Abstract
p53, a tumor suppressor and transcription factor, is a critical modulator in the cellular response to stress. Exposure of glutathione-deficient GCS-2 cells to arsenite significantly phosphorylated and stabilized p53. In addition, p53 transcriptionally repressed Hsp90β gene expression. Mutation analysis revealed a p53 binding site in the 5′flanking region responsible for the regulation of Hsp90β gene. Electrophoretic mobility shift assay showed that p53 is bound to Hsp90β promoter region. ATM kinase, a major determinant in the modulation of p53 specifically affected its phosphorylation at Ser-15. ATM kinase-mediated phosphorylation of p53 is regulated through phosphorylation of Chk2. Down-regulation of ATM and Chk2 by their small interfering RNAs (siRNAs) attenuated the arsenite-induced phosphorylation of p53 and restored Hsp90β mRNA levels. Taken together, these findings suggest that arsenite acts through ATM and Chk2 to induce phosphorylation of p53. This results in the transcriptional repression of Hsp90β, under GSH-deficient conditions which may play a role in arsenic-mediated pathogenesis.
Keywords: Glutathione, p53, arsenite, ATM, MAPK, Hsp90, phosphorylation, signal transduction, siRNA
INTRODUCTION
Arsenic, an environmental and occupational toxin, is a ubiquitous and well-documented human carcinogen [1, 2]. Environmental exposure to arsenic has been linked to the development of a variety of human cancers, diabetes, and cardiovascular disorders [1–3]. Molecular mechanisms responsible for potential health hazards associated with low levels of arsenic exposure have not been evaluated in detail.
Low molecular weight thiols play a critical role in many biological reactions. Of these compounds, glutathione (GSH)1, a major non-protein thiol, is present in millimolar concentrations in most cells [4, 5]. It is an important antioxidant that has been implicated in the detoxification of drugs and xenobiotics including arsenic by conjugation reactions [6, 7]. Cellular redox status is believed to regulate a number of processes including signal transduction, transcription, and apoptosis [8–10].
It has been shown that arsenic may induce oxidative DNA damage and inhibit DNA repair [11, 12]. Arsenic-induced formation of reactive oxygen species including O2•−, H2O2 and •OH are responsible, at least in part, for the observed DNA damage [13, 14]. It is well recognized that free radicals play a pivotal role in signal transduction pathways and transcription factor regulation [15, 16]. A recent report has shown that trivalent arsenicals induce lipid peroxidation, protein carbonylation, and oxidative DNA damage in human urothelial cells [17].
Accumulation of p53 usually occurs because of DNA strand breaks, and p53 plays a central role in the maintenance of genome integrity [18, 19]. Activation of p53 is involved in a variety of cellular processes including cell differentiation, cell cycle regulation, DNA repair, apoptosis, and different stress response programs triggered by endogenous and exogenous stressors [20, 21]. Studies on arsenic cytotoxicity have suggested that inorganic arsenic and its dimethylated derivatives could induce DNA damage and the subsequent activation of ATM followed by stabilization of p53 [13, 14, 22–24]. Recent findings indicate the involvement of ATR, PML, and Chk2 in arsenic trioxide-induced apoptosis [25].
Hsp90 is a major molecular chaperone that is involved in protecting cells against stress [26, 27]. It also plays a pivotal role in the proper folding and stability of a number of proteins and regulates signaling pathways [28, 29]. Hsp90β gene has been shown to be repressed by p53 in UV irradiation-induced apoptosis [30].
Our previous studies have found that GCS-2 cells can survive indefinitely when cellular GSH levels are maintained at ~2% (50–60 μM) of wild type (BDC-1) levels [31]. GSH levels in GCS-2 cells are undetectable after withdrawal of GSH from the medium for 24 h [31]. GCS-2 cells maintained in 2.5 mM GSH had about one-fourth the cysteine levels of BDC-1 cells and the levels did not change any further by withdrawal of GSH from the medium [ref. 32 and data not shown]. GCS-2 cells maintained in 2 mM NAC had about 25% of the cysteine levels of BDC-1 cells and NAC itself was undetectable in cells cultured in the presence of NAC [32]. Using GSH-deficient GCS-2 cells as an experimental system, we have previously shown that arsenite toxicity is responsible for the down-regulation of Hsp90 by ubiquitination [33]. In the present study, we demonstrate that arsenite represses Hsp90β transcription through activation of ATM and p53 in GCS-2 cells. A better understanding of arsenite-mediated signal transduction under conditions of low/no GSH levels would be necessary to develop better therapeutic strategies for arsenic toxicity.
MATERIALS and METHODS
Cell Culture
BDC-1 and GCS-2 cells are epithelial cells and are derived from 3.5-day postcoitus (dpc) embryos from both wild-type and Glutamate cysteine ligase catalytic subunit (Gclc)-deficient mice [32]. They have been passaged hundreds of times and they are morphologically stable. All cell culture studies used M15 complete medium and cultures were done essentially as described earlier [31, 32]. All GSH-dependent GCS-2 cells (mutant cells) were maintained in medium containing 2.5 mM GSH and NAC-dependent GCS-2 cell lines were cultured in medium supplemented with 2 mM NAC. BDC-1 cells (wild type controls) were maintained in medium without NAC or GSH.
Treatment of Cells
Sodium arsenite was purchased from Sigma (St. Louis, MO). A 100 μM stock solution was prepared in PBS, pH 7.4 and was used at final concentrations ranging from 0–2 μM. Briefly, the GCS-2 cells were seeded at 1.5 × 106 cells per 10 cm dish, allowed to grow for 48 h either in the presence of 2.5 mM GSH or 2 mM NAC, depleted of GSH or NAC for 24 h, and treated with various concentrations of arsenite for up to 21 h. BDC-1 cells were treated under similar conditions as GCS-2 cells except that they were grown in the absence of NAC or GSH.
Measurement of Thiols
Thiols were measured using the high-performance liquid chromatography-electrochemical detection method of Kleinman and Richie [34] with minor modifications.
Western Blotting
After various treatments, cells were collected by centrifugation and resuspended in lysis buffer [50 mM Tris-HCl, 150 mM NaCl, 1% Triton-X-100, 0.1% SDS, 1 MM EDTA, 1% sodium deoxycholate, 20 mM NaF, 1 mM Na3VO4, 50 μM Na2MoO4, 10 M Sodium pyrophosphate, supplemented with protease inhibitor mixture (Roche Applied Science, Indianapolis, IN)]. 100 μg of total protein was routinely used for Western blots, with the exception of p53, for which 30 μg was used. Protein was separated on 10% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, incubated with the indicated antibodies and developed by ECL technology (Amersham Biosciences Corp, Piscataway, NJ) according to manufacturer’s instructions. The blots were stripped and reprobed with mouse actin antibody. The band intensities on the films were quantified using a GS-800 calibrated scanning densitometer (Bio-Rad, Hercules, CA). Quantitative expression of specific proteins was normalized to the expression of actin.
