Abstract
Chronic exposure to arsenic-contaminated drinking water can lead to a variety of serious pathological outcomes. However, differential responsiveness within human populations suggests that interindividual genetic variation plays an important role. We are using Drosophila to study toxic metal response pathways because of unrivalled access to varied genetic approaches and significant demonstrable overlap with many aspects of mammalian physiology and disease phenotypes. Genetic analysis (via chromosomal segregation and microsatellite marker-based recombination) of various wild-type strains exhibiting relative susceptibility or tolerance to the lethal toxic effects of arsenite identified a limited X-chromosomal region (16D-F) able to confer a differential response phenotype. Using an FRT-based recombination approach, we created lines harboring small, overlapping deficiencies within this region and found that relative arsenite sensitivity arose when the dose of the glutathione synthetase (GS) gene (located at 16F1) was reduced by half. Knockdown of GS expression by RNA interference (RNAi) in cultured S2 cells led to enhanced arsenite sensitivity, while GS RNAi applied to intact organisms dramatically reduced the concentration of food-borne arsenite compatible with successful growth and development. Our analyses, initially guided by observations on naturally occurring variants, provide genetic proof that an optimally functioning two-step glutathione (GSH) biosynthetic pathway is required in vivo for a robust defense against arsenite; the enzymatic implications of this are discussed in the context of GSH supply and demand under arsenite-induced stress. Given an identical pathway for human GSH biosynthesis, we suggest that polymorphisms in GSH biosynthetic genes may be an important contributor to differential arsenic sensitivity and exposure risk in human populations.
Keywords: arsenite toxicity, glutathione synthetase, genetic variability, RNA interference, Drosophila
Arsenic-contaminated drinking water is widely distributed throughout both the developed and developing world and represents an extremely serious public health issue in many locations, particularly in Bangladesh (Parvez et al., 2006). Here, millions of tube wells were bored to discourage use of water from ponds, rivers, and open wells, often a source for cholera and other water-borne diseases (BGS and DPHE, 2001). Subsequently, it was discovered that tube-well water was frequently contaminated with arsenic (BGS and DPHE, 2001) and thus tens of millions in Bangladesh have been, and still are, ingesting arsenic via drinking water. Chronic arsenic ingestion can induce a myriad of pathologies (National Research Council, 2001), including malignancies of the bladder, kidney, and lung, as well as diabetes mellitus, neuropathy, vascular disease, respiratory effects, and various types of skin lesions. Owing to these worldwide health effects, a maximum contaminant limit for arsenic in water has been set to 10 μg/l (World Health Organization, 2006).
It has been suggested that variation in susceptibility occurring in populations chronically exposed to arsenic could have a genetic component. Differences in the metabolism of inorganic arsenic have been observed between native Andean women (Vahter et al., 1995) and children (Concha et al., 1998) chronically exposed to arsenic-contaminated drinking water when compared to other populations. Comparing total arsenic and arsenic metabolites in urine of individuals from Mexico, China, and Chile has supported the idea that interindividual differences in toxicity could be due to functional polymorphisms in genes involved in metabolizing the metal (Loffredo et al., 2003).
There are many studies of the cellular processes affected when arsenic interacts with biological tissues (reviewed in Kumagai and Sumi, 2007), but it is germane to consider genes in three particular categories: those that affect arsenic uptake, cellular elimination, and metabolism within the cell. Arsenite can be transported into mammalian cells via the aquaglyceroporin channels AQP7 and AQP9 (Liu et al., 2002b), but there are no data as yet regarding functional polymorphisms affecting individual sensitivity. Elimination of xenobiotics from cells frequently employs conjugation of glutathione (GSH) to substrate (via glutathione S-transferase [GST]), followed by ATP-dependent transporter-mediated export. Thus, a GSH-arsenic complex is secreted via the ABC multidrug resistant transporter family member MRP1 (Leslie et al., 2004). Differences in arsenic efflux might be associated with polymorphisms in GST-M1 and -T1 (Chiou et al., 1997), and a GST-P1 polymorphism was suggested to increase the odds of arsenic-induced skin lesions (McCarty et al., 2007). Knockout of the ABC transporter MDR1 in mice produced greater susceptibility to arsenic and higher arsenic accumulation in their tissues (Liu et al., 2002a).
When inside mammalian cells, inorganic arsenicals can undergo a series of metabolic reactions leading to mono- or dimethylated products of either the +3 or the +5 oxidation state. Different reaction schemes have been proposed: (1) inorganic arsenicals undergo a series of successive reduction and oxidative methylation reactions (Cullen et al., 1984) or (2) inorganic arsenicals (typically in the reduced +3 state) are conjugated to GSH and undergo a series of successive methylations, each intermediate providing a substrate for oxidation to the +5 state (Hayakawa et al., 2005). A GSH-requiring arsenate reductase (identical to glutathione S-transferase omega [GSTO]) has been implicated in scheme (1) (Zakharyan et al., 2001), while an arsenic(+3) methyltransferase (AS3MT) (Lin et al., 2002) can provide the methylating activity necessary in scheme (2) and could also function in scheme (1) by linking oxidative methylation with reduction of arsenic metabolites using GSH as a cofactor (Hayakawa et al., 2005; Thomas et al., 2004). Polymorphisms in GSTO (Marnell et al., 2003) and in AS3MT (Hernandez et al., 2008; Meza et al., 2005) have been correlated with individual differences in urinary metabolic arsenic profiles.
Such studies reinforce the notion that susceptibility to arsenic will have a strong genetic basis. To explore this further, we have taken an unbiased approach to the genetics of arsenic susceptibility through examination of natural variation present in geographically distinct populations. We have utilized Drosophila as our experimental organism owing to the ease and variety of genetic manipulations available (reviewed in Bier, 2005), as well as the high representation of genes homologous to those involved in many human disease pathways (Reiter et al., 2001), including cancer (Brumby and Richardson, 2005). This approach has revealed that, somewhat surprisingly, optimal activity of the glutathione synthetase (GS) gene is required for an effective physiological defense toward arsenite. The data obtained reinforce the notion that organismal tolerance toward long-term arsenic exposure requires a robust antioxidant system based on the GSH biosynthetic pathway and imply that allelic variation affecting the activity of any of the genes contributing to this pathway will likely be pertinent to individual susceptibility and risk.
