Abstract
To understand biological processes, biologists typically study how perturbations of protein functions affect the phenotype. Protein activity in living cells can be influenced in many different ways: by manipulation of the genomic information, by injecting inhibitory antibodies, or, more recently, by the use of ribonucleic acid-medicated interference (RNAi). All these methods have proven to be extremely helpful, as they possess a high degree of specificity. However, they are less suitable for experiments requiring precise timing and fast reversibility of the perturbation. The advantage of small molecules is that they specifically interact with their target on a fast time scale and often in a reversible manner. In the last 15 years, this approach, termed “chemical genetics,” has received a lot of attention. The term genetics pays tribute to the analogy between chemical genetics and the classic genetic approach, where manipulations at the gene level are used to draw conclusions about the function of the corresponding protein. Chemical genetics has only recently been used as a systematic approach in biology. The term was coined in the 1990’s, when combinatorial chemistry was developed as a fast method to synthesize large compound libraries [Mitchison (1994) “Towards a pharmacological genetics,” Chem. Biol. 1, 3–6; Schreiber (1998) “Chemical genetics resulting from a passion for synthetic organic chemistry,” Bioorg. Med. Chem. 6, 1127–1152].
The growth of the field in the last decade led to the publication of plenty of reviews on this field reflecting the wealth of information already available. Considering the scope of this journal and the existing literature, we decided to give a broad introduction for readers new to the field and focus on paradigmatic examples rather than trying to provide a complete listing of recent contributions to chemical genetics. Particularly, we would like to explain how the basic concepts in chemical genetics evolved and are connected to the history of drug discovery and then focus on the advances made and problems arising when starting with so called phenotype-based screenings for compound identification, which is an approach of growing importance.
THE ROOTS OF CHEMICAL GENETICS: DRUG DISCOVERY
The chemical genetic approach uses techniques that have been widely applied in drug discovery for a long time and adapts them to the study of biological problems. Chemical genetics comes in different “flavors,” which we will introduce with a short survey of the history of drug discovery. The earliest drugs, which have been used empirically for centuries, were mostly natural compounds that were discovered by chance. In most cases, humans or animals ingested, drank or were accidentally exposed to herbal or fungal substances and a useful effect was observed. Retrospectively, one could see this trial and error approach as an unbiased phenotype-based screen. The first progress toward a systematic approach to drug development was done in the late eighteenth century, when active agents were routinely purified from plant extracts (like opium from the opium poppy or digitalis, a drug used for heart failure, from the foxglove plant). At that time, the concept that the effect of plant extracts is mediated by their single constituents was shaped. In 1827, Emanuel Merck introduced morphine, a naturally occurring alkaloid, as the first commercially available pure drug in large-scale production. Another big step was made when Paul Ehrlich, influenced by the observation that tissues can be selectively stained with different dyes, formulated the idea that drug effects are mediated by “receptors” (Ehrlich, 1967). The receptor was a synonym for what is called nowadays “drug target,” i.e., the molecular structure the drug binds to and that mediates the effect of the drug. In fact, Ehrlich even attempted to analyze the drug-receptor interaction systematically and performed screens for drugs effective against trypanosomes, the pathogen causing sleeping disease. During the nineteenth century, chemists succeeded in synthesizing compounds that had been previously isolated from plants. In 1897, acetylsalicylic acid, the first synthetic drug to be commercially distributed (as Aspirin®), was generated. It was synthesized by Arthur Eichengrün and Felix Hoffmann at Bayer & Co. and was derived from salicylic acid, which had initially been purified from the willow bark and had been known as an anti-inflammatory agent for more than half a century. To reduce the side effects caused by the free phenolic group of salicylic acid Eichengrün and Hoffmann acetylated the free hydroxyl functional group yielding acetylsalicylic acid. In the body, the ester is hydrolyzed to the free active compound making aspirin the first synthetic drug that does not exist in nature. This was the starting point for the rise of pharmaceutical companies during the 20th century. By chemical synthesis, they accumulated giant “libraries” of compounds that were screened in different assays for their effectivity as drugs. This strategy for drug development dominated the 20th century and is what we know today as traditional medicinal chemistry:
(1) Choose a “lead” compound that is known to be effective and was found either by chance or during a dedicated screening. (2) Synthesize derivates of the lead compound and analyze these analogues in terms of potency, specificity, and chemical properties. (3) Assess compound effectivity in animal models and, if successful, use it in clinical trials.
