Abstract
Mammalian target of rapamycin (mTOR) plays a key role in determining how growth factor, nutrient and oxygen levels modulate intracellular events critical for the viability and growth of the cell. This is reflected in the impact of aberrant mTOR signalling on a number of major human diseases and has helped to drive research to understand how TOR is itself regulated. While it is clear that amino acids can affect TOR signalling, how these molecules are sensed by TOR remains controversial, perhaps because cells use different mechanisms as environmental conditions change. Even the question of whether they have an effect inside the cell or at its surface remains unresolved. This review summarises current ideas and suggests ways in which some of the models proposed might be unified to produce an amino acid detection system that can adapt to environmental change.
Keywords: PAT, SLC36A1, mTOR, Rag, amino acid transporter
mTOR: a hub for nutrient and growth factor regulation of cellular processes
Mammalian target of rapamycin (mTOR) initially came to light as the cellular target for the immunosuppressant drug rapamycin. Since then, interest in rapamycin has extended to a widespread array of human diseases, including cancer, cardiovascular disease, autoimmunity, neurodegenerative diseases and metabolic disorders like diabetes, e.g, [1, 2]. This reflects the emerging roles for mTOR as a key regulator of energy homeostasis, cell physiology and growth within all higher eukaryotes, (reviewed in [3]).
The initial characterisation of TOR relied on studies in yeast and mammals. However, subsequent work in flies has played a key role in establishing that the growth factor-regulated signalling cascade involving the lipid kinase PI3-kinase (PI3K) and Akt controls mTOR through the G-protein Rheb (Ras homologue enriched in brain) and the TSC (tuberous sclerosis complex) components TSC1 and TSC2 [4]. The demonstration that this regulatory mechanism is conserved in mammalian systems led to rapamycin being taken into clinical trials to treat patients with specific forms of cancer [5]. Indeed, therapies designed to block the activity of mTOR have already proved to have an effect in restricting tumour growth, while treatments directed at PI3K/Akt have been hampered by toxicity issues [6]. This has stimulated interest from pharmaceutical companies in developing more mTOR inhibitors [7].
TOR exists in two distinct complexes [8], TORC1 and TORC2, that appear to act downstream and upstream of Akt respectively. The TORC1 complex, which is controlled by Rheb, mediates anabolic processes that promote growth by increasing protein synthesis. TORC1 stimulates translation by directly phosphorylating p70S6 kinase (S6K) and 4E-BP1 to promote ribosome assembly and translation. TORC1 is inhibited by rapamycin, which was thought not to inhibit TORC2 [9, 10]. However, there is now evidence that rapamycin can inhibit TORC2 in some cell types [11]. While it is currently unclear how TORC2 is regulated, growth factor-regulated TORC1 activity has also been shown to respond to oxygen levels and cellular energy status, which partially reflects extracellular nutrient levels (see Figure 1). Understanding the molecular mechanisms by which these controls are achieved has the potential to provide significant therapeutic benefits.
Figure 1. Growth regulation from the inside.
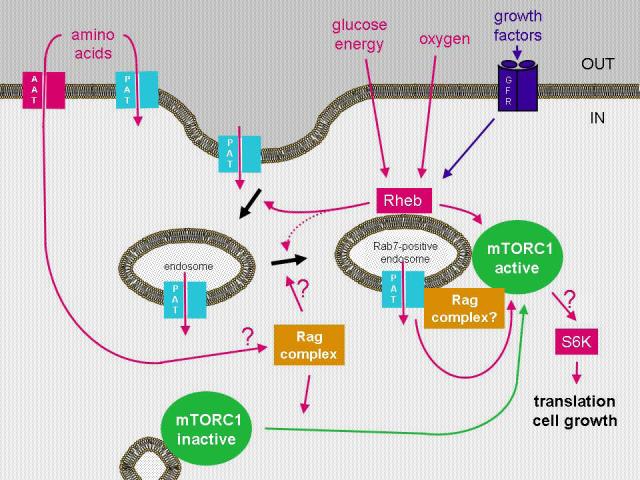
Rag GTPases are required to activate mTOR in response to amino acids [28, 29] and for mTOR to be shuttled to late (Rab7-positive) endosomes, which also contain the mTOR activator Rheb. The PAT amino acid transporters [35, 36] are TOR regulators [37] that can potentially operate in endosomal and lysosomal compartments, as well as at the cell surface. In this review, we suggest that the Rags might play a role in shuttling the PATs to late endosomes, from which they can help to recruit TOR, presumably leading to the activation of S6 kinase. The PATs would provide a means of utilising late endosomal versus extracellular amino acids, which may be particularly important under nutrient-deprived conditions.