RNA Isolation and Northern Blot Analysis
Total RNAs were isolated from cultured cells using TRIRzol reagent (InvitrogenLife Technologies, Carlsbad, CA) according to manufacturer’s instructions. Fifteen μg of total RNA from cells after various treatments were electrophoresed on 1% agarose and 2.2 M formaldehyde gels, transferred onto a Zeta-Probe nylon membrane (Bio-Rad), and hybridized to a 410-bp 32P-labeled cDNA probe corresponding to Hsp90β. The blot was then stripped and re-probed with β-actin cDNA. Signal intensities on the films were quantified using a Bio-Rad GS-800 calibrated scanning densitometer and normalized to the expression of actin.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA assays were performed using the Gel Shift Assay System (Promega) according to the manufacturer’s protocol. The double-stranded oligonucleotides containing the putative p53 binding sites (underlined) were as follows: 5′-GGTAGTGTGTCGGTATGGGTAAGCAAGGCT-3′. In the binding reactions, γ32P-labeled DNA probes were incubated with 10 μg of nuclear extract. Binding reactions were performed at room temperature for 20 min and the DNA-protein complexes were resolved by electrophoresis on 5% nondenaturing Tris-Borate-EDTA polyacrylamide gels and visualized by autoradiography. Signals were quantified using PhosphorImager and ImageQuant™ software. The p53 binding activity was further confirmed by coincubation with unlabeled p53 oligonucleotides (cold probe competition) or with antibodies of p53 (supershift assay).
Point Mutations of the p53 Binding Site in the Promoter of the Hsp90 Gene
Site-specific mutations of the p53 binding sites in the Hsp90β promoter were performed mainly according to QuikChangeR site-directed mutagenesis kit (Stratagene).
Plasmids
The Hsp90β promoter regions −194/+142 were amplified by PCR from the genomic DNA of BDC-1 cells using the following primers: forward primer, 5′-AGCTCACCGCCGGCTAAGTCG-3′; reverse primer, 5′-ACCAACCTACCGACTTACCCT-3′. PCR fragment was cloned into KpnI and BglII restriction sites of the Promega promoter less luciferase vector pGL3-Basic. Underlined bases in the primer sequences indicate nucleotides added to permit efficient restrictions of the PCR products by the KpnI (forward primer) and BglII (reverse primer) enzymes.
Transient DNA Transfection and Promoter Activity Assay
The pGL3 reporter plasmids were transfected into cells as described below. BDC-1 and GCS-2 cells were plated at a density of 5 × 104 cells in 60 mm plates and were allowed to grow for 48 h. GSH was then withdrawn from GCS-2 cells for 24 h and both BDC-1 and GCS-2 cells were transfected using the FuGene (Roche) transfection agent following the manufacturer’s protocol. Each transfection was performed using 1.5 μg of Firefly luciferase reporter construct DNA that contained various deletions and site-directed mutants of Hsp90 promoter gene plus 0.1 μg of an internal control Renilla luciferase reporter plasmid pRL-TK (Promega). 2.5 h after transfection, the medium was removed by aspiration and replaced with normal culture medium containing 15% knock-out serum and antibiotics. Following an overnight recovery period, cell extracts were prepared for luciferase determination according to the protocol accompanying the dual-luciferase reporter assay system (Promega). Firefly and Renilla luciferase activities were measured using a luminometer. Firefly luciferase activity was normalized to Renilla luciferase activity. Each experiment was performed in triplicate and repeated four times independently.
SiRNA-transfections
SiRNA duplexes were synthesized and purified by Invitrogen. Transfections of SiRNAs were performed using oligofectamine (Invitrogen).
Statistical Analysis
Data are expressed as mean ± S.D. Statistical evaluation of data was performed using Student’s t test, considering the p value of ≤ 0.05 as significant.
RESULTS
Arsenite down-regulates Hsp90β mRNA expression
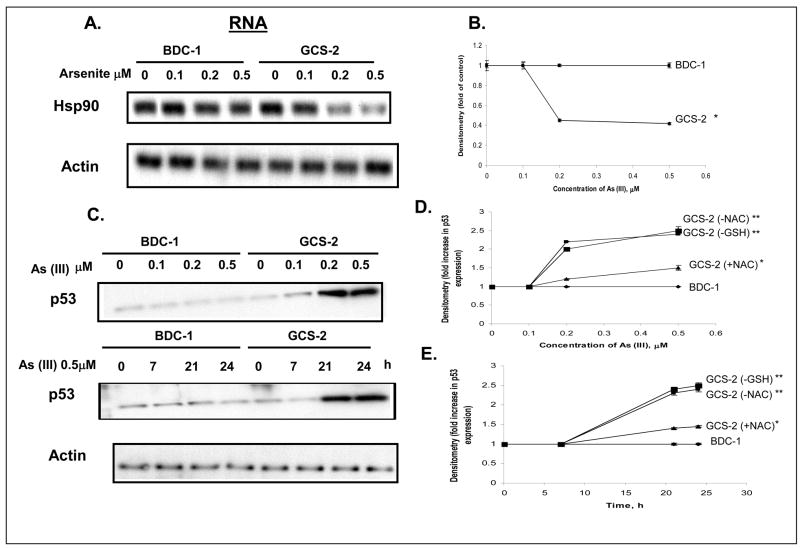
Availability of embryonic cell lines derived from Gclc-deficient mice (GCS-2) and wild-type mice (BDC-1) allowed us to examine the role of GSH depletion in arsenite-induced cytotoxicity. We have previously reported that Hsp90β protein is degraded by ubiquitination in GSH-deficient GCS-2 cells exposed to arsenite while Hsp90β levels remained unchanged in BDC-1 cells (Habib et al., 2007). Arsenite also down-regulates Hsp90β mRNA levels by 2.5 fold in GCS-2 cells at 0.5 μM at 21 h and it is time-and dose-dependent up to 24 h [(arsenite-induced regulation of Hsp90β has already been published in ref. 33)]; Figures 1A and 1B].
Figure 1.
Determination of the effect of arsenite on Hsp90β mRNA and p53 protein levels. A. Northern blot analysis of Hsp90β mRNA level. Culture and treatment of cells is done as described in ref. 33. GCS-2 and BDC-1 cells were incubated with increasing concentrations of arsenite (indicated along the top) for 21 h and total RNA was isolated. RNA was analyzed by Northern blot with Hsp90 (top) cDNA probe. The Hsp90 membrane was stripped and reprobed with actin (bottom) cDNA probe to verify equal loading. B. Quantification of Hsp90 expression by densitometry. Blots from A were quantified to measure the signal intensities corresponding to Hsp90 expression by densitometry. Each point is an average of three independent determinations. *, p ≤ 0.01 for 0.5 μM As (III) treated GCS-2 cells versus 0.5 μM arsenite treated BDC-1 cells. C. Dose- and time-dependent effect of arsenite on p53 protein levels. Cells were treated as in A, and cytosolic extract was separated by SDS-PAGE and analyzed by immunoblot with anti-p53 antibody. Equivalent loading was verified with an anti-actin antibody (bottom). D. Dose- dependent effect of arsenite on p53 levels in GSH-and NAC-dependent cells. Culture and treatment of cells is done as described in ref. 33. Cells were treated and resolved on SDS-PAGE as in C and quantified by densitometry. The results are expressed as mean ± S.D. of triplicate determinations. E. Time-dependent effect of arsenite on p53 levels in GSH- and NAC-dependent cells. Culture and treatment of cells is done as described in ref. 31. Cells were treated and resolved on SDS-PAGE as in C and quantified by densitometry. The results are expressed as mean ± S.D. of triplicate determinations. For both D and E, **, p ≤ 0.05 for 0.5 μM As (III) treated GCS-2 (-GSH) or GCS-2 (-NAC) cells versus 0.5 μM As (III) treated BDC-1 cells. *, p ≤ 0.01 for 0.5 μM As (III) treated GCS-2 (+NAC) cells vs 0.5 μM As (III) treated BDC-1 cells.
