöffl F. Hsf3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol. Gen. Genet. 1998;258:269–278. doi: 10.1007/s004380050731. [DOI] [PubMed] [Google Scholar]
- Raghothama KG, Maggio A, Narasimhan ML, Kononowicz AK, Wang G, D'Urzo MP, Hasegawa PM, Bressan RA. Tissue-specific activation of the osmotin gene by ABA, C2H4 and NaCl involves the same promoter region. Plant Mol. Biol. 1997;34:393–402. doi: 10.1023/a:1005812217945. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Manners JM, Anderson JP, Simpson RS, Wilson IW, Somerville SC, Maclean DC. Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol. 2003;132:999–1010. doi: 10.1104/pp.103.021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, Koskull-Döring P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 2006;60:759–772. doi: 10.1007/s11103-005-5750-x. [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Camme BPA, Thevissen K. Plant defensins. Planta. 2002;216:193–202. doi: 10.1007/s00425-002-0902-6. [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammune BPA, Broekaert W. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl Acad. Sci. U S A. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan P, Shockey J, Levesque CA, Cook RJ, Browse J. A role for jasmonate in pathogen defence of Arabidopsis. Proc. Natl Acad. Sci. U S A. 1998;95:7209–7214. doi: 10.1073/pnas.95.12.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Ann. Review Cell Dev. Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer FA. A model based background adjustment for oligonucleotide expression arrays. Working Paper 1, Dept. of Biostatistics Working Papers: Johns Hopkins University; 2004. [Google Scholar]
- Wunderlich M, Doll J, Busch W, Kleindt CK, Lohmann C, Schöffl F. Heat shock factors: regulators of early and late functions in plant stress response. Plant Stress. 2007;1:16–22. [Google Scholar]
- Zhou JM, Tang XY, Martin GB. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]