A variety of mechanisms exist to provide the building blocks for cellular growth. Nutrients can be brought into cells through classical transporters, as is the case for amino acids and sugars [12]. Endocytosis results in membrane and extracellular proteins being brought into the cell, some of which are broken down and recycled. Cytoplasmic material can also be reused to provide the supplies needed for growth. This is particularly important when extracellular nutrients are low and TORC1 signalling is reduced, which leads to enhanced autophagy [13, 14], where organelles are destroyed and their component macromolecules recycled through autophagolysosomes.
AMPK (AMP-activated protein kinase) has emerged as a key sensor of energy depletion. It not only acts on TSC2 to suppress TORC1 signalling [15], but also on a component of TORC1, Raptor [16]. AMPK is activated by cellular stresses that reduce ATP levels, such as exercise or reduction in nutrients like fatty acids and glucose (reviewed in [17]). In an exciting recent development, activation of AMPK has been shown to delay the progression of tumours in mice [18].
Importantly, work in cell culture has suggested that intracellular amino acids, in particular leucine, may be involved in regulating TOR signalling (reviewed in [19]). However, in vivo analysis has also highlighted the role of other amino acids outside the cell [20, 21]. Perhaps the most likely scenario is that multiple amino acid-dependent mechanisms that respond to different environmental and physiological conditions regulate TOR signalling. Recent studies have highlighted a number of candidate regulators and the challenge is now to assemble these into a coherent amino acid sensing model.
What intracellular players have been linked to amino acid sensing?
A number of different molecules have been implicated in amino acid sensing. The Ste20 kinase family member MAP4K3 has been highlighted as a regulator of S6K and 4E-BP1 activity that modulates cell growth in response to amino acids, but not to growth factors or insulin [22]. However, the functional significance of MAP4K3 has yet to be tested in vivo. In addition, the class III PI(3) kinase, Vps34, has been suggested to regulate mTOR in mammalian cell culture [23, 24], but in vivo analysis in flies with Vps34 mutants has not confirmed this role [25]. This either suggests that Vps34 operates in a mammalian-specific or cell type-specific fashion or that in vitro experiments in the altered environment of the culture dish are not fully reflecting the amino acid sensing mechanisms taking place in living animals.
Several studies have suggested that mTOR is associated with intracellular membranes within the cytoplasm, including endoplasmic reticulum, Golgi, endosome and mitochondrial membranes [26]. Localisation of Rheb to endomembranes has been shown to be critical for its functions [27]. However, until recently it was unclear whether any of the membrane-bound pools of TOR were essential for its activities. It has now been shown in human embryonic kidney (HEK)-293T cells that in response to increased amino acid levels, mTOR shifts to Rab7-positive compartments where its activator Rheb is located. Furthermore, this relocalisation of mTOR is dependent on the Rag GTPases, which have therefore been proposed to act as amino acid sensors transmitting the signal from extracellular amino acids to TOR [28, 29]. In flies, the Rags are potent growth regulators, suggesting that they play an evolutionarily conserved role in TOR regulation [29].
Regulation of growth and TOR signalling by amino acid transporters
In parallel with the identification of intracellular mediators of amino acid sensing discussed above, other groups have focused their energies on finding the cell surface molecules that might ‘detect’ extracellular amino acids. Since one of the most popular models for amino acid-dependent TOR activation involves sensing intracellular amino acids, amino acid transporters have been postulated to play an important part in this process. In the 1990s, multiple cellular ‘Systems’ responsible for transporting amino acids were characterised [30, 31]. With the advent of molecular cloning, families of genes were assigned to these different transport systems. These families have been genetically screened for growth regulatory functions in flies. Although transporters in several classes have been linked to amino acid sensing involved in endocrine signalling (Minidiscs [32], Slimfast [33] and some tumour cell growth in culture [34]), surprisingly, a different class, the proton-assisted amino acid (PAT/SLC36) transporters (reviewed in [35, 36]) were highlighted as particularly potent growth activators in growing tissues in vivo and shown to interact with components of the PI3K/Akt and TOR signalling cascades [37, 38]
The PATs were initially identified on the surface of mammalian lysosomes, which led to them being named as lysosomal amino acid transporters (LYAAT [39]) and also as proton-dependent amino acid transporters [40]. PATs are related to AVT transporters in yeast [41], which can transport amino acids out of the vacuole, a lysosomal structure. In keeping with a role in promoting transport from acidic lysosomal compartments, PAT-mediated amino acid transport is normally, but not always, enhanced by extracellular acidification [37, 39, 40]. It is now known that PATs are also found at the plasma membrane and in endosomal compartments [40, 42]. However, while the presence of different subcellular pools of PATs is intriguing, the roles of these pools are currently unclear. In mammalian cells the PATs have been suggested to transport amino acids from the apical surface of the gut [36]. However, some PATs are expressed throughout the body [35], suggesting that they are likely to have much more widespread functions. The regulation of TOR by PATs represents such a role and opens up the important question of which of the various subcellular pools of this molecule is involved in this process.