Arsenite stabilizes p53 protein levels
Preliminary studies from our laboratory on the p53 status (cloning and sequencing of p53) suggest that the p53 expressed in these cell lines is wild type. Immunochemical localization using anti-p53 antibody suggests that most of the p53 is localized in the nucleus and very little in the cytoplasm (unpublished observations). Since arsenite is known to induce oxidative damage and p53 is a well documented marker for DNA oxidative damage (Martindale and Holbrook, 2002; Kumagai and Sumi, 2007), we investigated further to see if arsenite cytotoxicity is associated with changes in p53 levels. To this end, we exposed BDC-1 and GCS-2 cells to different concentrations of arsenite for up to 21 h and found that there was a ~ 2.5 fold increase in dose-dependent accumulation of p53 protein levels in GCS-2 cells (Fig. 1C, top panel and Fig. 1D). We also determined the time-dependent accumulation of p53 and found that the p53 levels increased ~ 2 fold with time up to 24 h (Fig. 1C, middle panel and Fig. 1E).
Thiol antioxidant NAC restores p53 levels
To determine if other antioxidants such as NAC could reverse p53 to its original levels in GCS-2 cells, we treated GCS-2 cells previously rescued with NAC with arsenite. In this context, it is worth noting that we and others have found that NAC does not complex with arsenite [33, 35]. While p53 stabilized and accumulated to ~ 2.5 fold in GCS-2 cells treated with arsenite in the absence of NAC as in the case of GCS-2 cells treated with arsenite in the absence of GSH, we found that NAC could attenuate p53 accumulation in a dose- and time-dependent fashion (~ 1.4 fold) if it is present continuously during arsenite exposure (Figs. 1D and 1E).
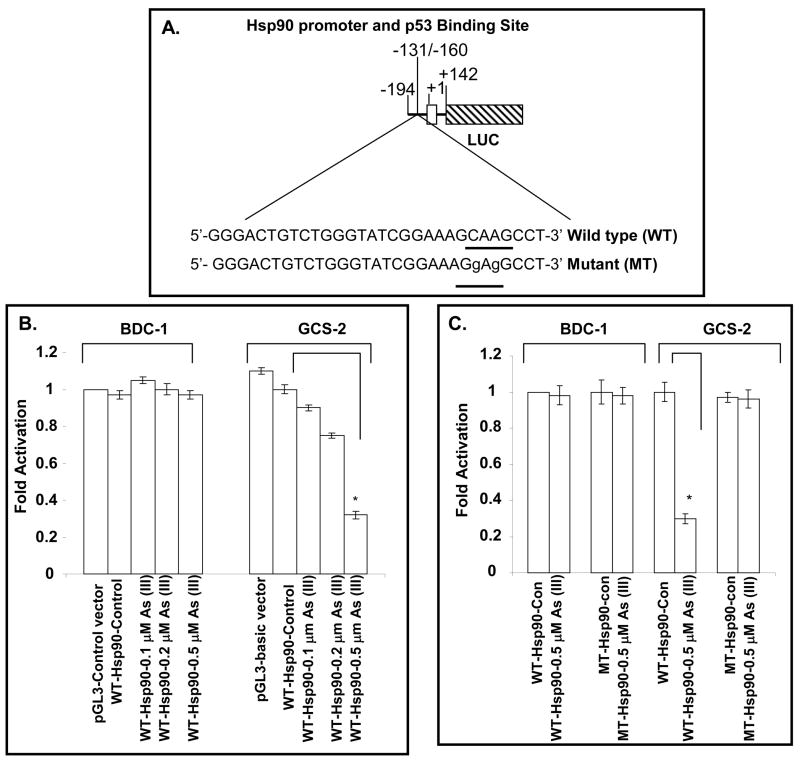
Expression of Hsp90 is regulated by p53 at the transcriptional level
To further examine the status of p53 binding to the Hsp90β promoter, we made constructs containing Hsp90 promoter driving luciferase reporter gene. Endogenous Hsp90 promoter contains DNA sequences for p53 binding (Fig. 2A). The putative p53 binding site in the mouse Hsp90β gene promoter region matches the p53 consensus half-site of PuPuC(A/T)(A/T)GPyPyPy [36]. To understand the mechanism of Hsp90β gene regulation by arsenite, the reporter was constructed with the promoter of Hsp90β preceding the luciferase (LUC) gene. To elucidate the importance of p53 binding site (BS) in the promoter region of the Hsp90β gene, the core sequence CAAG (−134/−137) of the p53 BS in WT-Hsp90-luciferase was mutated to GAGG and designated as MT-Hsp90-luciferase. These WT- and MT-Hsp90-luciferase plasmids were transfected into BDC-1 and GCS-2 cells. After transfection, endogenous p53 was induced by arsenite treatment. There was an arsenite dose-dependent down-regulation (~ 3 fold in the WT-Hsp90-LUC reporter activity in the GCS-2 cells treated with 0.5 μM arsenite compared to WT-Hsp90-LUC reporter activity in untreated GCS-2 cells) whereas the activity was unchanged in the BDC-1 cells (Fig. 2B). With mutated p53 BS, MT-Hsp90-LUC activity was comparable to WT-Hsp90-LUC control activity in GCS-2 cells (Fig. 2C). Taken together, these results suggest a direct involvement of p53 in Hsp90β promoter down-regulation.
Figure 2.
Impact of p53 BS in Hsp90β gene on the expression of gene in GCS-2 cells. A. Schematic diagram of the LUC reporter plasmid driven by Hsp90 promoter (−194/+142). A map and sequences of p53 BS at −131/−160 (wild type), mutated p53 BS (mutant), and the transcription initiation site at +1 are shown. For the sequence of Hsp90 promoter region, please see ref. 30. The first exon of the Hsp90 gene is denoted by an open box and the LUC reporter gene fused to the downstream region of the Hsp90 promoter is indicted by a hatched box. B. Effect of wild type p53 BS on Hsp90 promoter activity. The cells were transfected with the Hsp90-LUC reporter plasmid containing either wild type p53 BS (WT-Hsp90) and treated with and without various concentrations of arsenite for 21 h. The cell extracts were then assayed for LUC activity as described in “Materials and Methods”. The results are expressed as mean ± S.D. of triplicate determinations of four separate experiments. pGL3 control vector containing both the SV40 promoter and enhancers was used as a positive control. Empty pGL3 promoter-less basic vector which when used in the assay did show negative activation (data not shown). C. Effect of arsenite on wild type and mutant p53 BS in Hsp90 promoter. Cells were transfected with either WT-Hsp90-LUC or MT-Hsp90-LUC and treated with 0.5 μM arsenite for 21 h and the cell extracts were assayed for LUC activity. The results are expressed as mean ± S.D. of triplicate determinations of four separate experiments. For both B and C, *p ≤ 0.05.
