Abstract
In ribozyme catalysis, metal ions are generally known to make structural and∕or mechanistic contributions. The catalytic activity of a previously described Diels-Alderase ribozyme was found to depend on the concentration of divalent metal ions, and crystallographic data revealed multiple binding sites. Here, we elucidate the interactions of this ribozyme with divalent metal ions in solution using electron paramagnetic resonance (EPR) spectroscopy. Manganese ion titrations revealed five high-affinity Mn2+ binding sites with an upper Kd of 0.6±0.2 μM. In order to characterize each binding site individually, EPR-silent Cd2+ ions were used to saturate the other binding sites. This cadmium-induced EPR silencing showed that the Mn2+ binding sites possess different affinities. In addition, these binding sites could be assigned to three different types, including innersphere, outersphere, and a Mn2+ dimer. Based on simulations, the Mn2+-Mn2+ distance within the dimer was found to be ∼6 Å, which is in good agreement with crystallographic data. The EPR-spectroscopic characterization reveals no structural changes upon addition of a Diels-Alder product, supporting the concept of a preorganized catalytic pocket in the Diels-Alder ribozyme and the structural role of these ions.
Metal ions are of critical importance for the structure and function of biological macromolecules. Elucidating the function of individual ions is a challenging endeavor and particularly complicated in the case of multiple bound ions of one sort. First, it is often difficult to establish the precise number of bound ions, as their affinities may vary over several orders of magnitude. Second, spectroscopic techniques yield superpositions of all binding sites. Third, attempts to separate the individual binding sites by changing the experimental conditions may influence all sites, and therefore perturb the systems.
Most catalytically active RNAs are known to require divalent metal ions to establish full catalytic activity. This requirement, however, does not necessarily imply a mechanistic participation of the metal ions in catalysis, as their roles can also be purely structural. Examples for both cases are well described. For the extended hammerhead and hairpin ribozyme, crystallographic and mechanistic data suggest primarily a structural role of Mg2+ ions (Martick and Scott, 2006; Bevilacqua and Yajima, 2006), whereas in the group I intron, metal ions may be directly involved in the catalytic mechanism (DeRose, 2002). While these investigations provide an understanding of RNA’s strategies to catalyze phosphodiester chemistry, very little is known about how RNA accelerates other types of chemical reactions. One of the best-characterized artificial ribozymes catalyzes carbon-carbon bond formation by Diels-Alder reaction, a [4+2]-cycloaddition between an electron-rich anthracene diene and an electron-deficient maleimide dienophile, with high activity and selectivity (Seelig and Jäschke, 1999). This Diels-Alder ribozyme is active as a true catalyst exhibiting multiple turnover behavior and saturation-type kinetics, and further investigations revealed stereoselective formation of individual product enantiomers (Seelig et al., 2000). From the original ∼150 nucleotide long sequences, a 49-nt minimal ribozyme motif could be extracted, which was found to be fully active [Fig. 1a]. High catalytic activity required the presence of both monovalent and divalent cations (Seelig and Jäschke, 1999).
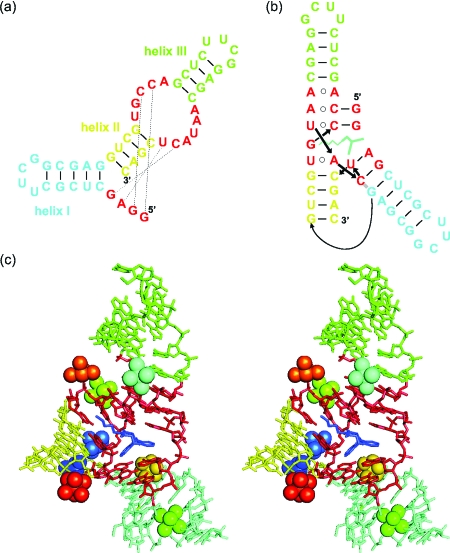
Figure 1. (a) Secondary structure of the 49-nt Diels-Alder ribozyme and (b) graphical representation of the tertiary fold in the crystal.
(c) Stereo view of the Mg2+-soaked crystal structure of the Diels-Alder ribozyme (Serganov et al., 2005, resolution 3.0 Å). Mg2+ ions are shown as spheres together with their coordinated water molecules.
The three-dimensional architecture of the Diels-Alder ribozyme was analyzed using mutation studies, a combination of probing techniques, and x-ray crystallography (Keiper et al., 2004; Serganov et al., 2005). The tertiary structure of the ribozyme is formed by an unusual tertiary interaction between the asymmetric internal loop and the 5′ terminus, resulting in the formation of a nested pseudoknot [Fig. 1b]. Moreover, the probing data indicated that the tertiary structure does not change upon substrate or product binding, thereby suggesting a preformed architecture of the Diels-Alder ribozyme. In the crystal structure of this ribozyme, eight Mg2+ ions were observed [Fig. 1c], out of which two are suggested to be due to crystal packing (Serganov et al., 2005). Interestingly, no Mg2+ ions were found in the immediate vicinity of the catalytic pocket, indicating that metal ions do not directly participate in catalysis, and a recent computational study attributes the proficiency of the ribozyme-catalyzed reaction to the stabilization of reactive ground state conformations in the ribozyme active site (Zhang and Bruice, 2007). However, the catalytic mechanism is still not fully understood, and metal(II) ions distant from the bond-breaking or bond-forming site have been shown to influence catalysis in other systems (Sigel and Pyle, 2007).
In the present study, we investigate the divalent metal ion binding sites of the 49-nt Diels-Alder ribozyme in solution using electron paramagnetic resonance (EPR) methods. EPR spectroscopy has been extensively utilized to probe metal ion binding sites in proteins (Reed and Markham, 1984; Ubbink et al., 2002; Calle et al., 2006) and recently also in nucleic acids, employing the paramagnetic Mn2+ instead of the physiological, but EPR spectroscopically silent Mg2+ (Vogt et al., 2006; Schiemann et al., 2003; Kisseleva et al., 2005). Due to the similar radii, Lewis acidity, and coordination chemistry of Mn2+ and Mg2+ ions, this substitution often causes no or only minor changes in the biological or chemical function. (Reed and Poyner, 2000). However, their coordination to RNAs shows some distinct differences (Freisinger and Sigel, 2007), making a case-by-case evaluation necessary. For the Diels-Alder ribozyme, this approach seems to be suited as the ribozyme was found to retain catalytic activity in the presence of Mn2+ (Seelig and Jäschke, 1999). To characterize the multiple metal ion binding sites in this system, we displace individual Mn2+ ions by EPR-silent Cd2+ ions, allowing the spectroscopic characterization of the remaining bound Mn2+ions.