PATs and intracellular amino acid sensing - towards a more unified model
The recent discovery that amino acids promote the shuttling of mTOR into Rheb-containing late endosomes via a Rag-dependent mechanism [28] has led to the proposal that Rags are involved in transmitting an amino acid-dependent signal from a sensor at the cell surface to these intracellular compartments. An alternate interpretation is that the Rags are involved in regulating a membrane-associated amino acid sensor that must shuttle from the cell surface to the Rheb-containing endosomes. Interestingly the yeast orthologues of Rags, Gtr1p and Gtr2p [43], are found in the late endosome and are required for proper shuttling of the amino acid permease Gap1p [44], suggesting that membrane protein trafficking is one of their key functions. This alternative model might also explain why Vps34, a Class III PI3-kinase involved in membrane trafficking, has also been implicated in amino acid sensing [25]. Furthermore, since Rheb and the TSC complex can influence protein trafficking [45-47], this might explain the synergistic interactions that take place between growth factor signalling and amino acid sensing.
The PATs are excellent candidate amino acid transporters to fulfil this shuttling-dependent amino acid sensing role, since they are found at the cell surface and in the endosomes and lysosomes, and they can function in acidic compartments like the late endosome (Figure 1). Furthermore, some data in flies suggest that one of the growth regulatory PATs, PATH, is a low capacity transporter and therefore may either function in a transport-independent fashion, acting as a so-called ‘transceptor’ (a receptor that is structurally related to a transporter [48] or activates TORC1 by transporting low levels of amino acids to a complex at a specific subcellular site [37]. Either model would fit nicely with a PAT/Rheb/TORC1 complex being specifically located at late endosomal membranes.
How could a model involving PATs and Rags fit with data suggesting cytosolic levels of leucine are important in controlling TORC1? One possibility is that amino acid-dependent regulation of TORC1 is multifaceted, involving TOR activation in subcellular compartments other than the late endosome, so that in some conditions, in cell culture for example, the import of amino acids like leucine into the cytosol can be a critical determining factor. In other conditions, signalling from the endosomes may be more important, and molecules like the PATs and Rags will play an increasingly central role. A more unifying alternative is that intracellular amino acids like leucine can modulate the activity of molecules like the PATs when they act via a transporter or transceptor mechanism. Fascinatingly, the activity of one of the transceptors characterised in yeast, the amino acid sensor Ssy1, is modulated in precisely this way by specific intracellular amino acids [49].
The presence of an endosomal amino acid sensing mechanism in higher eukaryotic cells might be particularly significant when extracellular nutrients or growth factors are sparse and/or when processes like autophagy are activated. There is evidence that autophagy has a positive effect on growth in some starvation conditions [50], and low level intracellular TOR signalling from late endosomal or lysosomal compartments might be essential for this. Equally important, subcellular localisation of TOR signalling in endosomes may become important in diseases like cancer, where cells need to survive and grow in low oxygen, nutrient and growth factor environments.
Thus, the finding that multiple mechanisms might be involved in linking amino acid sensing to TOR may really be a reflection of the cell’s versatility to adapt to different environments to which it is exposed. What is required now is a careful combination of in vivo and in vitro studies to define the different mechanisms that exist and the conditions in which they are employed. We are still some distance from understanding how amino acids are sensed by TOR. Recent work has, however, been critical in highlighting the importance of dissecting the protein shuttling mechanisms within specific parts of the cell, in addition to identifying the relevant signalling cascades.