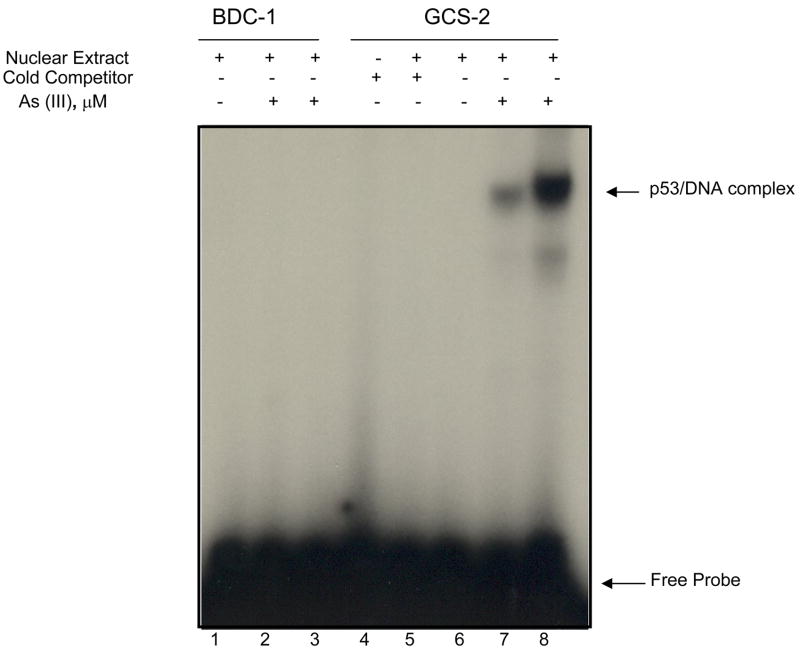
Arsenite enhances p53 binding to Hsp90β promoter
To determine if p53 can regulate Hsp90 gene expression in vitro, we end labeled a DNA fragment of 30 bp (−131/−160) consisting of the p53 binding site (BS) in the Hsp90β promoter region of the gene with [γ-32P] ATP and used it as a probe or as specific competitor without labeling in EMSA. Nuclear extracts prepared from either untreated or arsenite treated BDC-1 or GCS-2 cells were incubated with the probe in vitro. While nuclear extracts from untreated BDC-1, GCS-2, and arsenite treated BDC-1 cells were unable to bind the probe effectively, extracts from arsenite treated GCS-2 cells bound the probe more efficiently and the binding was dose-dependent (Fig. 3, compare lanes 1, 2, and 3 against 6, 7, and 8). Arsenite ().5 μM) was able to increase p53 binding to Hsp90β promoter sequence by 5 ± 1 fold (n=3; p ≤ 0.05) in GCS-2 cells (compare lanes 7 vs 8). In addition, the specific band could be competed out by excess unlabeled specific probe or when nuclear extract was omitted from the reaction (Fig. 3, lanes 4 and 5 respectively). Moreover, p53-specific antibody was able to super-shift the p53-bound DNA band confirming the specificity of the binding (data not shown). These results indicate that p53 BS in the proximal promoter region of Hsp90β gene participated in a more efficient constitutive expression of the Hsp90β gene and is important in the arsenite-induced response of the gene in GCS-2 cells.
Figure 3.
Effect of arsenite on p53 binding in GCS-2 cells. Gel mobility shift assay with nuclear extracts (BDC-1 and GCS-2) and 32P-labeled oligonucleotide containing GCAAG with excess cold competitor but no arsenite (lanes 1 and 6 respectively); BDC-1 and GCS-2 nuclear extracts without excess cold competitor and 0.2 μM arsenite treatment (lanes 2 and 7 respectively); 0.5 μM arsenite-treated BDC-1 and GCS-2 nuclear extracts without excess cold competitor (lanes 3 and 8 respectively); 32P-labeled oligonucleotide GCAAG with no nuclear extract and no arsenite (lane 4); GCS-2 Nuclear extract incubated with 32P-labeled oligonucleotide containing GCAAG and excess cold competitior (lane 5). For experimental conditions, see “Materials and Methods”. The data presented is a representative of three independent experiments.
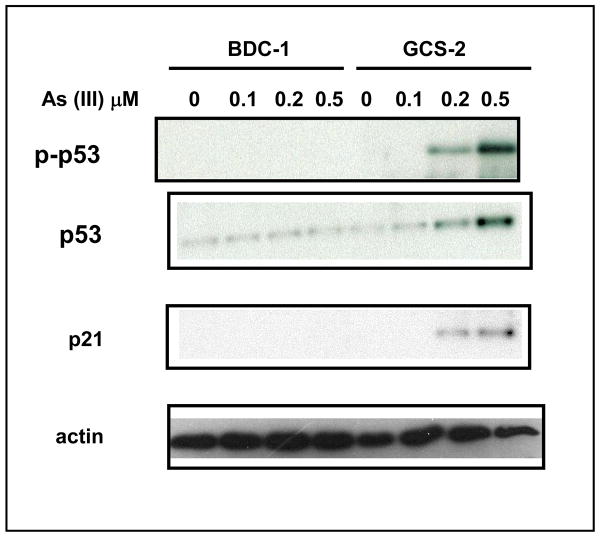
Arsenite Induces Phosphorylation of p53
A number of different stimuli are known to activate and stabilize p53 by post-translational modifications [20, 21]. These post-translational modifications include acetylation, phosphorylation, methylation, and ubiquitination [20, 21]. We investigated further to determine whether under our conditions p53 accumulation is also accompanied by its phosphorylation at Ser-15. We found that there was a dose-dependent increase in p53 phosphorylation at 21 h (3.5 ± 0.5 fold at 0.5 μM arsenite treated GCS-2 cells, n=3; p ≤ 0.05; Fig. 4, top panel). In addition to elevated p53 levels, we examined the role of p53 downstream effector gene encoding the cyclin/cdk inhibitor p21 involved in cell cycle arrest and found that its levels also increased with arsenite treatment in GCS-2 cells (2.2 ± 0.2 fold at 0.5 μM arsenite, n=3; p ≤ 0.05; Fig. 4).
Figure 4.
Effect of arsenite on p53 phosphorylation and p21 levels. Effect of arsenite dose response on p53 Ser-15 phosphorylation and p21 levels. Arsenite treated and untreated cell extracts were resolved by SDS-PAGE and immunoblotted with antibodies against phospho p53, total p53 or total p21. Equal loading was verified with anti-actin antibody (bottom panel). The blot presented is a representative of three independent experiments.
Arsenite stimulates different MAPK pathways
Arsenite has been shown to activate MAPK pathways which then transduce signals that lead to a wide array of cellular responses including cellular proliferation, stress responses, and apoptosis [37]. Phosphorylation-dependent activation of diverse transcription factors usually result from signaling through the MAPK pathways [37]. We examined if MAPK pathways are activated by arsenite in GCS-2 cells by western blotting (data not shown). Our results show that ERK, JNK and p38 MAPK were phosphorylated in a dose-dependent manner in GCS-2 cells upon exposure to arsenite (data not shown; ERK1/2, 11 ± 3 fold in 0.5 μM treated GCS-2 cells compared to untreated GCS-2 cells, n=3; p ≤ 0.05; JNK1/2, 12 ± 4 fold in 0.5 μM treated GCS-2 cells compared to untreated GCS-2 cells, n=3; p ≤0.05; p38 MAPK, 3 ± 0.05 fold in 0.5 μM treated GCS-2 cells compared to untreated GCS-2 cells, n=3; p ≤ 0.05).
Activation of MAP kinases is not required for arsenic-induced activation and stabilization of p53
We used specific inhibitors of ERKs, JNKs, and p38 MAPK to determine if the inhibition of their individual phosphorylation status had any effect on the arsenic-induced activation and stabilization of p53 (data not shown). As controls, we determined if specific inhibitors PD98059 (25 μM), SB600125 (25 μM), and SB203580 (10 μM) indeed disrupted ERK, JNK, and p38 MAP kinases respectively. The inhibition of ERK phosphorylation after pretreatment with PD98059, blocking JNK phosphorylation after treatment with SB600125, and inhibition of p38 MAP kinase phosphorylation after preincubation with SB203580 served as positive controls (data not shown). ERK1/2 phosphorylation was inhibited by PD98059 by more than 90% in 0.5 μM As (III) treated GCS-2 cells. Both SB600125 and SB203580 inhibited JNK1/2 and p38 MAPK phosphorylations by ~ 80% and 70% respectively (data not shown). We examined p53 phosphorylation and stabilization under the above conditions and found that it was unaffected (data not shown). Taken together, these findings indicate that the phosphorylation and stabilization of p53 by arsenic did not require the participation of any of the MAP kinases.