RESULTS AND DISCUSSION
Quantification of Mn2+ binding sites at room temperature
In aqueous solution, Mn2+ forms [Mn(H2O)6]2+ ions, which give rise to a six-line continuous wave (cw) EPR signal at room temperature (Abragam and Bleaney, 1986), with the signal intensity being proportional to the concentration of Mn2+. The binding of Mn2+ to RNA slows down the rotation of the complex and creates an asymmetric ligand field, which both lead to excessive line broadening and ultimately to the disappearance of the signal (Horton et al., 1998). This effect is exploited here to quantify the Mn2+ sites in the Diels-Alder ribozyme by monitoring the EPR signal intensity while titrating Mn2+ into the buffered ribozyme solution (Supplementary Fig. S1). For comparison, Mn2+ was also titrated to the same buffer solution in the absence of RNA, and the binding isotherm was constructed from the difference of both curves and fitted to Eq. 1 (see “Experimental” section). This analysis revealed that in solution, the Diels-Alder ribozyme possesses five high-affinity Mn2+ binding sites with an upper and apparent (Sigel and Griesser, 2005) dissociation constant Kd of 0.6±0.2 μM, which is within a typicalrange for ribozyme-bound Mn2+ ions (Horton et al., 1998; Schiemann et al., 2003; Kisseleva et al., 2005). Titrations of the Diels-Alder ribozyme at different monovalent salt concentrations ranging from 0.1 to 4.3 M always led to five Mn2+ binding sites with an unchanged Kd≤0.6 μM (data not shown), demonstrating the specificity of the binding sites for divalent ions. This specificity differs markedly from the behavior of the hammerhead ribozymes for which only a single high-affinity binding site with a Kd of 4 μM prevails at high monovalent ion concentrations.
Low temperature studies on the Mn2+ binding sites
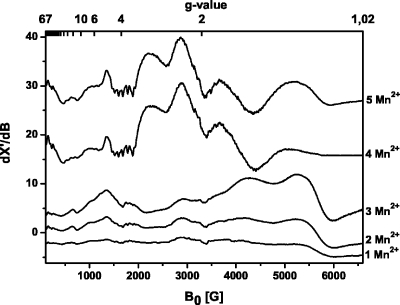
While the five high-affinity Mn2+ ions bound to the ribozyme show no EPR signal at room temperature (see above), lowering the temperature to 4.2 K slows the Mn2+ relaxation, and characteristic EPR spectra can be recorded (Fig. 2). The ribozyme with one Mn2+ ion bound ([ribozyme∕1 Mn2+] complex) yields an anisotropic Mn2+ spectrum with several features that spreads from a magnetic field value B0=100 to 6500 G. Similar Mn2+ spectra were previously observed for Mn2+-oxalate and -pyruvate complexes (Reed and Cohn, 1973), Mn2+ dioxygenase (Boldt et al., 1997), and Mn2+ superoxide dismutase and are distinctive for monomeric Mn2+ ions with a highly asymmetric coordination sphere, characterized by a large zero-field splitting D (Whittaker and Whittaker, 1991; Rusnak et al., 1999; White et al., 2001; Copik et al., 2005). One typical feature of such Mn2+ centers is a signal at g≈4.0 with a weakly resolved hyperfine splitting of roughly six lines and an average hyperfine coupling constant A≈90 G (Griscom and Griscom, 1967; Reed and Markham, 1984; Haddy et al., 1992; Ananyev and Dismukes, 1997), as observed for this ribozyme (Fig. 2). Although simulations of such manganese cw EPR spectra are complicated, the agreement between simulation and experiment (Supplementary Fig. S2) confirms the presence of a monomeric Mn2+ ion, with a large zero-field splitting D, here of ≈1500 G (see Supplementary Material for the complete set of parameters and Supplementary Fig. S2).
Figure 2. Cw X-band EPR spectra of the [Diels-Alder ribozyme∕ x Mn 2+ ] complexes with x =1, 2, 3, 4, 5 at 4.2 K.
The samples contained 0.2 mM Diels-Alder ribozyme in Tris-HCl buffer (pH 7.3, 0.3 M NaCl). EPR conditions: X-band frequency 9.424 GHz; microwave power 0.2 mW; modulation amplitude 22 G; scans 10.
The spectra of the [ribozyme∕1 Mn2+], [ribozyme∕2 Mn2+], and [ribozyme∕3 Mn2+] complexes appear to be identical to each other, only the EPR signal intensity increases with the number of bound Mn2+ ions, indicating that all three ions are monomeric and that their electronic structures are comparable (Fig. 2).
The spectrum changes dramatically when the ribozyme∕Mn2+ ratio reaches 1:4. The intense features at g=1.1 (6000 G) and 1.3 (5277 G) in the monomeric spectra are now strongly diminished and shifted by about 100 G to lower field. The resonances at g=1.6 (4265 G) and 2.17 (3095 G) disappear, whereas new broad and intense signals show up at g=1.8 (3665 G), 2.3 (2891 G), and 3.0 (2249 G). In addition, the resonances below 2000 G change their appearance and intensity. This new spectrum can be assigned to an electronically coupled dimer which is formed by two out of the four Mn2+ ions. The two remaining monomeric Mn2+ ions have weak spectral intensities and are hidden under the strong signal of the Mn2+ dimer.
Spectra with similar general appearance and intense broad resonances in the region between 2000 and 4600 G have been reported for model complexes of Mn2+-saldien (Mabad et al., 1986; Kitajima et al., 1991) and for rat liver arginase (Reczkowski and Ash, 1992; Khangulov, 1998) for which the dimeric Mn2+ centers are well established. A shift of the two high-field transitions, as observed here, has also been attributed to the formation of a dimeric Mn2+ center (Khangulov et al., 1995; White et al., 2001). In addition, dinuclear Mn2+ centers are often identified by a characteristic 11-line pattern with a splitting of 44 G (Epel et al., 2005), which is, however, not always observed (Golombek and Hendrich, 2003). In the case of the [ribozyme∕4 Mn2+] complex, the fine transitions at g≈1.6 and g≈1.85 may indicate such an average hyperfine splitting. However, the resolution of the splitting is too weak to be used as direct evidence for dimer formation. To further support the idea of a Mn2+-dimer formation, we have simulated the spectrum of the [ribozyme∕4 Mn2+] complex (see Supplementary Material for the complete set of parameters and Supplementary Fig. S3). The simulations resulted in an upper limit of the dipolar coupling constant Ddip of ∼360 MHz, which corresponds to a Mn2+-Mn2+ distance of ∼6 Å (Eaton and Eaton, 2000).