MATERIALS AND METHODS
Flies
Flies were maintained on standard cornmeal medium at room temperature. Most wild-type strains were obtained from the Indiana University Stock Center at Bloomington, IN, although several strains came from the now defunct stock center at Bowling Green State University, OH. For generation of deficiency lines, isogenic flies harboring inserted FRT elements that flank regions to be deleted were obtained from either the Exelixis collection at Harvard University Medical School or the Bloomington Drosophila Stock Center. FLP-induced X-chromosomal deletions were generated as previously described (Parks et al., 2004). Stocks were maintained by balancing over Binsinscy. GS RNA interference (RNAi) lines were obtained from the Vienna Drosophila RNAi Center (Dietzl et al., 2007). Since these lines were designed to target sequences toward the 5′ end (VDRC49801) or the 3′ end (VDRC49719) of the GS genes CG6835 and CG32495, we took advantage of their separate second and third chromosome insertion sites to create (via the use of chromosomal balancers) a compound line (designated GSRNAi[5′/3′]) containing both RNAi inserts in homozygous condition. The daughterless (da)-Gal4 line was obtained from the Bloomington stock center, and we received the GclmL0580 line from Dr Robert Saunders (Fraser et al., 2003).
Arsenite Sensitivity Assays
Wild-type Strains
Embryos were collected essentially as previously described (Polak et al., 2002). Briefly, flies were allowed to lay eggs on a small grape juice agar plate, seeded with yeast paste, inserted into the neck of an inverted culture bottle. In total, 150–200 embryos (0–8 h old) were transferred to a piece of sterilized grape juice–soaked filter paper. Filter paper with embryos was then laid down on 5 g of Instant Drosophila Medium (Carolina Biological, Burlington, NC) hydrated with 30 ml of H2O or sodium m-arsenite (Sigma, St. Louis, MO) solutions of various concentrations. Emerging adults were counted, and eclosion data were compared on arsenite-supplemented and nonsupplemented food.
Microsatellite-Based Recombination Mapping
A variety of microsatellite markers covering the X chromosome were identified either from previous literature descriptions (e.g., Kauer et al., 2002) or from BLAST analysis of the relevant Drosophila genomic sequence. Markers useful for further analysis were chosen by their size heterogeneity when comparing PCR products (using unique sequence primers flanking the particular repetitive regions) produced from Oregon R 1970 and PVM genomic DNA. For the recombination analysis, F1 virgin females derived from an Oregon R-PVM cross were mated to PVM males, F2 embryos collected and placed on arsenite-free or arsenite-containing food as described, and individual eclosing adult progeny collected for PCR analysis of specific microsatellite markers. PCR products from experimental and control parental flies were sized on 3% MetaPhor agarose (Cambrex, Rockland, ME) gels run in 1× tris-acetate-EDTA at 4°C. For any given marker, the percentage of flies carrying one or the other parental allele was calculated.
X-Chromosome Deficiency Lines
Since most X-chromosomal deficiencies (Df) generated were lethal when homozygous, we maintained stocks as heterozygotes over the X-chromosome balancer Binsinscy. In order to compare the arsenite sensitivity of deficiency lines with otherwise genetically identical control lines, we had to create females heterozygous for the non-deficiency parental X chromosome (w1118) balanced over Binsinscy. We crossed these w1118/Binsinscy females to Binsinscy/Y males and collected the resulting embryos. These were placed on either arsenite-free or arsenite-supplemented food, and eclosing female adults of the genotype w1118/Binsinscy were counted. We performed an arsenite dose-response assay to identify a threshold concentration where the control female flies (w1118/Binsinscy) showed no obvious effects of arsenite on relative viability and then used this concentration (0.25mM sodium arsenite) to assess the comparative viability of each deficiency line. For these experiments, isogenic deficiency females of the various lines (w1118, Df/Binsinscy) were crossed to Binsinscy/Y males, the resulting embryos were collected and exposed to arsenite-free and arsenite-supplemented food, and eclosing female adults of the genotype w1118, Df/Binsinscy were counted.
Deficiency line data analysis
A viability ratio was calculated for the average of eclosing w1118/Binsinscy females from three bottles of 0.25mM arsenite-supplemented food to that from three bottles of arsenite-free food. This ratio was compared to the ratio of the average of w1118, Df/Binsinscy females eclosing from three bottles of arsenite-supplemented food to that from three bottles of arsenite-free food. If the chromosome deficiency produced sensitivity toward arsenite, then the viability ratio of deficiency lines should be significantly lower than that of control lines. On the other hand, if there is no effect of the chromosomal deficiency toward arsenite, the ratios should not be significantly different.
GSRNAi[5′/3′] Lines
GSRNAi[5′/3′] flies were crossed to a strain expressing Gal4 under the control of the da regulatory element. As controls, nontransgenic w1118 parental flies were crossed to the Gal4-expressing strain and GSRNAi[5′/3′] flies were crossed to a non-Gal4 expressing w1118 line. In all cases, progeny embryos were collected and the relative adult eclosion on arsenite-supplemented or arsenite-free food was determined. Quantitative reverse transcriptase–PCR (RT-PCR) was performed on adults as described below to demonstrate knockdown of GS transcripts via RNAi.
Cell Culture
Schneider's S2 cells were maintained at 25°C in Schneider's Drosophila Medium (1×) (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% Antibiotic/Antimycotic mix (Gibco).
Production of Double-Stranded RNA for RNAi in Tissue Culture
Production of double-stranded RNA (dsRNA) was performed as described previously (Clemens et al., 2000) with some adjustments. A ∼900- to 1000-bp fragment corresponding to a segment of the target gene was amplified from fly genomic DNA via PCR. A second round of PCR was performed to add the T7 promoter sequence to either end of a ∼500- to 700-bp segment within this fragment. Each T7 fragment was produced individually. PCR products were purified by using the MinElute PCR Purification kit (Qiagen, Valencia, CA). Two micrograms of each T7 fragment was used as template to produce dsRNA using the MEGAscript RNAi kit (Ambion, Austin, TX). Primers used in the generation of T7 constructs are described in the Supplementary Data.
Conditions for RNAi in Drosophila Cell Culture
We followed the basic RNAi conditions, with some modifications, of those described previously (Clemens et al., 2000). Drosophila S2 cells were diluted to a final concentration of 6.75 × 105 cells/ml in medium containing 10% FBS and 1% antibiotic/antimycotic. dsRNA (1 μg) was added directly to corresponding wells of a 96-well plate. Aliquots of cells (15 μl, 1 × 104 cells) were pipetted into wells containing either dsRNA or not. FBS-free Schneider's medium (50 μl) was added to the wells, and plates were then shaken vigorously at room temperature for 30 min, followed by addition of 100 μl of arsenite-free or arsenite-supplemented (35, 45, or 80 μM) Schneider's medium supplemented with 15% FBS. Cells were incubated for 0, 24, 48, and 72 h at 25°C prior to testing for arsenite sensitivity by cell viability. Quantitative RT-PCR was performed as described below to confirm knockdown of GS mRNA transcripts.