Arsenite Induces Phosphorylation of ATM
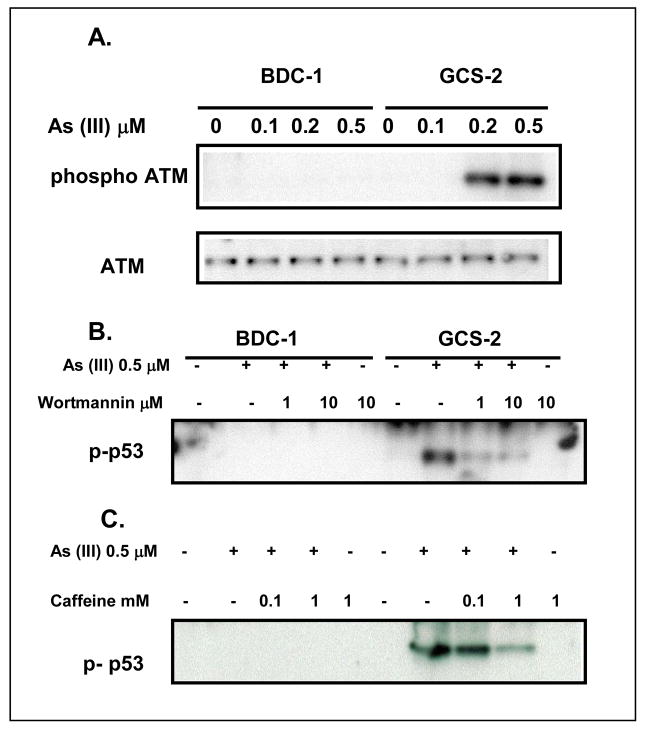
There are several kinases that are known to sense DNA damage and respond by initiating signaling pathways through p53 phosphorylation at the serine residues. Among the kinases identified to date, ATM is known to play a principal role in the transmission of oxidative DNA damage signals through phosphorylation of [13, 14, 22, 23, 37]. ATM is believed to be activated by its autophosphorylation at Ser-1981. In response to DNA damage, activated ATM is generally responsible for phosphorylating p53 at Ser-15. We assessed the possibility that arsenite- mediated toxicity in GCS-2 cells occurs via ATM-induced signaling events. As shown in Fig. 5A, arsenite induced ATM phosphorylation (5 ± 1 fold, n=3; p ≤ 0.05) in GCS-2 cells but not in BDC-1 cells. Pretreatment and continued presence of ATM inhibitors caffeine and wortmannin in these cells abolished ATM phosphorylation by 90% (data not shown). Under the same conditions, wortmannin or caffeine partially attenuated arsenite-induced phosphorylation of p53 (by 60–70% in 10 μM wortmannin and 1 mM caffeine treated GCS-2 cells compared to inhibitor untreated GCS-2 cells, Fig. 5B and C), implicating ATM in the p53 response to arsenite toxicity.
Figure 5.
Phosphorylation of ATM is required for arsenite-dependent p53 phosphorylation and stabilization in GCS-2 cells. A. Dose response of arsenite-dependent phosphorylation of ATM. Cells were treated with and without arsenite (0.5 μM) for 21h. Cell extracts were prepared and analyzed as described under “Materials and Methods” and immuno blotted with anti-phospho ATM (Ser 1981) or total ATM antibody. B and C. Effect of ATM inhibitor wortmannin or caffeine on arsenite-dependent phosphorylation of ATM. Cells were pretreated with the indicated concentrations of wortmannin or caffeine for 30 min prior to incubation with or without arsenite (0.5 μM) for 21 h. Cells were processed and immunoblotted with anti-phospho p53 antibody. The blots shown are representative of three independent experiments.
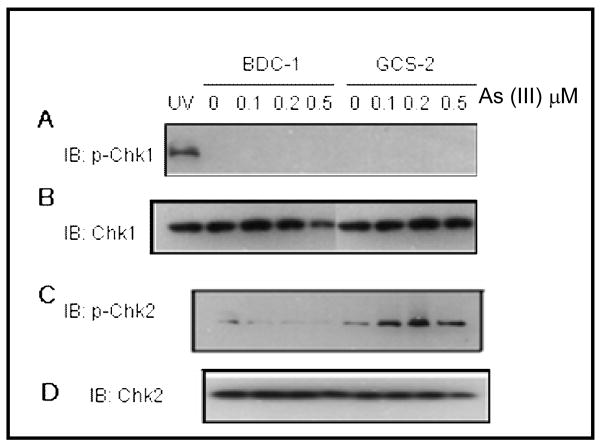
Arsenite induces phosphorylation of Chk2
Following the induction of DNA damage, phosphorylated ATM normally signals downstream to checkpoint kinases Chk1, Chk2, and p53. We assessed the phosphorylation of Chk1 (Ser-345) and Chk2 (Thr-68) in GCS-2 and BDC-1 cells exposed to arsenite. While phosphorylated Chk1 (Ser-345) was not detected in either control or treated BDC-1 or GCS-2 cells, phosphorylated Chk2 (Thr-68) was readily detectable in GCS-2 cells exposed to arsenite (Fig. 6A and C respectively; fold activation of Chk2 phosphorylation 2 ± 0.2, n=3, p ≤ 0.05). Taken together, these data show that oxidative stress caused by arsenite exposure under GSH deficient conditions induces phosphorylation of checkpoint proteins ATM, Chk2, and p53 through a caffeine- and wortmannin-sensitive pathway.
Figure 6.
Chk2, but not Chk1, is phosphorylated in GCS-2 cells in response to arsenite. Cells were treated with various concentrations of arsenite for 21 h and the cell extracts were processed and immunoblotted with anti-phospho Chk1, total Chk1, anti-phospho Chk2 or total Chk2 antibody. For Chk1 blots, UV induced phospho Chk1 was used as a positive control. The blots shown are representative of three independent experiments.
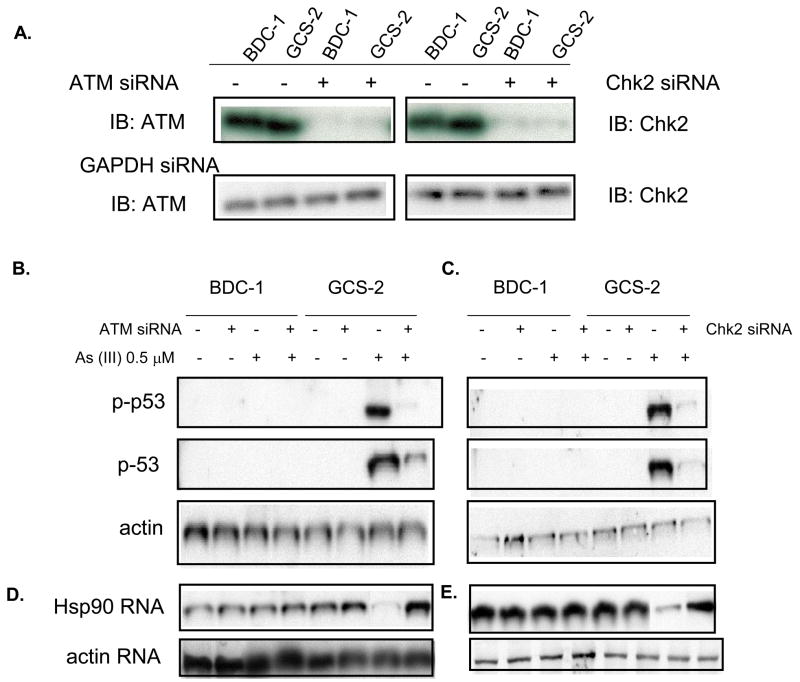
Inhibition of ATM and Chk2 prevent activation and stabilization of p53
To provide evidence that there is a link between ATM, Chk2 signaling and p53 stability in response to arsenite, we examined whether inhibition of ATM signaling through Chk2 blocked arsenite-induced phosphorylation and stabilization of p53. To achieve this, we utilized siRNAs to inhibit the expression of ATM and Chk2 in GCS-2 and BDC-1 cells. As shown in Fig. 7A, both BDC-1 and GCS-2 cells expressing the ATM and Chk2 siRNAs had reduced ATM and Chk2 levels by 90% and 80% respectively. The reduced expression of ATM and Chk2 were accompanied by attenuated p53 phosphorylation in response to arsenite in GCS-2 cells (4.3 ± 0.2 fold in 0.5 μM arsenite treated GCS-2 cells without ATM siRNA vs 1.1 ± 0.3 with ATM siRNA compared against BDC-1 cells treated with 0.5 μM arsenite without ATM siRNA, n=4, p ≤ 0.05 in Fig. 8B and 4.6 ± 0.3 fold in 0.5 μM arsenite treated GCS-2 cells without Chk2 siRNA vs 1.2 ± 0.3 with Chk2 siRNA compared against BDC-1 cells treated with 0.5 μM arsenite without Chk2 siRNA, n=4, p ≤ 0.05 in Fig. 7C). Thus, these results suggest that arsenite phosphorylates p53 via ATM and Chk2 signaling.