Addition of a fifth Mn2+ ion yields the [ribozyme∕5 Mn2+] complex with a spectrum that is only slightly different compared to the spectrum of the [ribozyme∕4 Mn2+] complex (Fig. 2). The intensity of the two high-field resonances increases, and they are shifted back to higher field [g=1.3 (5277 G) and g=1.1 (6000 G)] as found for the monomeric Mn2+ spectra. Therefore, the fifth Mn2+ does not engage in an additional dimer but is more likely a magnetically isolated Mn2+ ion whose signal is masked by the intense dimer spectrum.
These low-temperature studies indicate that the five Mn2+ ions possess different affinities and that the obtained Kd of 0.6 μM is indeed only an upper value.
Competition experiments with Cd2+ ions
Detailed characterizations of metal ion binding sites are typically performed on the sites of highest affinity, while the characterization of the lower-affinity sites is hampered by the presence of the occupied high-affinity sites. To characterize the lower-affinity sites while at the same time having the high-affinity sites populated (as would be the case in the catalytically active structure) we devise here a method for the silencing of the high-affinity sites by using EPR-inactive Cd2+ ions.
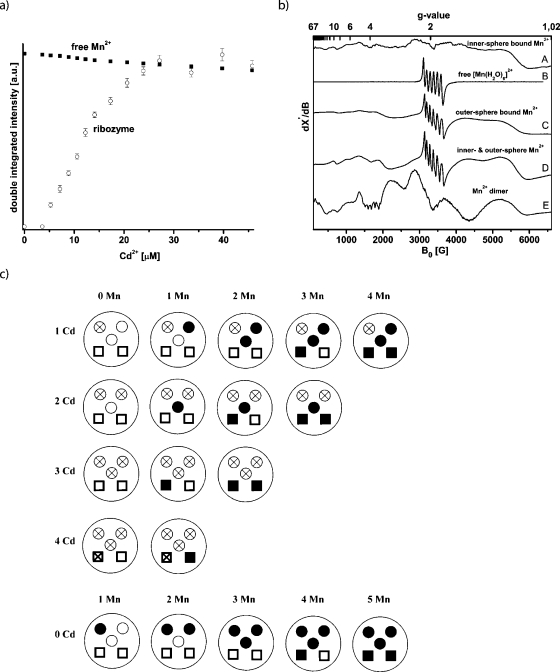
In a first step, the ability of Cd2+ to replace Mn2+ ions was probed. The [Diels-Alder ribozyme∕5 Mn2+] complex was prepared, showing no cw EPR signal at room temperature. The titration of Cd2+ into this solution led to the reappearance of the [Mn(H2O)6]2+ cw-EPR signal with an intensity that is within the experimental error comparable to the signal intensity of a sample of 40 μM Mn2+ [Fig. 3a]. This demonstrates that Cd2+ replaces all five high-affinity Mn2+ ions and that roughly one Cd2+ ion replaces one Mn2+ ion from the ribozyme. The other way around, the titration of the [ribozyme∕5 Cd2+] complex with Mn2+ showed no binding of Mn2+ ions up to a Mn2+ concentration of 1 mM (data not shown). Consequently, it can be concluded that Cd2+ binds stronger to the high-affinity binding sites than Mn2+, and that Cd2+ cannot be substituted by Mn2+.
Figure 3. (a)Cd 2+ titration curve of a sample containing 40 μMMn 2+ in 30 mM Tris-HCl buffer, pH 7.3, 0.3 M NaCl (squares) and of a sample containing.
8 μM of the Diels-Alder complex [ribozyme∕5Mn2+] in the same buffer (open circles). (b) Schematic of the Cd2+∕Mn2+ distribution in the Diels-Alder ribozyme. Open circle: nonoccupied innersphere binding site; Open circle with cross: innersphere binding site occupied by Cd2+; Solid circle: innersphere binding site occupied by Mn2+; Open square: nonoccupied outersphere binding site; Square with cross: outersphere binding site occupied by Cd2+; Solid square: outersphere binding site occupied by Mn2+. (c) cw X-band EPR spectra of the following complexes recorded at 4.2 K. (A) [ribozyme∕1 Mn2+], (B) free [Mn(H2O)6]2+ ion in water, (C) [ribozyme∕3 Cd2+∕1 Mn2+], (D) [ribozyme∕1 Cd2+∕4 Mn2+], and (E) [ribozyme∕4 Mn2+].
This ability of Cd2+ was used to prepare the following ribozyme∕metal complexes: one Cd2+ and no Mn2+ (1Cd2+-0Mn2+), 1Cd2+-1Mn2+, 1Cd2+-2Mn2+, 1Cd2+-3Mn2+, and 1Cd2+-4Mn2+. Then complexes with 2Cd2+, 3Cd2+, and 4Cd2+ ions were made and the free metal binding sites subsequently filled with Mn2+ ions. In every preparation the Cd2+ ions were added first so that the Mn2+ ions can occupy only the free binding sites. The occupation of the binding sites can be followed by the schematic representation of the experiment as depicted in Fig. 3b. The cw EPR spectra of these ten [ribozyme∕x Cd2+∕y Mn2+] complexes were recorded at 4.2 K and are summarized in Fig. 3c together with the spectrum of free [Mn(H2O)6]2+.
For the [ribozyme∕x Cd2+∕y Mn2+] complexes with a total amount of metal(II) ions of no more than three (1 Mn2+, 2 Mn2+, 3 Mn2+, 1Cd2+-1Mn2+, 1Cd2+-2Mn2+, 2Cd2+-1Mn2+), the spectral features are identical to the spectrum of the [ribozyme∕1 Mn2+] complex [Fig. 3c, trace A] but are distinctively different from free [Mn(H2O)6]2+ with octahedral symmetry [Fig. 3c, trace B]. This confirms the finding that the first three binding sites have comparable chemical environments. It furthermore indicates that the addition of Cd2+ to one binding site does not lead to significant structural perturbations at the other binding sites. The large anisotropy of these spectra is interpreted to be due to an asymmetric innersphere coordination of these first three monomeric Mn2+ ions.