Cell Viability Assay
Cell viability was determined using the CyQUANT NF Cell Proliferation Assay kit (Invitrogen, Carlsbad, CA) following the manufacturer's directions. After reagent addition, the microplate was covered and incubated at 25°C for 30 min. Fluorescence was measured with excitation at 485 nm and emission at 530 nm using a BioTek FL600 Microplate Fluorescence Reader, and data were collected using the BioTek KC4 version 3.01 computer software program.
Quantitative RT-PCR
First strand synthesis was performed using the iScript cDNA Synthesis kit using 1 μg total RNA according to manufacturer's directions (Bio-Rad, Hercules, CA). Real-time PCR was performed as previously described with variations (Schweitzer and DeKoter, 2004). Briefly, 2 μl cDNA was used as template in a 25-μl reaction including 100 ng of primers, 1mM MgCl2, 0.2mM deoxynucleoside triphosphates (Fisher, Pittsburgh, PA), 1× ThermoPol Taq Buffer (NEB, Ipswich, MA), and 1U Taq polymerase (NEB) in filter-sterilized MilliQ H2O. SYBR green (Invitrogen) was used at a final concentration of 0.5× from a 10,000× stock. All amplification protocols used a 2-min melting step at 95°C followed by 40 cycles of amplification. Each cycle used a 15-s melting step at 95°C, an annealing step of 15 s at 62.1°C for actin, and 61.4°C and 61.7°C for the 5′ and 3′ region of CG6835 and CG32495, respectively, followed by an extension step at 72°C for 15 s. Each cycle ended with measurement of fluorescence at 85°C for 10 s. Primers used in RT-PCR are described in the Supplementary Data. Data analysis was performed using a relative quantification of Ct values to calculate the expression of GS (CG6835 and CG32495) relative to actin control expression using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
RESULTS
Identification of “Arsenite-Sensitive” and “Arsenite-Tolerant” Drosophila Strains
Several dozen geographic variants of Drosophila melanogaster were obtained from stock centers and individual investigators and examined for their ability to eclose successfully as adults after seeding of embryos on to food containing sodium arsenite at a variety of concentrations. The average percent survivability of a representative sample of 35 such strains (over 60 were tested) as compared to those reared on control food is depicted graphically in Figure 1. On the basis of such data, we decided to further test Oregon R 1970 and PVM as examples of strains that showed relative tolerance and relative susceptibility to arsenite, respectively.
FIG. 1.
Viability of Drosophila melanogaster strains in sodium arsenite. Seeded embryos were scored for percent adult eclosion on arsenite-containing food and normalized to values obtained when seeded on arsenite-free food. Two strains that showed relative resistance (R) and sensitivity (S) to arsenite are marked by arrows and were selected for further analysis (though others could also have been investigated).
Contribution of an X-linked Component to Differential Arsenite Sensitivity
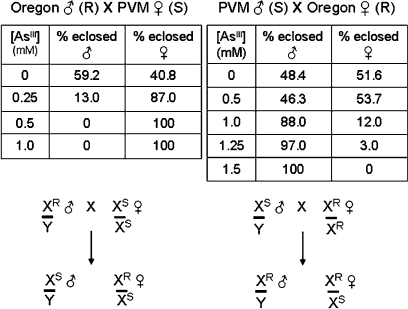
Embryonic offspring of reciprocal crosses between Oregon R 1970 and PVM adults were collected, reared on arsenite-free or arsenite-containing food as described above, and scored for percent adult eclosion relative to their male or female siblings at that arsenite concentration (Fig. 2). The most notable result from these studies was that relative tolerance to dietary arsenite segregated to a remarkable extent with parental origin of the X chromosome. Hence, males hemizygous for the Oregon R–derived X (XR) were quite tolerant to arsenite, whereas those males that carried the PVM-derived X chromosome (XS) were rather sensitive (see segregation scheme in Fig. 2, lower part). While this differential tolerance/sensitivity phenotype was clearly apparent in this first generation cross, its magnitude and X-chromosome segregation rapidly diminished upon subsequent crosses into F2 and later generations (data not shown). We infer that a quantitatively significant component(s) related to the observed differential arsenite sensitivity is/are encoded on the X chromosome but that other segregating loci contributing to the phenotype are present on the autosomes such that quantifiable Mendelian segregation is apparently lost in subsequent generations.
FIG. 2.
Relative adult eclosion percentages for male and female progeny resulting from reciprocal crosses of the Oregon R 1970 and PVM strains. Embryos were seeded on arsenite-free or arsenite-containing food and the sex of eclosing adults scored as a percentage of the total flies hatching at that concentration. Strains are designated as resistant (R) or sensitive (S) based on the data in Figure 1; the crossing scheme shown below represents the expected genotype of progeny from the reciprocal crosses if the arsenite tolerance/sensitivity gene(s) were X-linked.
Mapping an X-linked Arsenite-Tolerance Component to Subdivision 16
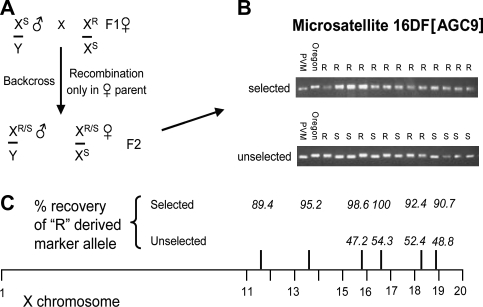
Since it seemed clear that at least one region of the X chromosome was associated with a differential arsenite sensitivity phenotype, we attempted to gain some preliminary genomic location information. To achieve this, we made use of a variety of microsatellite markers mapped to different locations throughout the X chromosome (see Materials and Methods) which showed size heterogeneity in direct comparisons between the Oregon R 1970 and PVM strains. In these experiments, the size of microsatellite-specific PCR fragments obtained from arsenite-tolerant F2 progeny of an Oregon R-PVM F1 heterozygous female crossed back to PVM males (see Fig. 3A) was compared with the parental PVM and Oregon R fragments on high-resolution agarose gels. This procedure allows for recombination occurring on the X chromosome in the F1 hybrid female to be detected in the F2 progeny. With the crossing scheme employed, only F2 progeny that receive the relevant arsenite tolerance-encoding X-chromosomal region from the F0 Oregon R 1970 parent would be expected to survive the selection on high arsenite concentration food (0.5–1mM). As the percentage of surviving flies that carry individual Oregon R–specific microsatellite marker fragments increased toward 100%, we could infer increasingly tight linkage of the arsenite-tolerance region to the particular marker being scored. A representative example of such data for the tightly linked marker 16DF[AGC9] is shown for surviving male F2 progeny (Fig. 3B). Note that on nonselecting, arsenite-free food, eclosing flies have an approximately 1:1 distribution of the parental marker, as would be expected from randomly located recombination events in the F1 maternal X chromosome. The sum of these studies (Fig. 3C) led us to identify chromosome subdivision 16 as a likely location for an arsenite-tolerance region, and subsequent experiments were aimed at more closely defining (via deficiency analysis) a region where potential candidate genes could be identified and subsequently tested for their involvement in arsenite sensitivity.