Acknowledgements
Work on nutrient sensing from the D.C.I.G, C. W. and C.A.R.B groups is currently supported by grants from Cancer Research Technology (grant number C19591/A9093), Cancer Research UK (grant numbers C7713/A6174 and C19591/A6181) and previously by the Biotechnology and Biological Sciences Research Council. We apologise to those authors whose work we have been unable to cite due to limitations on the number of references.
Abbreviations used
- mTOR
mammalian target of rapamycin
- TORC
target of rapamycin complex
- Rheb
Ras homologue enriched in brain
- PAT
proton-assisted amino acid transporter
- LYAAT
lysosomal amino acid transporter
- PI3K
PI3-kinase
- TSC
tuberous sclerosis complex
- p70 S6K
p70 S6 kinase
- 4E-BP1
eukaryotic initiation factor 4E-binding protein
- AMPK
AMP-activated protein kinase
- ATP
adenosine triphosphate
- HEK
human embryonic kidney
References
- 1.Easton JB, Houghton PJ. mTOR and cancer therapy. Oncogene. 2006;25:6436–46. doi: 10.1038/sj.onc.1209886. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. doi: 10.1038/cdd.2008.110. 18 July 2008 epub. [DOI] [PubMed] [Google Scholar]
- 3.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Hafen E. Cancer, type 2 diabetes, and ageing: news from flies and worms. Swiss Med. Wkly. 2004;134:711–9. doi: 10.4414/smw.2004.09885. [DOI] [PubMed] [Google Scholar]
- 5.LoPiccolo J, Blumenthal GM, Bernstein WB, Dennis PA. Targeting the PI3K/Akt/mTOR pathway: effective combinations and clinical considerations. Drug Resist. Updat. 2008;11:32–50. doi: 10.1016/j.drup.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marone R, Cmiljanovic V, Giese B, Wymann MP. Targeting phosphoinositide 3-kinase: moving towards therapy. Biochim Biophys Acta. 2008;1784:159–85. doi: 10.1016/j.bbapap.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat. Rev. Cancer. 2006;6:729–34. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 8.Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 10.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell. Biol. 2004;6:1122–8. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 11.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 2006;22:159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 12.Edinger AL. Controlling cell growth and survival through regulated nutrient transporter expression. Biochem. J. 2007;406:1–12. doi: 10.1042/BJ20070490. [DOI] [PubMed] [Google Scholar]
- 13.Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–23. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardie DG. The AMP-activated protein kinase pathway-new players upstream and downstream. J. Cell. Sci. 2004;117:5479–87. doi: 10.1242/jcs.01540. [DOI] [PubMed] [Google Scholar]
- 16.Fruman DA, Edinger AL. Cancer therapy: staying current with AMPK. Biochem. J. 2008;412:e3–5. doi: 10.1042/BJ20080823. [DOI] [PubMed] [Google Scholar]
- 17.Kimball SR. The role of nutrition in stimulating muscle protein accretion at the molecular level. Biochem. Soc. Trans. 2007;35:1298–301. doi: 10.1042/BST0351298. [DOI] [PubMed] [Google Scholar]
- 18.Huang X, Wullschleger S, Shpiro N, McGuire VA, Sakamoto K, Woods YL, McBurnie W, Fleming S, Alessi DR. Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. Biochem. J. 2008;412:211–21. doi: 10.1042/BJ20080557. [DOI] [PubMed] [Google Scholar]
- 19.Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino Acid Regulation of TOR Complex 1. Am. J. Physiol. Endocrinol. Metab. 2008 doi: 10.1152/ajpendo.90645.2008. 2 Sept 2008 epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortimore GE, Khurana KK, Miotto G. Amino acid control of proteolysis in perfused livers of synchronously fed rats. Mechanism and specificity of alanine co-regulation. J. Biol. Chem. 1991;266:1021–8. [PubMed] [Google Scholar]
- 21.Bohe J, Low A, Wolfe RR, Rennie MJ. Human muscle protein synthesis is modulated by extracellular, not intramuscular amino acid availability: a dose-response study. J Physiol. 2003;552:315–24. doi: 10.1113/jphysiol.2003.050674. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol Chem, 2005;280:33076–82. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 24.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. 2005;102:14238–43. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–66. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Zheng XF. Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol. Biol. Cell. 2007;18:1073–82. doi: 10.1091/mbc.E06-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun. 2006;344:869–80. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- 28.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008;10:935–45. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christensen HN. Role of amino acid transport and countertransport in nutrition and metabolism. Physiol. Rev. 1990;70:43–77. doi: 10.1152/physrev.1990.70.1.43. [DOI] [PubMed] [Google Scholar]