In order to select the last two high-affinity binding sites we occupied the first three sites with Cd2+ ions and added one Mn2+ (3Cd2+-1Mn2+). The resulting spectrum with a dominant sextet at g=2 (3400 G) is shown in Fig. 3c, trace C. This spectrum is totally different compared with the spectra of the first three monomeric innersphere bound Mn2+ ions and of the free [Mn(H2O)6]2+. To exclude the possibility that the spectrum is a superposition of free and bound Mn2+, the solution was warmed to room temperature and showed no EPR signal, thereby verifying that all Mn2+ is bound. The intense sextet at g=2 suggests a more symmetric octahedral coordination geometry (Antanaitis et al., 1987) and indicates that this fourth high-affinity binding site binds a manganese-hexa-aqua ion through outersphere contacts (Reed and Poyner, 2000). The same spectrum was obtained for the [ribozyme∕ 4Cd2+∕1 Mn2+] complex, suggesting that the fifth Mn2+ is an outersphere bound [Mn(H2O)6]2+ too. Consequently, the [ribozyme∕3 Cd2+∕2 Mn2+] complex shows the same spectrum but with roughly doubled intensity. All the other combinations of metal(II) ion ratios (1Cd2+-3Mn2+, 1Cd2+-4Mn2+, 2Cd2+-2Mn2+, 2Cd2+-3Mn2+) show only superpositions of the spectra of the innersphere and outersphere bound Mn2+ ions in the respective ratios [trace D, Fig. 3c].
Interestingly, the [ribozyme∕1 Cd2+∕3 Mn2+] complex in which one of the three innersphere high-affinity binding sites is occupied by Cd2+ shows no Mn2+ dimer spectrum as observed for the [ribozyme∕4 Mn2+] complex. Similarly, no dimer spectrum was obtained in the case of the [ribozyme∕1 Cd2+∕4 Mn2+] complex [Fig. 3c]. We also turned the experiment around in the sense that first the four Mn2+ ions were added yielding the dimer spectrum, followed by the addition of one Cd2+, which led to the loss of the dimer spectrum and the appearance of the superposition of the monomeric innersphere and outersphere Mn2+ spectra. In fact, none of the spectra with at least one Cd2+ show the Mn2+ dimer spectrum.
These data lead us to conclude that the first and fourth ion selectively occupy sites critical for the formation of the dimer. Different scenarios can be considered to explain the observations. One possibility is that metal ion 4 is directly involved in the dimer and couples with one of the other three, or the binding of the fourth Mn2+ ion induces a conformational change, and the dimer is in fact formed from any combination out of the first three ions. According to scenario 1, the highest-affinity site occupied by the first Cd2+ ion belongs to the dimer actually formed from metal ions 1 and 4. Thus, the metal-metal dimer is still formed, but does not give rise to an electronically coupled dimer EPR spectrum as one of its ions is diamagnetic. In case of a conformational change (scenario 2), the first Cd2+ still occupies one of the dimer sites but pairs after the conformational change with either ion 2 or 3. Again, the formed dimer would not be electronically coupled.
The fact that the sextet at g=2 characteristic for outersphere bound [Mn(H2O)6]2+ is not visible in the [ribozyme∕4 Mn2+] spectrum supports the idea that the dimer is indeed formed by sites 1 and 4, since the fourth ion would no longer be a monomeric outersphere ion but part of the dimer with the characteristic dimer features. In case of the other scenario, the fourth ion would remain the monomeric outersphere ion and should show the sextet, which is, however, not observed in the experiment. Finally, we can rule out the possibility that the binding of Cd2+ to site 1 inhibits the conformational change induced by metal ion binding to site 4 and thereby prevents the formation of a dimer between ions 2 and 3, because the ribozyme remains catalytically active in the presence of cadmium ions, which it should not if the structural change did not take place (see below).
Based on these data, we conclude that the binding sites possess different affinities in the following order: site 1>site 2≅site 3>site 4>site 5. The order in which the binding sites are occupied seems to be the same for Mn2+ and Cd2+, since the sequence in which the outersphere and innersphere EPR spectra appear remains unchanged [compare Figs. 23c]. Yet it should be mentioned that we cannot fully rule out the possibility that the different metal binding sites are to a certain extent also statistically occupied. However, the percentage of this statistical occupation appears to happen on a minor scale according to all EPR spectroscopic evidence outlined above.
Influence of AMDA on the Mn2+ binding sites
For the minimal hammerhead ribozyme the cationic organic ligand neomycin was found to inhibit the catalytic self-cleavage of this RNA (Clouet-d’Orval et al., 1995) and to displace the high-affinity metal(II) ion (Schiemann et al., 2003). In the case of the Diels-Alder ribozyme the charge-neutral product analog AMDA binds to the catalytic site and inhibits the multiple-turnover reaction (Keiper et al., 2004). Therefore, we were interested in the question whether this inhibition is also accompanied by metal(II) displacement.
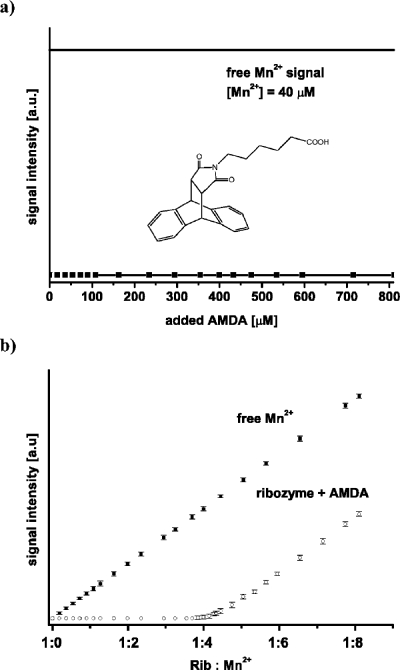
The titration of the [Diels-Alder ribozyme∕5 Mn2+] complex with AMDA up to an excess of 100-fold at room temperature does not lead to the appearance of a signal corresponding to free Mn2+ ions [Fig. 4a]. This indicates that AMDA does not replace any of the five high affinity Mn2+ ions from the Diels-Alder ribozyme molecule.
Figure 4. (a) AMDA titration curves of the [Diels-Alder ribozyme∕5Mn 2+ ].