FIG. 3.
Recombination mapping between Oregon R 1970 and PVM using strain-specific microsatellite markers on the X chromosome to locate an arsenite tolerance/sensitivity locus. (A) Crossing scheme shows X-chromosomal constitution of F1 heterozygous females backcrossed to PVM males—recombination on the female X leads to male progeny that contain either the resistant (R) or the sensitive (S) arsenite-response allele. (B) Microsatellite mapping in the 16DF region of the X chromosome shows mobility difference depending on parental source (R or S), allowing genotypic frequency to be scored in F2 males either selected for survival on 1mM arsenite or not selected (i.e., raised on normal food). (C) “R”-derived allele frequencies were scored for a variety of markers located along the X chromosome. As expected, when not selected for arsenite resistance, markers segregated in an approximately 1:1 ratio. When the R allele approaches 100% representation in resistant males, very close linkage to the arsenite-responsive allele(s) is anticipated.
Creation of Targeted X-Chromosomal Deficiency Lines
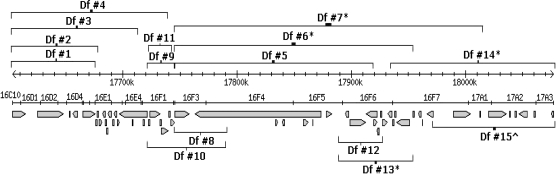
To create chromosomal deficiencies in this region, we took advantage of the Exelixis library of FRT-transformed lines that allow easy production (via FLP-FRT recombination) of a series of overlapping deficiencies of varying sizes (Parks et al., 2004). Using this approach, we were able to produce numerous deficiency lines within the chromosomal subdivision; several that contain overlapping deficiencies spanning the 16C10–17A3 region of the X chromosome are depicted in Figure 4. All deficiency lines were confirmed by PCR analysis of genomic DNA using primers flanking the predicted deficiency limits (as deduced from coordinates available in FlyBase—data not shown). Most of these deficiencies were homozygous lethal and so were maintained as heterozygotes over the X-chromosome balancer Binsinscy. Interestingly, a region toward the proximal end of that investigated here displays haploinsufficiency, while a region situated just beyond this could be deleted and homozygotes remained viable and fertile (Df #15).
FIG. 4.
X-chromosomal overlapping deficiencies created using the FLP-FRT recombination system to aid in identification of an arsenite response locus. Recovered deficiencies are shown both above and below a physical map of the 16C–17A chromosomal region, while the annotated genetic organization of the region from FlyBase is shown directly below it. *Haploinsufficient; ^Homozygous viable.
Arsenite Sensitivity of X-Chromosomal Deficiency Lines
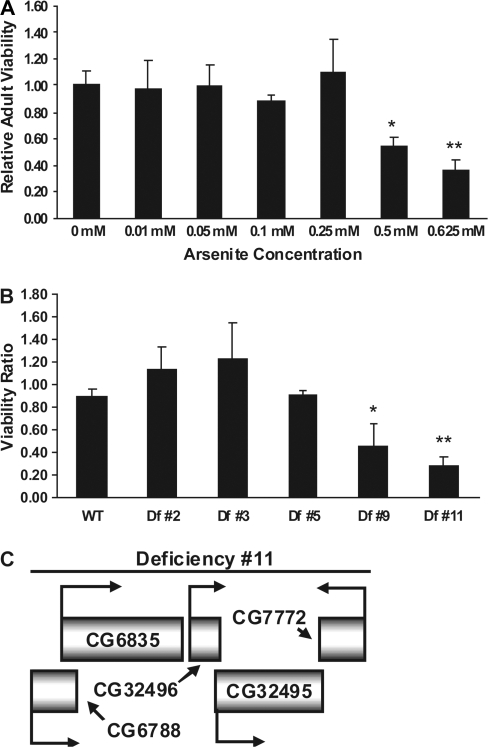
To determine the appropriate arsenite concentration for use in comparative sensitivity studies, we exposed nondeficiency (but otherwise isogenic) embryos to various concentrations, ranging from 0 to 0.625mM, of sodium arsenite (Fig. 5A). In this way, we chose an experimental toxicity testing threshold of 0.25mM arsenite, a concentration at which the relative viability of emerging w1118/Binsinscy female adults compared to those not exposed to arsenite was still one, but a concentration beyond which the relative survival rate fell substantially. In order to conclude that a particular X-chromosomal deficiency line was sensitive to arsenite compared to its nondeficient, but otherwise isogenic, parent, the ratio of emerging female adults exposed to 0.25mM arsenite versus those nonexposed should be significantly less than one. When tested in this way, the analysis showed that deficiency lines #9 and #11 were significantly more sensitive than all others tested (Fig. 5B). Inspection of the sequences removed in these lines, using annotated data derived from FlyBase, disclosed that both lines are deficient in five genes located at cytological subdivision 16F1, two of which, CG32496 and CG32495, appear to have resulted from an ancestral genomic duplication of sequences encompassing CG6788 and CG6835, respectively (see Fig. 5C). Intriguingly, given previous indications of the importance of GSH to arsenic metabolism, the duplicated genes CG6835 and CG32495 were found to encode GS, an enzyme which condenses γ-glutamylcysteine with glycine to create GSH in the second (and terminal) step of its biosynthesis.
FIG. 5.
Arsenite sensitivity of X-chromosomal deficiency lines. (A) Arsenite dose-response assay on parental w1118/Binsinscy strain to identify an experimental concentration threshold for testing toxic effects on Df lines. We chose to use 0.25mM arsenite for the sensitivity assays. Each bar represents the relative viability of emerging w1118/Binsinscy adults exposed to the specified concentration of arsenite-supplemented food when compared to emerging w1118/Binsinscy adults exposed to nonsupplemented food. *p < 0.01, **p < 0.005. (B) Viability ratio of various Df lines compared to that of the isogenic parental strain (w1118/Binsinscy) tested on 0.25mM arsenite-supplemented food. *p < 0.05, **p < 0.01. (C) Deficiency #11 encompasses a region containing five annotated genes: CG6835 and CG32495 encode the enzyme GS, CG6788 and CG32496 are genes encoding cell adhesion molecules, and CG7772 encodes a protein with carbonate dehydratase activity.