The last piece of the puzzle to make chemical genetics useful for biological research consisted of the biochemical and molecular genetic tools necessary for target (receptor) identification and characterization. As soon as molecular biology matured and protein expression and purification techniques became widely available, it became possible to characterize the activity profile of compounds by in vitro screens. This development resulted in the first commercial success with the registration of imatinib (Gleevec®) by Novartis, which is effective in chronic myeloid leukaemia (CML). Based on the discovery that bcr-abl is the main oncogene causing CML, development started with an in vitro screen for inhibitors of abl-kinase activity. Bioactive compounds were derivatized resulting in the synthesis of imatinib in 1992 and the registration as Gleevec® in 2001 (Rang, 2006).
The strong correlation between scientific progress and the advancements made in drug discovery is nicely exemplified by the timeline of breakthroughs related to acetylsalicylic acid and its usage as a drug (Vane and Botting, 2003):
Antiquity: first mentions of willow bark extract as an effective antipyretic and analgesic.
1820–1840: isolation of salicylic acid from the willow bark
1874: commercial organic synthesis and distribution by Heyden Chemical Company
1853: Charles Frederic Gerhardt synthesizes for the first time acetylsalicylic acid
1897: resynthesis of acetylsalicylic acid by Felix Hoffmann at Bayer & Co.
1899: commercial distribution of acetylsalicylic acid as Aspirin® by Bayer & Co.
1971: discovery of mechanism of action (inhibition of prostaglandin synthesis) of Aspirin® (Vane, 1971)
1975: identification of cyclooxygenase as inhibited enzyme involved in prostaglandin synthesis (Roth and Majerus, 1975; Roth et al., 1975)
1976: purification of active cyclooxygenase 1, one of its target enzymes (Hemler et al., 1976)
It took a long time for chemical genetics to evolve into a systematic strategy to answer biological questions because it requires a known protein target. One of the first examples for successful target identification in biological research was the discovery of tubulin as the target protein for colchicine in the 1960’s (Borisy and Taylor, 1967a; Borisy and Taylor, 1967b). This accomplishment was achieved by labeling colchicine with 3H and biochemically purifying the protein binding the radioactive compound, which turned out to be tubulin. There are several other examples for early successful target identifications, but it was only in the 1990’s that compounds used in biological research were identified by systematic screening and the term “chemical genetics” was coined.
CHEMICAL GENETICS: ADVANTAGES AND CAVEATS
As mentioned before, other well established methods exist to modulate protein activity both in vitro and in vivo, so how can chemical genetics help to gain new insights and how does it complement or improve the existing tools? The most prominent advantage of using compounds is that they act on a fast time scale and that their effect can be easily controlled through titration. Other methods, like mutagenesis or RNAi interference, cause either permanent effects or have a temporal resolution at the scale of days. Moreover, the effect of small molecules is usually equally strong in all cells treated, while methods like antibody microinjection and RNAi are highly variable. These properties of small molecules become relevant in the study of dynamic processes (e.g., cell cycle), because high time resolution and synchrony of the effect in all cells is essential here. The most common application of chemical genetics in this context is synchronization and release of cells at a certain cell cycle stage with molecules like aphidicolin, nocodazole, taxol, or thymidine. This cannot be achieved with any of the other mentioned approaches and is very efficient and cheap. Another gap filled by chemical genetics is the possibility to inhibit protein activity without physically removing the protein from the system studied. Gene deletion and RNAi eliminate the protein of interest and thus both characteristics that define protein function are affected: its (catalytic) activity and its potential relevance as a part of protein complexes. The phenotype generated by genetic manipulation can, therefore, often be different from the result of a chemical genetic experiment (Knight and Shokat, 2007). Compounds can also be used across species and in different systems (e.g., in vitro, tissue culture cells, or multicellular organisms), if the target protein is well conserved, whereas mutants have to be generated one by one, which is a tedious and time-consuming process. In summary, in many situations, chemical genetics can provide a very precise tool, which allows for otherwise impossible fine tuning of experimental conditions.