Figure 7.
SiRNA-mediated inhibitions of ATM and Chk2 can abolish p53 phosphorylation and restoration of Hsp90β mRNA levels. A. ATM and Chk2 siRNAs silence the expression of ATM and Chk2 in BDC-1 and GCS-2 cells. Cells were transiently transfected with ATM or Chk2 or GAPDH siRNA for 6 h and the cells were then washed and incubated in normal medium for an additional 18 h. Cells were then processed and analyzed by immunoblotting against ATM or Chk2 antibody. B and C. Inhibitions of ATM and Chk2 prevent phosphorylation and stabilization of p53. Transfected cells were treated with and without 0.5 μM arsenite for 21 h. Cell extracts were immunoblotted with anti-phospho p53 antibodies. D and E. Silencing of ATM and Chk2 restore Hsp90β mRNA levels. Cells were treated as described above under “B and C” and RNA was analyzed by Northern blotting using Hsp90 cDNA probes. The blots presented are representative of four independent experiments.
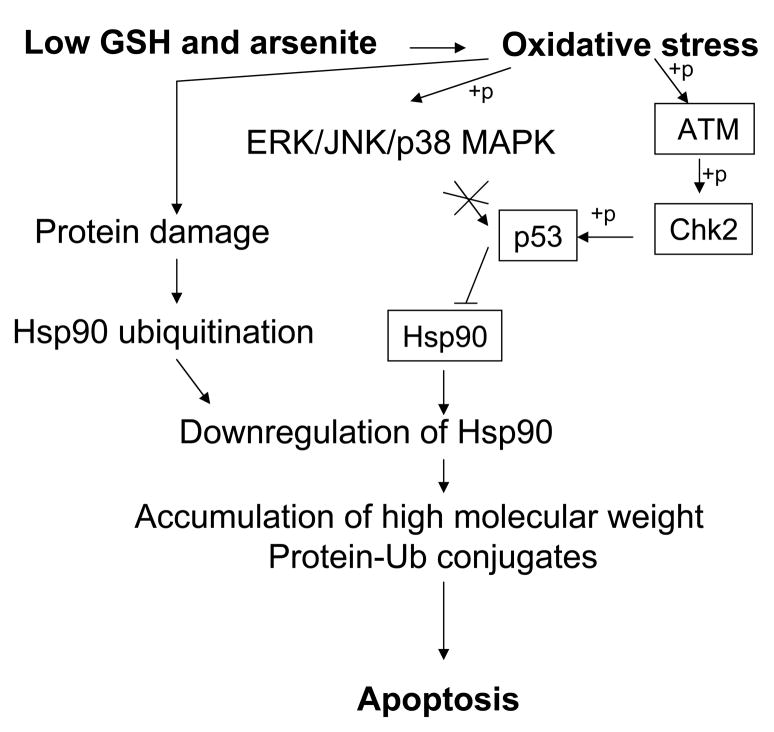
Figure 8.
Proposed model showing the signaling events induced by arsenite in the absence of GSH, culminating in apoptotic cell death. The processes indicated in this figure are a composite of events supported by data from this study and from our previous studies [33]. +p indicates phosphorylation.
Inhibition of ATM and Chk2 restore Hsp90β mRNA levels
We have previously shown that treatment of GCS-2 cells with arsenite resulted in the downregulation of Hsp90β mRNA levels and enhanced its ubiquitination [Fig. 1 and ref. 33]. Experiments were carried out to assess the possibility if inhibition of ATM and Chk2 expression in GCS-2 cells restores Hsp90β expression to wild type levels. Our results indicate that reduced expression of ATM and Chk2 in GCS-2 cells restored Hsp90β mRNA levels (Figs. 7D and E).
DISCUSSION
In the current study, we have taken advantage of GSH-deficient GCS-2 cells as an experimental system to examine the effects of very low/absent GSH levels on arsenite-induced Hsp90β regulation and the upstream mediators involved in the signaling process. A number of new observations have emerged from this work. First, ATM and Chk2 are activated by arsenite. Second, p53 is the downstream target of ATM activation. Third, activated and stabilized p53 trans-represses Hsp90β.
To achieve its cellular function as a molecular chaperone, it is essential to tightly control the level of Hsp90β. A delicate balance between transcription, translational efficiency, and posttranslational stability of the Hsp90 protein is critical for coordinating the level of expression of the gene. Our data show that Hsp90 RNA levels are downregulated in GCS-2 cells exposed to arsenite, suggesting that it is regulated at the transcriptional level (Fig. 1A).
In NAC-dependent GCS-2 cells, thiol antioxidant NAC could only partially reverse arsenite-induced p53 stabilization (Fig. 1D and 1E). This concurs with our earlier findings on arsenite-induced ubiquitination in GCS-2 cells cultured in the presence of NAC [33]. This suggests that even though the cysteine concentrations are comparable in GSH and NAC rescued GCS-2 cells, NAC may not be able to substitute for all the functions of GSH in the cell.
Arsenic has been implicated in the generation of hydroxyl radicals and induction of oxidative base lesions and oxidative DNA damage in mammalian cells [13, 14]. Most previous studies on the stabilization and activation of p53 in cells with normal GSH levels required exposure to high concentrations of arsenite ranging from 5 μM to 50 μM [14, 22, 24, 38]. However, under GSH deficient conditions in GCS-2 cells, p53 activation could be achieved by 10–60 fold lower arsenic levels (Fig. 1). Low GSH levels per se had no effect on the activation of the signaling pathways.
Levels of p53 are known to be stabilized by different stresses [18, 19]. Our observations on p53 stabilization by arsenite (Figs 1 and 4) are supported by a number of reports that have implicated the involvement of increased accumulation of p53 in arsenic toxicity in various cell types [14, 22, 24, 25). The cell cycle regulator, p21, is a transcriptional target of p53, and arsenite induces the accumulation of both p21 and p53 in GCS-2 cells (Fig. 4) suggesting that arsenite modulates the functions of both the proteins.