Genetic Manipulation of the GSH Biosynthetic Pathway Drastically Alters Arsenite Sensitivity
Previous work in mammalian tissue culture has supported a role for GSH in the cellular response to arsenic administration. GSH is synthesized in a two-step pathway that involves the initial condensation of glutamic acid with cysteine by the rate-limiting enzyme glutamate-cysteine ligase (GCL) to produce γ-glutamylcysteine, followed by the action of GS as described above. In order to determine if this biosynthetic pathway plays a critical role in arsenite sensitivity, we decided to test the effects of genetic manipulation of its components. Fraser et al. (2003) described a viable line containing a P element insertion in the 5′ untranslated region of the gene encoding the modifier subunit of GCL (GclmL0580) that reduced cellular GSH levels to approximately 50% of wild type. We induced precise excision of the P element in this line (Supplementary Fig. 1A) and then measured the arsenite sensitivity of both. The analysis showed that the GclmL0580 mutant line is substantially more sensitive to arsenite than the wild-type revertant (Supplementary Fig. 1B) and confirms data previously obtained in mouse cells that have sustained a Gclm knockout (Kann et al., 2005).
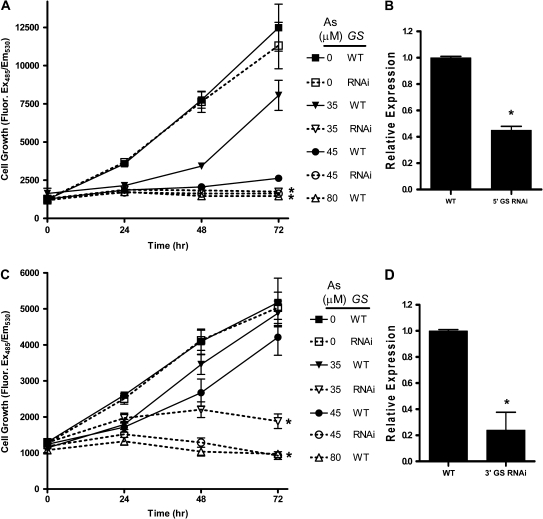
Since it is generally accepted that GCL-catalyzed synthesis of γ-glutamylcysteine represents the rate-limiting step in GSH biosynthesis, it might be expected that a reduction in activity of GS to 50% of wild-type levels, as anticipated for heterozygous deficiency lines #9 and #11, would have little overall consequence for GSH levels in the cell. To confirm whether GS activity could be important for arsenite sensitivity, as suggested by the chromosomal deficiency results, we initiated a series of RNAi studies, directed toward GS, to independently confirm the importance of its quantitative levels of activity in arsenite toxicity. In initial studies conducted in Drosophila S2 tissue culture cells, we targeted both an upstream (Fig. 6A) and a downstream (Fig. 6C) exon of the CG6835 and CG32495 genes via transfection of appropriately located double-stranded RNA (dsRNA) oligomers. These GS-compromised S2 cells showed heightened sensitivity toward arsenite as compared to cells exposed to the same arsenite concentration but expressing wild-type amounts of the enzyme. To determine the magnitude of RNAi-induced silencing of GS, we performed quantitative real-time RT-PCR of GS transcripts. Those transcripts targeted by the upstream region dsRNA were knocked down by approximately 50% (Fig. 6B), whereas transcripts targeted by the downstream region dsRNA were knocked down by approximately 80% (Fig. 6D).
FIG. 6.
RNAi-induced knockdown of GS expression in S2 cells. (A) dsRNA was targeted to the 5′ region of CG6835 and CG32495 and cell viability measured under differing concentrations of arsenite-supplemented growth medium. *p < 1.0 × 10−6 for 35 and 45μM As WT versus 35 and 45μM As GS RNAi. (B) RT-PCR analysis of GS transcript levels after targeting the 5′ region. Results have been normalized to actin. *p < 0.01. (C) dsRNA was targeted to the 3′ region of CG6835 and CG32495 and viability measured as described. *p < 1.0 × 10−7 for 35 and 45μM As WT versus 35 and 45μM As GS RNAi. (D) Real-time RT-PCR analysis of GS transcript levels after targeting the 3′ region. Results have been normalized to actin. *p < 0.01.
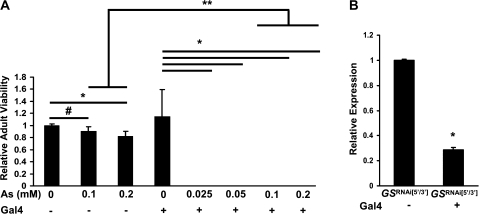
To test whether similar effects could be seen in the whole organism, we procured two lines of transgenic Drosophila engineered to inducibly express (under Gal4 regulatory control) dsRNA hairpin transcripts derived from either the 5′ or the 3′ regions of the two GS genes (Dietzl et al., 2007). In order to more efficiently target GS transcripts in vivo, we combined these two independent transgenes into a single homozygous line, GSRNAi[5′/3′] (see Materials and Methods). After crossing these compound RNAi flies to a line expressing the Gal4 protein under control of the ubiquitously expressed da regulatory element, we tested progeny for their sensitivity to food-borne arsenite, as previously described. Animals expressing the Gal4-induced dsRNA GS hairpins displayed no adult hatching as compared to control non-Gal4–expressing GSRNAi[5′/3′] animals when tested on food containing as little as 0.025mM arsenite (Fig. 7A). In fact, we failed to observe any feeding third instar larvae under these conditions indicating that a very strong developmental toxicity had been induced. In contrast, control non-Gal4–expressing GSRNAi[5′/3′] embryos not only formed larvae and pupae (tested at arsenite concentrations up to 0.2mM) but also developed into viable adults at high frequency. Quantitative real-time RT-PCR data showed that GS transcripts were indeed significantly reduced in the Gal4-expressing GSRNAi[5′/3′] larvae compared to those in the non-Gal4 GSRNAi[5′/3′] larvae when tested on arsenite-free food (Fig. 7B). Additional control experiments demonstrated that expression of the yeast Gal4 protein by itself does not affect the development of embryos into adulthood under arsenite-free or arsenite-supplemented conditions (Supplementary Fig. 2). We therefore conclude that, somewhat surprisingly, the sensitivity of whole organisms to ingested arsenite depends critically on the level and/or activity of the GS enzyme and not solely on that of the supposedly rate-limiting enzyme GCL.
FIG. 7.