(b) Mn2+ titration curves of samples containing only buffer (30 mM Tris-HCl, pH 7.3, 0.3 M NaCl) (squares) and of samples containing 8 μM Diels-Alder ribozyme, 0.8 mM AMDA in the same buffer (open circles).
To investigate whether the presence of AMDA influences the binding of Mn2+ ions to the ribozyme, a sample containing the Diels-Alder ribozyme and AMDA in ratios of 1:1, 1:40, 1:100 was titrated with Mn2+ [Fig. 4b]. These titrations showed that five Mn2+ ions were bound per ribozyme with an upper Kd of 0.6 μM as in the titration without AMDA. Additionally, the [ribozyme∕5 Mn2+] complex shows the same cw X-band EPR spectrum at 4.2 K in the presence of bound AMDA (complex:AMDA=1:100) as without AMDA. Thus, AMDA has no influence on the number, affinity, and general structure of the Mn2+ binding sites. This agrees with the crystal structure in which no metal(II) ions are located in the catalytic pocket and which shows no changes in the Mg2+ binding sites with AMDA bound.
Correlation of the EPR data with crystallographic and kinetic information
The crystal structure shows a total of eight Mg2+ ions [Serganov et al., 2005; Fig. 1c]. Out of these eight, the metal(II) ions Mg7 (orange) and Mg8 (red) make contacts between two RNA molecules in the crystal lattice. However, these two ions should not be present in solution, since the ribozyme acts—according to all available biochemical evidence (Wombacher et al., 2006)—as a monomeric enzyme. Furthermore, Mg4 (dark green, outersphere) apparently binds to a stretch of regular A-RNA (blue helix I), rendering a high-affinity interaction in solution less likely. This leaves a total of five potential metal ion binding sites, which agrees well with the five high-affinity binding sites found EPR spectroscopically.
Of the remaining five metal(II) ions in the crystal, two ions (Mg1 and Mg2, shown in dark blue and light blue, respectively) are innersphere coordinated, involving phosphate oxygens, and are located in close proximity to each other (7.4 A distance). Mg3 (cyan) is outersphere coordinated at the interface of the catalytic pocket and helix III. Mg5 (light green)—although only outersphere coordinated—seems to stabilize the back of the catalytic pocket by intricate coordination. Mg6 (gold, outersphere) sits at the interface of helix I and one of the pseudoknot-forming minihelices. Thus, the crystal structure and the EPR experiments reveal both innersphere and outersphere bound ions. However, an assignment of the outersphere and innersphere ions found in the EPR studies to the ions in the crystal structure must be done with caution because the EPR experiments were performed with Mn2+ whereas Mg2+ was used for the crystal structure. As known from the crystal structures of the minimal hammerhead ribozyme, Mn2+ ions can be shifted within the binding sites compared to Mg2+ ions (Pley et al., 1994; Scott et al., 1995; Scott et al., 1996), which is also in agreement with the hard and soft acids and bases (HSAB) concept. Furthermore, differences could arise from the ambiguities associated with the 3.0 Å resolution of the crystal structure. Hence, it is not surprising that the amount of the respective metal(II) ion types differs, 2 innersphere and 3 outersphere in the crystal structure and 3 innersphere and 2 outersphere in solution by EPR.
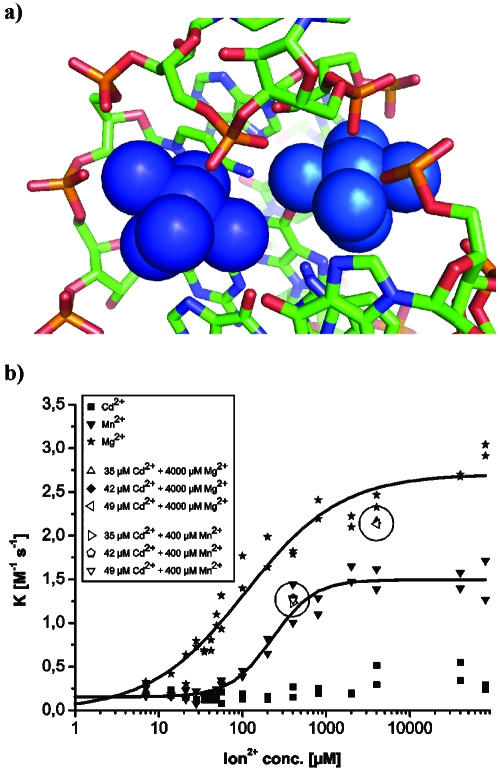
According to all evidence, the electronically coupled dimer identified by EPR corresponds to Mg1-Mg2 in the crystal structure. The distance of ∼6 Å inferred from the EPR spectrum agrees well with the distance of 7.4 Å measured between Mg1 and Mg2 in the crystal, especially considering the exchange of Mg2+ by Mn2+. A close-up view of the chemical environment [Fig. 5a] shows a shared binding pocket in which two neighboring phosphates, connected by one ribose, provide the innersphere ligands and various N- and O-ligands provide outersphere coordination at distances between 3.9 and 4.4 A. These two metal ions are obviously important for folding into the catalytically active structure and in particular for packing one of the pseudoknot minihelices tightly into the major grove of helix II. Furthermore, in the crystal, Mg2 stabilizes a crucial reversed Hoogsteen base pair. This is, to our knowledge, the first demonstration of an electronically coupled metal ion dimer in RNA.
Figure 5. Kinetic and structural correlations.
(a) Close-up view of the structure of the metal ion dimer formed by Mg1 (dark blue, left) and Mg2 (light blue, right) and its binding site as observed in the crystal structures (PDB code 1YLS). (b) Dependence of the initial rate on the metal(II) ion concentration. Highlighted spots (circled) represent metal ion mixtures.
In order to correlate the EPR data with catalytic activity, the initial rate of the ribozyme-catalyzed multiple-turnover Diels-Alder reaction was studied as a function of the concentration of Mg2+, Mn2+, and Cd2+ ions [Fig. 5b]. With Mg2+, we observe a sigmoidal dependence with a K1∕2 of 150 μM, corresponding to 21 Mg ions per RNA molecule. With Mn2+, the catalytic performance is reduced by about half, and K1∕2 increases slightly to 200 μM. These figures indicate that high-affinity binding alone is not sufficient for full catalytic activity, and further RNA-metal ion interactions are required.