- 31.Christensen HN, Albritton LM, Kakuda DK, MacLeod CL. Gene-product designations for amino acid transporters. J. Exp. Biol. 1994;196:51–7. doi: 10.1242/jeb.196.1.51. [DOI] [PubMed] [Google Scholar]
- 32.Martin JF, Hersperger E, Simcox A, Shearn A. minidiscs encodes a putative amino acid transporter subunit required non-autonomously for imaginal cell proliferation. Mech. Dev. 2000;92:155–67. doi: 10.1016/s0925-4773(99)00338-x. [DOI] [PubMed] [Google Scholar]
- 33.Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–49. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 34.Kim CH, Park KJ, Park JR, Kanai Y, Endou H, Park JC, Kim do K. The RNA interference of amino acid transporter LAT1 inhibits the growth of KB human oral cancer cells. Anticancer Res. 2006;26:2943–8. [PubMed] [Google Scholar]
- 35.Bermingham JR, Jr., Pennington J. Organization and expression of the SLC36 cluster of amino acid transporter genes. Mamm. Genome. 2004;15:114–25. doi: 10.1007/s00335-003-2319-3. [DOI] [PubMed] [Google Scholar]
- 36.Thwaites DT, Anderson CMH. Deciphering the mechanisms of intestinal imino (and amino) acid transport: the redemption of SLC36A1. Biochim. Biophys. Acta. 2007;1768:179–9. doi: 10.1016/j.bbamem.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Goberdhan DC, Meredith D, Boyd CA, Wilson C. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development. 2005;132:2365–75. doi: 10.1242/dev.01821. [DOI] [PubMed] [Google Scholar]
- 38.Reynolds B, Laynes R, Ogmundsdottir MH, Boyd CA, Goberdhan DC. Amino acid transporters and nutrient-sensing mechanisms: new targets for treating insulin-linked disorders? Biochem. Soc. Trans. 2007;35:1215–7. doi: 10.1042/BST0351215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sagne C, Agulhon C, Ravassard P, Darmon M, Hamon M, El Mestikawy S, Gasnier B, Giros B. Identification and characterization of a lysosomal transporter for small neutral amino acids. Proc. Natl. Acad. Sci. 2001;98:7206–11. doi: 10.1073/pnas.121183498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boll M, Foltz M, Rubio-Aliaga I, Kottra G, Daniel H. Functional characterization of two novel mammalian electrogenic proton-dependent amino acid cotransporters. J. Biol. Chem. 2002;277:22966–73. doi: 10.1074/jbc.M200374200. [DOI] [PubMed] [Google Scholar]
- 41.Russnak R, Konczal D, McIntire SL. A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J. Biol. Chem. 2001;276:23849–57. doi: 10.1074/jbc.M008028200. [DOI] [PubMed] [Google Scholar]
- 42.Rubio-Aliaga I, Boll M, Vogt Weisenhorn DM, Foltz M, Kottra G, Daniel H. The proton/amino acid cotransporter PAT2 is expressed in neurons with a different subcellular localization than its paralog PAT1. J. Biol. Chem. 2004;279:2754–60. doi: 10.1074/jbc.M305556200. [DOI] [PubMed] [Google Scholar]
- 43.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152:853–67. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 2006;8:657–67. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 45.Hennig KM, Colombani J, Neufeld TP. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J. Cell Biol. 2006;173:963–74. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urano J, Tabancay AP, Yang W, Tamanoi F. The Saccharomyces cerevisiae Rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J. Biol. Chem. 2000;275:11198–206. doi: 10.1074/jbc.275.15.11198. [DOI] [PubMed] [Google Scholar]
- 47.van Slegtenhorst M, Carr E, Stoyanova R, Kruger WD, Henske EP. Tsc1+ and tsc2+ regulate arginine uptake and metabolism in Schizosaccharomyces pombe. J. Biol. Chem. 2004;279:12706–13. doi: 10.1074/jbc.M313874200. [DOI] [PubMed] [Google Scholar]
- 48.Forsberg H, Ljungdahl PO. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 2001;40:91–109. doi: 10.1007/s002940100244. [DOI] [PubMed] [Google Scholar]
- 49.Wu B, Ottow K, Poulsen P, Gaber RF, Albers E, Kielland-Brandt MC. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J. Cell. Biol. 2006;173:327–31. doi: 10.1083/jcb.200602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–78. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]