Studies on p53-regulated transcription have mainly focused on genes trans-activated by p53 [39, 40]. The tumor suppressor, p53, has also been shown to mediate the transcriptional repression of a number of cellular genes including c-myc, Cdk1, Cdc25C, and Cdc2 upon stabilization [41–45] but the molecular basis for this trans-repression remains unclear. Previous reports have implicated Hsp90 as the chaperone for wild type p53 and that Hsp90 is required to maintain the folded and active state of p53 and thus regulate its activity [46, 47]. Our findings indicate that Hsp90 is the target for arsenite-induced transcriptional repression by p53 (Fig. 2). This suggests that the molecular interaction between p53 and Hsp90β is more complex than previously thought, each playing an important role in the regulation of the other. In this context it is worth noting that Zhang et al have shown that p53 trans-represses Hsp90β in UV irradiation-induced apoptosis but the mechanism responsible for the activation of p53 after DNA damage was not elucidated [30].
Functional studies indicated that arsenite downregulated wild type p53 binding to p53 BS in the Hsp90β promoter region only under low GSH conditions (Fig. 3). Our data show that this inhibition could contribute to the repression of Hsp90β expression in GCS-2 cells. The p53 protein has been known to make significant contribution to antioxidant mechanisms that regulate genetic stability but its function as a transcription factor is highly susceptible to oxidative stress by pathophysiological conditions that generate products such as H2O2 and hydroxyl radicals. Several lines of evidence have indicated that exposure to arsenic and the generation of reactive oxygen species affects signal transduction cascades and thus activates or inactivates transcription factors and kinases [15, 16]. We recently reported that there was enhanced lipid peroxidation in GCS-2 cells depleted of GSH and NAC [33].
Although redox signaling is well accepted as a cellular mechanism, there are still uncertainties about how these signals are transmitted, and the critical targets of oxidant action are not well characterized. Perturbations in the cellular redox status are critical events that result either in cellular proliferation or cell death through the activation of a number of downstream signaling cascades. Previous studies have suggested the involvement of MAPK signaling pathways in the determination of cellular response to p53 activation [48]. Filomeni et al reported that in neuroblastoma cells, glutathione disulfide activated c-Jun-N-terminal kinase and p53 signaling and triggered apoptotic cell death [49]. Even though, arsenite preferentially activated MAPK pathways in the GCS-2 cells, it had no effect on p53 phosphorylation and stabilization indicating that p53 activation by arsenite in these cells is independent of MAPKs activation (data not shown).
Several lines of evidence support a role for ATM in IR-, drugs-, and UV- induced DNA damage. ATM and ATM-dependent signaling pathways have been investigated in the studies examining the effects of oxidative stress-induced DNA damage response [50]. Our results (Fig. 5) on ATM activation in GCS-2 cells are in general agreement with the studies on p53 accumulation in an ATM-dependent fashion in response to arsenite in human fibroblasts and porcine aortic endothelial cells [15, 24]. We also investigated the involvement of Chk1 and Chk2 as possible regulators of p53 downstream of ATM (Fig. 6). Despite the functional similarity of Chk1 and Chk2 in cell cycle checkpoint regulation, only Chk2 was found to be active under our cellular context indicating that arsenite-induced oxidative stress in GCS-2 cells activates ATM and subsequently Chk2 but not Chk1 (Fig. 6).
We have presented several lines of evidence to indicate that ATM is the kinase that phosphorylates Chk2 which in turn phosphorylates p53 (Figs. 5, 6, and 7). Our data thus indicate that both ATM and Chk2 are directly involved in the arsenite-induced phosphorylation and stabilization of p53 and thus p53 functions downstream of ATM and Chk2. These observations add to our knowledge of an intricate and tightly regulated signaling mechanism that is important for the control of Hsp90β function under oxidative stress. At present we can not rule out the existence of additional pathways regulating p53 mediated Hsp90 signaling.
Phosphorylation of p53 by multiple stress-activated kinases has been proposed to be essential for its stabilization, interaction with various transcriptional co-activators, and trans-activation or trans-repression of its target genes. Protein p53 target genes can be divided into two categories: i) genes that are induced or repressed independently of phosphorylation within the trans-activation or trans-repression domains and ii) genes that are dependent on such phosphorylation [51–53]. Our results indicate that the trans-repression of Hsp90 belongs to the latter category. Regulation of Hsp90 by p53 in GCS-2 cells appears to be a complex interplay between phosphorylation (Figs. 4 and 7) and possibly, the oxidation of critical cysteines present in the DNA binding domain of p53.
In summary, we have shown for the first time that arsenite toxicity mediates Hsp90 down-regulation through the ATM, Chk2, and p53-dependent pathway. These results and our previous work [33] demonstrate that arsenite regulates Hsp90β both at transcriptional and post-translational levels under GSH deficient conditions. These changes could lead to intracellular aggregation of damaged proteins culminating in cell death (Fig. 8). The intricate regulatory interplay between p53 and Hsp90 and their connection to stress signaling pathways will likely be critical for the cellular homeostasis.
Acknowledgments
This work was supported by NIH grants ES-08668. I thank Dr. Michael Lieberman for his support during the initial stages of this investigation; Dr. Mohamed Habib for critically reading the manuscript; and Ms. Weili Liu for expert technical assistance.
Footnotes
The abbreviations used are: ATM, ataxia-telangiectasia-mutated; ChIP, chromatin immunoprecipitation; GSH, glutathione; Hsp, heat shock protein; LUC, luciferase; NAC, N-acetyl cysteine; SiRNA, small interfering RNA;
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kitchin KT. Toxicol Appl Pharmacol. 2001;172:249–261. doi: 10.1006/taap.2001.9157. [DOI] [PubMed] [Google Scholar]
- 2.Simeonova PP, Wang S, Toriuma W, Kommineni V, Matheson J, Unimye N, Kayama F, Harki D, Ding M, Vallyathan V, Luster MI. Cancer Res. 2000;60:3445–3453. [PubMed] [Google Scholar]
- 3.Lau ATY, He Q-Y, Chiu J-F. Biochem J. 2004;382:641–650. doi: 10.1042/BJ20040224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meister A. J Biol Chem. 1988;263:17205–17208. [PubMed] [Google Scholar]