Cd2+ ions alone do not induce catalytic activity. However, the addition of Cd2+ to a catalytically active ribozyme mixture containing either Mg2+ or Mn2+ had only minor effects on the catalytic activity [the highlighted spots in Fig. 5b]. These latter experiments were performed under conditions identical to those in the respective EPR measurements, indicating that the high-affinity binding sites are occupied by Cd2+ in these enzyme assays too. The high-catalytic activity of the ribozyme in Mn2+∕Cd2+ mixtures confirms that the use of Cd2+ in the EPR experiments did not lead to the formation of catalytically incompetent RNA structures and that cadmium-induced EPR silencing is apparently a valid method for the investigation of multiple metal ion-RNA interactions.
CONCLUSIONS
To conclude, the Diels-Alder ribozyme in solution contains five high-affinity Mn2+ binding sites with an upper Kd of 0.6±0.2 μM irrespective of the monovalent ion concentration. This renders Mg2+ ions 4, 7, and 8 found in the crystal structure as likely being due to solid state interactions. Out of the five metal(II) ions, two are outersphere and three are innersphere bound, where the latter ones possess higher affinities than the two outersphere bound ions. The competition experiments with Cd2+ showed that the affinities are also different within the two classes, allowing the five sites to be occupied consecutively. Two Mn2+ ions were found to form an electronically coupled dimer with a metal-to-metal distance of ∼6 Å, which is proposed to correspond to Mg1 and Mg2 in the crystal structure with a measured intermetal distance of 7.4 Å. The EPR measurements in the presence of the product analog inhibitor AMDA provide further support for the concept of a preformed catalytic pocket (Keiper et al., 2004) and suggests furthermore that the high-affinity metal(II) ions are not directly involved in the catalytic carbon-carbon bond formation but are involved in the stabilization of the RNA fold.
To obtain more detailed structural information about the different binding sites, temperature-dependent high-field EPR measurements and pulsed EPR experiments combined with site-directed spin labeling are underway. However, the methodology of cadmium-induced EPR silencing developed within this project was already found to be a useful tool for the investigation of RNA molecules interacting with multiple metal ions of one sort.
EXPERIMENTAL SECTION
RNA oligonucleotides and chemicals
The 49-nt Diels-Alder ribozyme (for sequence, secondary, and tertiary structure, see Fig. 1) was purchased from CSS Chemical Synthesis Service (Craigavon, UK). For the EPR experiments, the RNA was dissolved in 30 mM Tris-HCl buffer solution (pH 7.3) containing 0.1–4.3 M NaCl (>99.999%). MnCl2 and CdCl2 (>99.99%) were obtained from Sigma-Aldrich and were dissolved in the same buffer to a final concentration of 10 mM. The product analog inhibitor AMDA was synthesized as described previously (Stuhlmann and Jäschke, 2002), and 10 mM solutions of AMDA were prepared in Tris-HCl buffer with 10% acetonitrile and 10% ethanol as organic modifiers. Anthracene hexaethylene glycol (AHEG) for activity measurements was synthesized according to Fiammengo et al. (2005). All other chemicals were obtained from Sigma-Aldrich, Fluka, and Carl Roth.
EPR experiments
Cw X-band EPR spectra were recorded on a Bruker ESP 500e EPR spectrometer, equipped with an ER 4103TM cylindrical mode resonator for room temperature measurements in aqueous solutions. The titrations of the ribozyme with Mn2+ or ADMA were performed at 295 K using a sterilized flat cell with a volume of 150 μl and the following spectrometer parameter: center field 3480, sweep width 1000 G, 1024 points, microwave power 2 mW, modulation amplitude 12 G, modulation frequency 100 kHz, conversion time 41 ms, time constant 41 ms, 20 scans. The concentration of the ribozyme was in each case 8 μM. The resulting Mn2+ EPR signals were base-line corrected, doubly integrated, and the magnitude of the integral was plotted against the concentration of added Mn2+. Binding isotherms for Mn2+ were constructed by plotting the concentration of bound Mn2+ divided by the concentration of ribozyme in solution (∕[ribozyme]) versus the concentration offree Mn2+. The concentration of bound Mn2+ was determined by comparing the Mn2+-signal intensity of a Mn2+-ribozyme sample with a Mn2+ standard sample containing 8 μM of free Mn2+. The difference in intensity between both is assigned to the concentration of bound Mn2+. The binding isotherms were then fitted according to Eq. 1 assuming j classes of n independent noninteracting binding sites to determine the dissociation constants Kd:
| (1) |
Each experiment was repeated at least three times and the results were in each case reproducible (the error bars are shown on the graphs).
Low-temperature experiments at 4.2 K were performed on samples containing 0.2 mM Diels-Alder ribozyme in Tris-HCl buffer with 0.3 M NaCl and 20% sucrose. The cw-EPR spectra were recorded at this temperature with an ER 4102ST rectangular resonator and a cryostat from Oxford using the following parameters: center field 3350, sweep width 6500 G, 2048 points, microwave power 0.2 mW, modulation amplitude 23 G, modulation frequency 100 kHz, conversion time 82 ms, time constant 82 ms, 10 scans. The simulations of cw-EPR spectra were performed using XSophe software (University of Queensland, Brisbane, Australia and Bruker Biospin GmbH).
Ribozyme activity measurements
All substances were dissolved in water, except for N-pentyl maleimide (NPM) which was dissolved in ethanol (10 mM stock solution). The assay was conducted in a 7 μl cuvette (Hellma) at room temperature in a Cary 50 UV spectrometer (Varian). Components were added in the following order, with the final concentrations in brackets: Tris-HCl buffer pH 7.4 (30 mM), NaCl (300 mM), ribozyme (7 μM), divalent ions (Mg2+, Mn2+, Cd2+) (range of 0–80000 μM), AHEG (100 μM). Reactions were started by adding NPM (1 mM). The decrease in anthracene absorbance at 365 nm was recorded over 10 min.
For the determination of the initial rate Vini [μM∕min], only the first 5% decrease of absorbance was analyzed by linear regression. The second-order rate constant was calculated from Eq. 2:
| (2) |
ACKNOWLEDGMENTS
The financial support of the DFG (SFB 579 and Ja 794∕3), the Center of Biomolecular Magnetic Resonance, and HFSP is gratefully acknowledged.