Effects of RNAi-induced knockdown of GS in flies. (A) Embryos containing the Gal4-inducible GSRNAi[5′/3′] transgene were tested for their ability to eclose as adults in the presence or absence of ubiquitously expressed Gal4 on differing concentrations of arsenite-supplemented food. Survival is expressed relative to the non-Gal4–expressing GSRNAi[5′/3′] line on control (0mM arsenite) food. # p = 0.05, * p < 0.05, ** p < 0.01. (B) Real-time RT-PCR analysis of GS transcription in GSRNAi[5′/3′] flies in the presence or absence of a Gal4 transgene. *p < 0.01.
DISCUSSION
The long-term ingestion of arsenic-contaminated ground water represents one of the worst environmental calamities in history. Such water is extensively consumed not only in developing nations, such as Bangladesh (Parvez et al., 2006) and parts of India (Guha Mazumder et al., 1998), but also in more highly developed countries such as China (Yu et al., 2007) and the United States (Steinmaus et al., 2006). Its health effects range widely from conditions such as diabetes mellitus, peripheral vascular disease (e.g., Blackfoot disease), and neuropathy to a large variety of cancers. Interindividual genetic variations have been proposed as contributing to differences in susceptibility and response—for example, polymorphisms in either the AS3MT (Hernandez et al., 2008) or the GSTO (Marnell et al., 2003) genes have been associated with increased likelihood in developing arsenic-related diseases. Given the pleiotropic nature of disease outcomes upon arsenic exposure and the large number of pathways implicated in its biological interactions (reviewed in Kumagai and Sumi, 2007; Rossman, 2003), an unbiased approach to determining genes and/or pathways involved in differential susceptibility to its toxic effects at the whole organismal level seems warranted. Here, we have used natural geographic variants of D. melanogaster as a model system to test for genetic factors present in wild-type populations that may possibly predispose to arsenic susceptibility. Historically, Drosophila has been used to shed light on many fundamental biological processes, such as organismal development, owing to its ease of genetic manipulation and analysis. However, it has become an increasingly used model system to study human disease processes and signal transduction, owing to its unexpectedly high content of cognates to many genes involved in human genetic disease and metabolic pathways (see Introduction).
By employing a combination of classical chromosomal segregation and microsatellite marker-based recombination analyses, together with the creation of a series of overlapping deficiencies in the inferred region of interest, we identified a small region of the X chromosome (defined by Df #11 and encompassing cytological location 16F1) as being of particular interest with regard to arsenic susceptibility. The genomic sequence information available in FlyBase for this region, together with the inferred annotation of its genetic function, shows that a direct sequence duplication has apparently occurred, such that two copies of the GS gene are present, along with two copies of an adjacent gene encoding a cell adhesion protein and a single gene encoding a protein with carbonate dehydratase activity.
Given well-documented observations that GSH appears to play a role in the defense of cells against arsenic toxicity (Brambila et al., 2002), it was obviously of interest that an enzyme involved in the biosynthesis of GSH was implicated by our genetic analysis. On the other hand, that this enzyme might be GS seemed somewhat surprising in light of the prevailing view that it is the enzyme that precedes GS in the two-step GSH biosynthetic pathway, namely GCL, that provides the rate-limiting step (reviewed in Griffith and Mulcahy, 1999). Indeed, when we examined a fly line with a P-element insert in the 5′ UTR of the Gclm gene (encoding the regulatory subunit of the heterodimeric GCL enzyme) that causes about twofold reduction in GSH levels, it displayed very high sensitivity to arsenite, confirming previous studies performed in Gclm knockout mouse embryo fibroblasts (Kann et al., 2005). Thus, one prediction would be that moderately reduced GS expression, as anticipated in Df #11 owing to its 50% reduction in GS gene dose, would affect neither overall GSH levels in the cell nor its arsenite sensitivity, as long as its substrate (γ-glutamyl cysteine derived from the GCL-catalyzed step) was present at normal levels. The first part of this prediction was true, since we found that GSH levels in Df line #11 appeared similar to those of its nondeficient parent (data not shown). However, such deficiency flies showed distinct arsenite sensitivity and encouraged us to investigate the role of GS in arsenite sensitivity in greater detail.
RNAi-based knockdown analysis in S2 tissue culture cells amply confirmed that reduction in the expression of GS produced sensitivity to arsenite. Most strikingly, however, whole-organism knockdown of GS (to approximately 30% of normal levels) induced extreme sensitivity, with complete developmental toxicity occurring at up to 10-fold lower concentrations (and potentially even less) than those at which the first conspicuous effects on adult eclosion typically start to occur. Particularly noticeable was the fact that this toxicity appeared to occur very early in development (presumably shortly after embryo hatching), since very few active larvae could be observed under these conditions. Though these data seemed highly contradictory based on the prediction outlined above (in support of which Drosophila GCL has been shown to be rate limiting, Fraser et al., 2002, as in other organisms), they make a good deal more sense when the pathway for GSH biosynthesis is viewed in a broader context. The key to this is understanding that GSH is not a static component in the cell under conditions of arsenite-induced stress. This is because it is actively bound by arsenite in a stable As(GSH)3 complex (Kobayashi et al., 2005), which not only ties up free GSH from participating in its role as an antioxidant and regulator of the redox state of the cell (many studies have shown high levels of ROS in the presence of arsenic—see Kumagai and Sumi, 2007) but also provides the substrate for active transport of arsenic out of the cell by the multidrug resistance proteins (Leslie et al., 2004). It is in this situation of both synthesis and active consumption of GSH that the rate-limiting properties of GCL are likely to be compromised because changes in flux through the pathway (as would be produced under arsenite stress conditions when GSH is being consumed at a much higher rate) become sensitive to other steps in both the supply and demand pathways. Furthermore, though GCL is feedback inhibited by GSH under zero or low GSH consumption conditions (forming the basis for its reported rate-limiting behavior), this inhibition will be substantially relieved in a high GSH consumption situation, allowing other control points (such as the GS-catalyzed step) to contribute to a correspondingly greater extent. Such supply and demand considerations, inherent in the biochemical approach known as metabolic control analysis, have been recently discussed in great detail, both for the GSH pathway (Mendoza-Cozatl and Moreno-Sanchez, 2006) and for metabolic pathways in general (Moreno-Sanchez et al., 2008). In the present case, it provides an extremely plausible rationale for why reduced GS activity sensitizes cells that are experiencing chronic stress from arsenite exposure. The fact that this effect is particularly strong when considering developmental susceptibility as compared to cultured cell susceptibility emphasizes the importance of a whole-animal model in studying mechanisms and pathways of toxicity.