- 5.Kosower NS, Kosower EM. Int Rev Cytol. 1978;54:109–160. doi: 10.1016/s0074-7696(08)60166-7. [DOI] [PubMed] [Google Scholar]
- 6.Kala SV, Kala G, Prater CI, Sartorelli AC, Lieberman MW. Chem Res Toxicol. 2004;17:243–249. doi: 10.1021/tx0342060. [DOI] [PubMed] [Google Scholar]
- 7.Kala SV, Neely MW, Kala G, Prater CI, Atwood DW, Rice JS, Lieberman JMW. J Biol Chem. 2000;275:33404–33408. doi: 10.1074/jbc.M007030200. [DOI] [PubMed] [Google Scholar]
- 8.Dickinson DA, Forman HJ. Biochem Pharmacol. 2002;64:1019–1026. doi: 10.1016/s0006-2952(02)01172-3. [DOI] [PubMed] [Google Scholar]
- 9.Allen RG, Tresini M. Free Rad Biol Med. 2000;28:463–499. doi: 10.1016/s0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson DA, Forman HJ. Ann NY Acad Sci. 2002;973:488–504. doi: 10.1111/j.1749-6632.2002.tb04690.x. [DOI] [PubMed] [Google Scholar]
- 11.Mei N, Lee J, Sun X, Xing JZ, Hanson J, Le XC, Weinfeld M. FASEB J. 2003;17:2310–2312. doi: 10.1096/fj.02-0093fje. [DOI] [PubMed] [Google Scholar]
- 12.Yih LH, Lee TC. Cancer Res. 2000;60:6346–6352. [PubMed] [Google Scholar]
- 13.Martindale JL, Holbrook NJ. J Cell Physiol. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- 14.Kumagai Y, Sumi D. Annu Rev Pharmacol Toxicol. 2007;47:243–262. doi: 10.1146/annurev.pharmtox.47.120505.105144. [DOI] [PubMed] [Google Scholar]
- 15.Liu SX, Athar M, Lippai I, Waldren C, Hei TK. Proc Natl Acad Sci USA. 2001;98:1643–1648. doi: 10.1073/pnas.031482998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H, Hudson GG, Ding W, Wang S, Cooper KL, Liu S, Chen Y, Shi X, Liu KJ. Chem Res Toxicol. 2004;17:871–878. doi: 10.1021/tx049939e. [DOI] [PubMed] [Google Scholar]
- 17.Wang TC, Jan KY, Wang ASS, Gurr JR. Mutat Res. 2007;615:75–86. doi: 10.1016/j.mrfmmm.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Lavin MF. N Gueven Cell Death Diff. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 19.Oren M. Harvey Lect. 2001–2002;97:57–82. [PubMed] [Google Scholar]
- 20.Bates S, Vousden KH. Curr Opin Genet Dev. 1996;6:12–18. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 21.Hansen R, Oren M. Curr Opin Genet Dev. 1997;7:46–51. doi: 10.1016/s0959-437x(97)80108-6. [DOI] [PubMed] [Google Scholar]
- 22.Filippova M, Duerksen-Hughes PJ. Chem Res Toxicol. 2003;16:423–431. doi: 10.1021/tx025606a. [DOI] [PubMed] [Google Scholar]
- 23.Vogt BL, Rossman TG. Mut Res. 2001;478:159–168. doi: 10.1016/s0027-5107(01)00137-3. [DOI] [PubMed] [Google Scholar]
- 24.Tsou TC, Tsai FY, Yeh SC, Chang LW. Arch Toxicol. 2006;80:804–810. doi: 10.1007/s00204-006-0110-4. [DOI] [PubMed] [Google Scholar]
- 25.Joe Y, Jeong J–H, Yang S, Kang H, Motoyama N, Pandolfi PP, Chung JH, Kim MK. J Biol Chem. 2006;281:28764–28771. doi: 10.1074/jbc.M604392200. [DOI] [PubMed] [Google Scholar]
- 26.Pratt WB, Toft DO. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 27.Young JC, Barral JM, Hartl FU. Trends Biochem Sci. 2003;28:541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Blank M, Mandel M, Keisari Y, Meruelo D, Lavie G. Cancer Res. 2003;63:8241–8247. [PubMed] [Google Scholar]
- 29.Zhao Q, Boscheli F, Caplan A, Arndt KT. J Biol Chem. 2004;279:12560–12564. doi: 10.1074/jbc.M308242200. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Wang J–S, Chen L–L, Zhang Y, Cheng X–K, Heng F–Y, Wu N–H, Shen Y–F. J Biol Chem. 2004;279:42545–42551. doi: 10.1074/jbc.M314213200. [DOI] [PubMed] [Google Scholar]
- 31.Valverde M, Rojas E, Kala SV, Kala G, Lieberman MW. Mutat Res. 2006;594:172–180. doi: 10.1016/j.mrfmmm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Shi Z–Z, Osei-Frimpong J, Kala G, Kala SV, Barrios RJ, Habib GM, Lukin DJ, Danney CM, Matzuk MM, Lieberman MW. Proc Natl Acad Sci USA. 2000;97:5101–5106. doi: 10.1073/pnas.97.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Habib GM, Shi Z–Z, Lieberman MW. Free Radic Biol Med. 2007;42:191–201. doi: 10.1016/j.freeradbiomed.2006.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleinman WA, Richie JP., Jr J Chromatogr B Biomed Appl. 1995;672:73–80. doi: 10.1016/0378-4347(94)00194-a. [DOI] [PubMed] [Google Scholar]
- 35.Hirano S, Cui X, Li S, Kanno S, Kobayashi Y, Hayakawa T, Shraim A. Arch Toxicol. 2003;77:305–312. doi: 10.1007/s00204-003-0447-x. [DOI] [PubMed] [Google Scholar]
- 36.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 37.Tsou TC, Yeh SC, Tsai FY, Chang LW. Chem Res Toxicol. 2004;17:208–217. doi: 10.1021/tx034202v. [DOI] [PubMed] [Google Scholar]
- 38.Peng Z, Fan Y, Zandi E, Shertzer HG, Xia YA. J Biol Chem. 2007;282:21487–21496. doi: 10.1074/jbc.M702510200. [DOI] [PubMed] [Google Scholar]
- 39.Kannan K, Amariglio N, Rechavi G, Jakob-Hirsch J, Kela I, Kaminski N, Getz G, Domany E, Givol D. Oncogene. 2001;20:2225–2234. doi: 10.1038/sj.onc.1204319. [DOI] [PubMed] [Google Scholar]
- 40.Polyak K, Xia Y, Zweier J, Kinzler KW, Vogelstein B. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 41.Le Gac G, Estève PO, Ferec C, Pradhan S. J Biol Chem. 2006;281:24161–24170. doi: 10.1074/jbc.M603724200. [DOI] [PubMed] [Google Scholar]
- 42.Koumenis C, Alarcon A, Hammond E, Sutphin P, Hoffman W, Murphy M, Derr J, Taya Y, Lowe SW, Kastan M, Giaccia A. Mol Cell Biol. 2001;21:1297–1310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho JS, Ma W, Mao DYL, Benchimol S. Mol Cell Biol. 2005;25:7423–7431. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.St Clair S, Giono L, Varmeh-Ziaie S, Resnick-Silverman L, Liu W–J, Padi A, Dastidar J, DaCosta A, Mattia M, Manfredi J. Mol Cell. 2004;16:725–736. doi: 10.1016/j.molcel.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Ho J, Benchimol S. Cell Death Differ. 2003;10:404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- 46.Müller L, Schaupp A, Walerych D, Wegele H, Buchner J. J Biol Chem. 2004;279:48846–48854. doi: 10.1074/jbc.M407687200. [DOI] [PubMed] [Google Scholar]
- 47.Walerych D, Kudla G, Gutkowska M, Wawrzynow B, Muller L, King FW, Helwak A, Boros J, Zylicz A, Zylicz M. J Biol Chem. 2004;279:48836–48845. doi: 10.1074/jbc.M407601200. [DOI] [PubMed] [Google Scholar]
- 48.Brown L, Benchimol S. J Biol Chem. 2006;281:3832–3840. doi: 10.1074/jbc.M507951200. [DOI] [PubMed] [Google Scholar]
- 49.Filomeni G, Aquilano K, Civitareale P, Rotilio G, Ciriolo MR. Free Radic Biol Med. 2005;39:345–354. doi: 10.1016/j.freeradbiomed.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 50.Kurz EU, Douglas P, Lees-Miller SP. J Biol Chem. 2004;279:53272–53281. doi: 10.1074/jbc.M406879200. [DOI] [PubMed] [Google Scholar]
- 51.Ohki R, Kawase T, Ohta T, Ichikawa H, Taya Y. Cancer Sci. 2007;98:189–200. doi: 10.1111/j.1349-7006.2006.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Giono L, Manfredi JJ. Mol Cell Biol. 2007;27:4166–4178. doi: 10.1128/MCB.01967-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson T, Tovar C, Yang H, Carvajal D, Vu BT, Xu Q, Wahl GM, Heimbrook DC, Vassilev LT. J Biol Chem. 2004;279:53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]