References
- Abragam, A and Bleaney, B (1986). Electron Paramagnetic Resonance of Transition Ions, Dover, New York. [Google Scholar]
- Ananyev, G M and Dismukes, G C (1997). “Calcium induces binding and formation of a spin-coupled dimanganese(II,II) center in the apo-water oxidation complex of photosystem II as precursor to the functional tetra-Mn∕Ca cluster.” Biochemistry 10.1021/bi970626a 36, 11342–11350. [DOI] [PubMed] [Google Scholar]
- Antanaitis, B C, Brown, R D, Chasteen, N D, Freedman, J H, Koenig, S H, Lilienthal, H R, Peisach, J, and Brewer, C F (1987). “Electron paramagnetic resonance and magnetic susceptibility studies of dimanganese concanavalin A. Evidence for antiferromagnetic exchange coupling.” Biochemistry 10.1021/bi00398a058 26, 7932–7937. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P C and Yajima, R (2006). “Nucleobase catalysis in ribozyme mechanism.” Curr. Opin. Chem. Biol. 10.1016/j.cbpa.2006.08.014 10, 455–464. [DOI] [PubMed] [Google Scholar]
- Boldt, Y R, Whiting, A K, Wagner, M L, Sadowsky, M J, Que, L, and Wackett, L P (1997). “Manganese(II) active site mutants of 3,4-dihydroxyphenylacetate 2,3-dioxygenase from Arthrobacter globiformis strain CM-2.” Biochemistry 10.1021/bi962362i 36, 2147–2153. [DOI] [PubMed] [Google Scholar]
- Calle, C et al. (2006). “Pulse EPR methods for studying chemical and biological samples containing transition metals.” Helv. Chim. Acta 10.1002/hlca.200690229 89, 2495–2521. [DOI] [Google Scholar]
- Clouet-d’Orval, B, Stage, T K, and Uhlenbeck, O C (1995). “Neomycin inhibition of the hammerhead ribozyme involves ionic interactions.” Biochemistry 10.1021/bi00035a025, 34, 11186–11190. [DOI] [PubMed] [Google Scholar]
- Copik, A J, Nocek, B P, Swierczek, S I, Ruebush, S, Jang, S B, Meng, L, D’souza, V M, Peters, J W, Bennett, B, and Holz, R C (2005). “EPR and x-ray crystallographic characterization of the product-bound form of the MnII-loaded methionyl aminopeptidase from Pyrococcus furiosus.” Biochemistry 10.1021/bi048123+ 44, 121–129. [DOI] [PubMed] [Google Scholar]
- DeRose, V (2002). “Two decades of RNA catalysis.” Chem. Biol. 10.1016/S1074-5521(02)00217-X 9, 961–969. [DOI] [PubMed] [Google Scholar]
- Eaton, S S and Eaton, G R (2000). “Distance measurements by cw and pulsed EPR.” In Biological Magnetic Resonance, Berliner, L J, Eaton, G R, and Eaton, S S, eds., 19, 2–21, Academic∕Plenum, New York. [Google Scholar]
- EPAPS Document No. E-HJFOA5-1-005702 for manganese(II) titration curve and binding isotherm, simulated cw X-band EPR spectra and the corresponding parameters, as well as a description of the difficulties in simulating Mn2+ EPR spectra. This document can be reached through a direct link in the online article’s HTML reference section or via the EPAPS homepage (http://www.aip.org/pubservs/epaps.html).
- Epel, B, Schaefer, K-O, Quentmeier, A, Friedrich, C, and Lubitz, W (2005). “Multifrequency EPR analysis of the diamagnese cluster of the purivative sulfate thiohydrolase SoxB of Paracoccus pantotrophus.” J. Biol. Chem. 10, 636–642. [DOI] [PubMed] [Google Scholar]
- Fiammengo, R, Musilek, K, and Jäschke, A (2005). “Efficient preparation of organic substrate-RNA conjugates via in vitro transcription.” J. Am. Chem. Soc. 127, 9271–9276 [DOI] [PubMed] [Google Scholar]
- Freisinger, E and Sigel, R KO (2007). “From nucleotides to ribozymes—a comparison of their metal ion binding properties.” Coord. Chem. Rev. , 251, 1834–1851. [Google Scholar]
- Golombek, A P and Hendrich, M P (2003). “Quantitative analysis of dinuclear manganese(II) EPR spectra.” J. Magn. Reson. 10.1016/j.jmr.2003.07.001 165, 33–48. [DOI] [PubMed] [Google Scholar]
- Griscom, D L and Griscom, R E (1967). “Paramagnetic resonance of Mn2+ in glasses and compounds of the lithium borate system.” J. Chem. Phys. 10.1063/1.1712288 47, 2711–2722. [DOI] [Google Scholar]
- Haddy, A, Dunham, W R, Sands, R H, and Aasa, R (1992). “Multifrequency EPR investigations into the origin of the S2-state signal at g=4 of the O2-evolving complex.” Biochim. Biophys. Acta 10.1016/0005-2728(92)90183-3 1099, 25–34. [DOI] [PubMed] [Google Scholar]
- Horton, T E, Clardy, D R, and DeRose, V J (1998). “Electron paramagnetic resonance spectroscopic measurement of Mn2+ binding affinities to the hammerhead ribozyme and correlation with cleavage activity.” Biochemistry 10.1021/bi981425p 37, 18094–18101. [DOI] [PubMed] [Google Scholar]
- Keiper, S, Bebenroth, D, Seelig, B, Westhof, E, and Jäschke, A (2004). “Architecture of a Diels-Alderase ribozyme with a preformed catalytic pocket.” Chem. Biol. 10.1016/j.chembiol.2004.06.011 11, 1217–1227. [DOI] [PubMed] [Google Scholar]
- Khangulov, S V (1998). “L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of the guanidinium group to the dimanganese(II,II) center.” Biochemistry 10.1021/bi972874c 37, 8539–8550. [DOI] [PubMed] [Google Scholar]
- Khangulov, S V, Pessiki, P J, Barynin, V V, Ash, D E, and Dismukes, G C (1995). “Determination of the metal ion separation and energies of the three lowest electronic states of dimanganese(II,II) complexes and enzymes: catalase and liver arginase.” Biochemistry 10.1021/bi00006a023 34, 2015–2025. [DOI] [PubMed] [Google Scholar]