These results on the effects of reduced GS expression suggested that the differential sensitivity displayed by the PVM and Oregon R 1970 strains toward arsenite might be due to sequence polymorphisms in the CG6835 and/or CG32495 genes between the two strains. Such polymorphisms could lead to differences in levels of the enzyme, differences in the levels of transcripts, differences in transcript splice variants, or differences in enzyme activity, any or all of which might then lead to differential availability of GSH under the sustained stress of arsenite intoxication. While it is clear that both strains do contain the duplicated GS genes, we have not yet sequenced the genes and their flanking regions in the two strains in order to address these possibilities; preliminary analyses of multiple GS transcripts (i.e., splice variants) and their levels have shown that a good deal of complexity is present (data not shown). We have also measured GSH levels in these two strains under both control and arsenite-stressed conditions and have not found obvious differences (data not shown). However, these data might easily be compromised by the fact that typical GSH assays (including the one that we used—Senft et al., 2000) cannot distinguish between GSH and the substrate for the GS reaction (i.e., γ-glutamyl cysteine), so this distinction needs to be made in further investigations.
According to the results described here, even though the GCL heterodimer is the rate-limiting enzyme in the production of GSH under normal conditions, in the presence of arsenite (and potentially other heavy metal toxicants) optimal GS activity is required to sustain high enough levels of bioavailable GSH to protect cells, and thus an organism, against the effects of the chronically ingested toxicant. The HapMap consortium has reported single nucleotide polymorphisms in the GS gene of individuals from different regions worldwide (The International HapMap Consortium, 2003), and GS deficiency (whole or partial) is a well-described inherited autosomal recessive human condition (reviewed in Njalsson, 2005). Since it is clear that the synthesis and use of GSH in defense against arsenic intoxication is a common feature of both the invertebrate model studied here and the mammalian situation, we suggest that future studies of genetic polymorphism in human populations exposed to arsenic should consider potential associations between variant alleles of genes in the GSH biosynthetic pathway (such as GCL and GS) and disease susceptibility.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/ that include additional detailed methodological descriptions as well as arsenite sensitivity data on Gclm mutant and control Gal4-expressing lines.
FUNDING
National Institutes of Health (ES11009) to I.L.C.; National Institutes of Health-funded Center for Environmental Genetics (ES06096).
Acknowledgments
Thanks are expressed to Dr Chad Price, now sadly deceased, who provided invaluable aid in genomic BLAST analysis. We appreciate the help of Mayank Patel for providing the flies with nourishment.
References
- Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Genet. Rev. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- Brambila EM, Achanzar WE, Qu W, Webber MM, Waalkes MP. Chronic arsenic-exposed human prostate epithelial cells exhibit stable arsenic tolerance: Mechanistic implications of altered cellular glutathione and glutathione S-transferase. Toxicol. Appl. Pharmacol. 2002;183:99–107. [PubMed] [Google Scholar]
- BGS and DPHE. (2001). Arsenic contamination of groundwater in Bangladesh (D. Kinniburgh, and P. Smedley, Eds.), BGS Technical Report WC/00/19, British Geological Survey, Keyworth, UK. [Google Scholar]
- Brumby AM, Richardson HE. Using Drosophila melanogaster to map human cancer pathways. Nat. Rev. Cancer. 2005;5:626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- Chiou HY, Hsueh YM, Hsieh LL, Hsu LI, Hsu YH, Hsieh FI, Wei ML, Chen HC, Yang HT, Leu LC, et al. Arsenic methylation capacity, body retention, and null genotypes of glutathione S-transferase M1 and T1 among current arsenic-exposed residents in Taiwan. Mutat. Res. 1997;386:197–207. doi: 10.1016/s1383-5742(97)00005-7. [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha G, Nermell B, Vahter MV. Metabolism of inorganic arsenic in children with chronic high arsenic exposure in northern Argentina. Environ. Health Perspect. 1998;106:355–359. doi: 10.1289/ehp.98106355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen WR, McBride BC, Reglinski J. The reaction of methylarsenicals with thiols: Some biological implications. J. Inorg. Biochem. 1984;21:179–193. [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Kansagra P, Kotecki C, Saunders RD, McLellan LI. The modifier subunit of Drosophila glutamate-cysteine ligase regulates catalytic activity by covalent and noncovalent interactions and influences glutathione homeostasis in vivo. J. Biol. Chem. 2003;278:46369–46377. doi: 10.1074/jbc.M308035200. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Saunders RD, McLellan LI. Drosophila melanogaster glutamate-cysteine ligase activity is regulated by a modifier subunit with a mechanism of action similar to that of the mammalian form. J. Biol. Chem. 2002;277:1158–1165. doi: 10.1074/jbc.M106683200. [DOI] [PubMed] [Google Scholar]
- Griffith OW, Mulcahy RT. The enzymes of glutathione synthesis: Gamma-glutamylcysteine synthetase. Adv. Enzymol. Relat. Areas Mol. Biol. 1999;73:209–267. doi: 10.1002/9780470123195.ch7. [DOI] [PubMed] [Google Scholar]
- Guha Mazumder DN, Haque R, Ghosh N, De BK, Santra A, Chakraborty D, Smith AH. Arsenic levels in drinking water and the prevalence of skin lesions in West Bengal, India. Int. J. Epidemiol. 1998;27:871–877. doi: 10.1093/ije/27.5.871. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Kobayashi Y, Cui X, Hirano S. A new metabolic pathway of arsenite: Arsenic-glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch. Toxicol. 2005;79:183–191. doi: 10.1007/s00204-004-0620-x. [DOI] [PubMed] [Google Scholar]
- Hernandez A, Xamena N, Surralles J, Sekaran C, Tokunaga H, Quinteros D, Creus A, Marcos R. Role of the Met(287)Thr polymorphism in the AS3MT gene on the metabolic arsenic profile. Mutat. Res. 2008;637:80–92. doi: 10.1016/j.mrfmmm.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Kann S, Estes C, Reichard JF, Huang MY, Sartor MA, Schwemberger S, Chen Y, Dalton TP, Shertzer HG, Xia Y, et al. Butylhydroquinone protects cells genetically deficient in glutathione biosynthesis from arsenite-induced apoptosis without significantly changing their prooxidant status. Toxicol. Sci. 2005;87:365–384. doi: 10.1093/toxsci/kfi253. [DOI] [PubMed] [Google Scholar]
- Kauer M, Zangerl B, Dieringer D, Schlotterer C. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics. 2002;160:247–256. doi: 10.1093/genetics/160.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Cui X, Hirano S. Stability of arsenic metabolites, arsenic triglutathione [As(GS)3] and methylarsenic diglutathione [CH3As(GS)2], in rat bile. Toxicology. 2005;211:115–123. doi: 10.1016/j.tox.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Kumagai Y, Sumi D. Arsenic: Signal transduction, transcription factor, and biotransformation involved in cellular response and toxicity. Annu. Rev. Pharmacol. Toxicol. 2007;47:243–262. doi: 10.1146/annurev.pharmtox.47.120505.105144. [DOI] [PubMed] [Google Scholar]
- Leslie EM, Haimeur A, Waalkes MP. Arsenic transport by the human multidrug resistance protein 1 (MRP1/ABCC1). Evidence that a tri-glutathione conjugate is required. J. Biol. Chem. 2004;279:32700–32708. doi: 10.1074/jbc.M404912200. [DOI] [PubMed] [Google Scholar]
- Lin S, Shi Q, Nix FB, Styblo M, Beck MA, Herbin-Davis KM, Hall LL, Simeonsson JB, Thomas DJ. A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. J. Biol. Chem. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- Liu J, Liu Y, Powell DA, Waalkes MP, Klaassen CD. Multidrug-resistance mdr1a/1b double knockout mice are more sensitive than wild type mice to acute arsenic toxicity, with higher arsenic accumulation in tissues. Toxicology. 2002a;170:55–62. [Google Scholar]
- Liu Z, Shen J, Carbrey JM, Mukhopadhyay R, Agre P, Rosen BP. Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP9. Proc. Natl. Acad. Sci. U.S.A. 2002b;99:6053–6058. doi: 10.1073/pnas.092131899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loffredo CA, Aposhian HV, Cebrian ME, Yamauchi H, Silbergeld EK. Variability in human metabolism of arsenic. Environ. Res. 2003;92:85–91. doi: 10.1016/s0013-9351(02)00081-6. [DOI] [PubMed] [Google Scholar]
- Marnell LL, Garcia-Vargas GG, Chowdhury UK, Zakharyan RA, Walsh B, Avram MD, Kopplin MJ, Cebrian ME, Silbergeld EK, Aposhian HV. Polymorphisms in the human monomethylarsonic acid (MMA V) reductase/hGSTO1 gene and changes in urinary arsenic profiles. Chem. Res. Toxicol. 2003;16:1507–1513. doi: 10.1021/tx034149a. [DOI] [PubMed] [Google Scholar]
- McCarty KM, Ryan L, Houseman EA, Williams PL, Miller DP, Quamruzzaman Q, Rahman M, Mahiuddin G, Smith T, Gonzalez E, et al. A case-control study of GST polymorphisms and arsenic related skin lesions. Environ. Health. 2007;6:5. doi: 10.1186/1476-069X-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Cozatl DG, Moreno-Sanchez R. Control of glutathione and phytochelatin synthesis under cadmium stress. Pathway modeling for plants. J. Theor. Biol. 2006;238:919–936. doi: 10.1016/j.jtbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Meza MM, Yu L, Rodriguez YY, Guild M, Thompson D, Gandolfi AJ, Klimecki WT. Developmentally restricted genetic determinants of human arsenic metabolism: Association between urinary methylated arsenic and CYT19 polymorphisms in children. Environ. Health Perspect. 2005;113:775–781. doi: 10.1289/ehp.7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Sanchez R, Saavedra E, Rodriguez-Enriquez S, Olin-Sandoval V. Metabolic control analysis: A tool for designing strategies to manipulate metabolic pathways. J. Biomed. Biotechnol. 2008;2008:597913. doi: 10.1155/2008/597913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. Arsenic in Drinking Water: 2001 Update. Washington, DC: National Academy Press; 2001. [PubMed] [Google Scholar]
- Njalsson R. Glutathione synthetase deficiency. Cell. Mol. Life Sci. 2005;62:1938–1945. doi: 10.1007/s00018-005-5163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Parvez F, Chen Y, Argos M, Hussain AZ, Momotaj H, Dhar R, van Geen A, Graziano JH, Ahsan H. Prevalence of arsenic exposure from drinking water and awareness of its health risks in a Bangladeshi population: Results from a large population-based study. Environ. Health Perspect. 2006;114:355–359. doi: 10.1289/ehp.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak M, Opoka R, Cartwright IL. Response of fluctuating asymmetry to arsenic toxicity: Support for the developmental selection hypothesis. Environ. Pollut. 2002;118:19–28. doi: 10.1016/s0269-7491(01)00281-0. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001;11:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman TG. Mechanism of arsenic carcinogenesis: An integrated approach. Mutat. Res. 2003;533:37–65. doi: 10.1016/j.mrfmmm.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Schweitzer BL, DeKoter RP. Analysis of gene expression and Ig transcription in PU.1/Spi-B-deficient progenitor B cell lines. J. Immunol. 2004;172:144–154. doi: 10.4049/jimmunol.172.1.144. [DOI] [PubMed] [Google Scholar]
- Senft AP, Dalton TP, Shertzer HG. Determining glutathione and glutathione disulfide using the fluorescence probe o-phthalaldehyde. Anal. Biochem. 2000;280:80–86. doi: 10.1006/abio.2000.4498. [DOI] [PubMed] [Google Scholar]
- Steinmaus C, Bates MN, Yuan Y, Kalman D, Atallah R, Rey OA, Biggs ML, Hopenhayn C, Moore LE, Hoang BK, et al. Arsenic methylation and bladder cancer risk in case-control studies in Argentina and the United States. J. Occup. Environ. Med. 2006;48:478–488. doi: 10.1097/01.jom.0000200982.28276.70. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Waters SB, Styblo M. Elucidating the pathway for arsenic methylation. Toxicol. Appl. Pharmacol. 2004;198:319–326. doi: 10.1016/j.taap.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Vahter M, Concha G, Nermell B, Nilsson R, Dulout F, Natarajan AT. A unique metabolism of inorganic arsenic in native Andean women. Eur. J. Pharmacol. 1995;293:455–462. doi: 10.1016/0926-6917(95)90066-7. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Guidelines for Drinking-Water Quality: Incorporating First Addendum. Vol. 1, Recommendations. 3rd ed. Geneva, Switzerland: World Health Organization; 2006. [Google Scholar]
- Yu G, Sun D, Zheng Y. Health effects of exposure to natural arsenic in groundwater and coal in China: An overview of occurrence. Environ. Health Perspect. 2007;115:636–642. doi: 10.1289/ehp.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyan RA, Sampayo-Reyes A, Healy SM, Tsaprailis G, Board PG, Liebler DC, Aposhian HV. Human monomethylarsonic acid (MMA(V)) reductase is a member of the glutathione-S-transferase superfamily. Chem. Res. Toxicol. 2001;14:1051–1057. doi: 10.1021/tx010052h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.