- Kisseleva, N, Khvorova, A, Westhof, E, and Schiemann, O (2005). “Binding of manganese(II) to a tertiary stabilized hammerhead ribozyme as studied by electron paramagnetic resonance spectroscopy.” RNA 10.1261/rna.7127105 11, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima, N, Singh, U P, Amagai, H, Osawa, M, and Morooka, Y (1991). “Oxidative conversion of a Mn(μ-OH)2Mn to a Mn(μ-O)2Mn moiety. Synthesis and molecular structures of a (μ-hydroxo)-dimanganese(II,II) and (μ-oxo)dimanganese(III,III) complex with a hindered N3 ligand.” J. Am. Chem. Soc. 10.1021/ja00020a045 113, 7757–7758. [DOI] [Google Scholar]
- Mabad, B, Cassoux, P, Tuchagues, J-P, and Hendrickson, D N (1986). “Manganese(II) complexes of polydentate Schiff bases. 1. Synthesis, characterization, magnetic properties, and molecular structure.” Inorg. Chem. 10.1021/ic00229a025 25, 1420–1431. [DOI] [Google Scholar]
- Martick, M and Scott, W G (2006). “Tertiary contacts distant from the active site prime a ribozyme for catalysis.” Cell 10.1016/j.cell.2006.06.036 126, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley, H W, Flaherty, K M, and MacKay, D B (1994). “Three-dimensional structure of a hammerhead ribozyme.” Nature (London) 10.1038/372068a0 372, 68–72. [DOI] [PubMed] [Google Scholar]
- Reczkowski, R S and Ash, D E (1992). “EPR evidence for binuclear manganese(II) centers in rat liver arginase.” J. Am. Chem. Soc. 10.1021/ja00053a064 114, 10992–10994. [DOI] [Google Scholar]
- Reed, G H and Cohn, M (1973). “Electron paramagnetic studies of manganese(II)–pyruvate kinase-substrate complexes.” J. Biol. Chem. 248, 6436–6442. [PubMed] [Google Scholar]
- Reed, G H and Markham, G D (1984). “EPR of Mn(II) complexes with enzymes and other proteins.” Biol. Magn. Reson. 6, 73–142. [Google Scholar]
- Reed, G H and Poyner, R R (2000). Metal Ions in Biological Systems Siegel, A. and Siegel, H., eds., Dekker, New York, Vol. 37, pp. 192–193. [PubMed] [Google Scholar]
- Rusnak, F, Yu, L, Todorovic, S, and Mertz, P (1999). “Interaction of bacteriophage lambda protein phosphatase with Mn(II): evidence for the formation of a [Mn(II)]2 cluster.” Biochemistry 10.1021/bi982606u 38, 6943–6952. [DOI] [PubMed] [Google Scholar]
- Schiemann, O, Fritscher, J, Kisseleva, N, Sigurdsson, S T, and Prisner, T F (2003). “Structural investigation of a high-affinity Mn2+ binding site in the hammerhead ribozyme by EPR spectroscopy and DFT calculations. Effects of neomycin B on metal-ion binding.” ChemBioChem 10.1002/cbic.200300653 4, 1057–1065. [DOI] [PubMed] [Google Scholar]
- Scott, W G, Finch, J T, and Klug, A (1995). “The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage.” Cell 10.1016/S0092-8674(05)80004-2 81, 991–1002. [DOI] [PubMed] [Google Scholar]
- Scott, W G, Murray, J B, Arnold, J RP, Stoddard, B L, and Klug, A (1996). “Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.” Science 10.1126/science.274.5295.2065 274, 2065–2069. [DOI] [PubMed] [Google Scholar]
- Seelig, B and Jäschke, A (1999). “A small catalytic RNA motif with Diels-Alderase activity.” Chem. Biol. 10.1016/S1074-5521(99)89008-5 6, 167–176. [DOI] [PubMed] [Google Scholar]
- Seelig, B, Keiper, S, Stuhlmann, F, and Jäschke, A (2000). “Enantioselective ribozyme catalysis of a bimolecular cycloaddition reaction.” Angew. Chem., Int. Ed. 39, 4576–4579. [DOI] [PubMed] [Google Scholar]
- Serganov, A et al. (2005). “Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.” Nat. Struct. Mol. Biol. 10.1038/nsmb906 12, 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigel, H and Griesser, R (2005). “Nucleoside 5-triphosphates: self-association, acid–base, and metal ion-binding properties in solution.” Chem. Soc. Rev. 10.1039/b505986k 34, 875–900. [DOI] [PubMed] [Google Scholar]
- Sigel, K O and Pyle, A M (2007). “Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry.” Chem. Rev. (Washington, D.C.) 10.1021/cr0502605 107, 97–113. [DOI] [PubMed] [Google Scholar]
- Stuhlmann, F and Jäschke, A (2002). “Characterization of an RNA active site: interactions between a Diels-Alderase ribozyme and its substrates and products.” J. Am. Chem. Soc. 124, 3238–3244. [DOI] [PubMed] [Google Scholar]
- Ubbink, M, Worrall, J AR, Canters, G W, Groenen, E JJ, and Huber, M (2002). “Paramagnetic resonance of biological metal centers.” Annu. Rev. Biophys. Biomol. Struct. 10.1146/annurev.biophys.31.091701.171000 31, 393–422. [DOI] [PubMed] [Google Scholar]
- Vogt, M, Lahiri, S, Hoogstraten, C G, Britt, R D, and DeRose, V J (2006). “Coordination environment of a site-bound metal ion in the hammerhead ribozyme determined by 15N and 2H ESEEM spectroscopy.” J. Am. Chem. Soc. 10.1021/ja057035p 128, 16764–16770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, D J, Reiter, N J, Sikkink, R A, Yu, L, and Rusnak, F (2001). “Identification of the high affinity Mn2+ binding site of bacteriophage lambda phosphoprotein phosphatase: effects of metal ligand mutations on electron paramagnetic resonance spectra and phosphatase activities.” Biochemistry 10.1021/bi010637a 40, 8918–8929. [DOI] [PubMed] [Google Scholar]
- Whittaker, J W and Whittaker, M M (1991). “Active site spectral studies on manganese superoxide dismutase.” J. Am. Chem. Soc. 10.1021/ja00015a003 113, 5528–5540. [DOI] [Google Scholar]
- Wombacher, R, Keiper, S, Suhm, S, Serganov, A, Patel, D J, and Jäschke, A (2006). “Control of stereoselectivity in an enzyme-catalyzed reaction by backdoor access.” Angew. Chem., Int. Ed. 10.1002/anie.200503280 45, 2469–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X and Bruice, T C (2007). “Diels-Alder ribozyme catalysis: a computational approach.” J. Am. Chem. Soc. 10.1021/ja067416i 129, 1001–1007. [DOI] [PubMed] [Google Scholar]