CONCEPTS AND DESIGN OF A CHEMICAL GENETIC PROJECT
The basic outline of a chemical genetic project is as follows:
design and synthesis of the library of compounds to be screened
screening of the library in the system of choice
target identification∕validation including specificity tests.
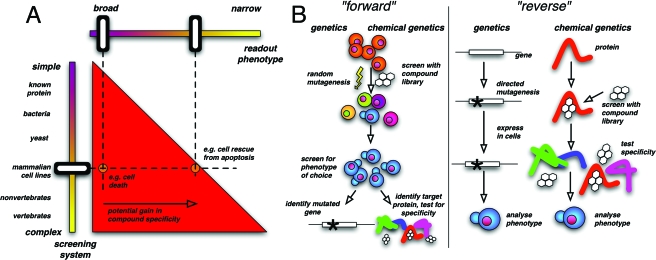
Conceptually, the most important part is the design of the screening strategy, since it has fundamental implications for the entire screening procedure and outcome of the screen [Fig. 1A]. In general, it is possible to perform a screen in systems ranging from pure proteins to whole organisms. Using more complex systems has the advantage that the screening settings mirror more closely or even match the conditions under which the sought-after compound has to fulfill its designated function, i.e., to specifically modulate the activity of one particular protein within a complex mixture of molecules. Thus, the more complex the screening conditions are, the more likely it is that the identified compounds meet the required criteria of, e.g., specificity, membrane permeability and stability. Additionally, unbiased more complex screens have the potential to identify inhibitors for cellular targets hitherto not known to be involved in the process of choice. However, to fully exploit the advantage of phenotype-based screens, it is essential to choose a very well defined phenotype. A priori, a search for small molecules that improve rather than impair an existing situation will yield more specific compounds. For example, a cell-based screen for compounds that induce cell death will most likely identify a large number of nonspecifically acting molecules, whereas the search for molecules that restore cell viability, e.g., under cell-death promoting conditions, has a much greater potential to identify suitable molecules.
Figure 1. Basic concepts in chemical genetics.
A. Overview of screening strategies in chemical genetics. There are two basic variables defining a chemical genetic screen. One is the screening system, which can range from purified proteins (reverse approach) to cells or multicellular organisms (forward approach). The second variable is the readout phenotype of the screen. The red triangle indicates the area of possible combinations of screening system and phenotype. The triangular shape results from the larger choice of possible readout phenotypes offered by more complex screening systems. B. Comparison of the forward and reverse approach in “classic” and chemical genetics. For details see text.
As always, there is also the other side of the picture, which in the case of phenotype-based screens is the challenge of target identification. In the past, affinity purification and photoaffinity labeling of compound targets have proven to be extremely powerful. Examples are the identification of the cellular binding partners of FK506 (Harding et al., 1989), Trapoxin (Taunton et al., 1996), lactacystin (Fenteany et al., 1995) and ilimaquinone (Radeke and Snapper, 1998). With the recent advancements in molecular biology more strategies evolved to find the binding partner of bioactive compounds and these approaches are discussed in detail by Burdine and Kodadek (2004). The task of target identification can be lessened by simplifying the screening system. The smaller the genome and the simpler the organization of the screening organism, the easier it is to identify the target protein. The simplest conceivable system is a single, purified protein (or a protein fragment), which is tested in an in vitro assay. Previously, these assays were mainly focused on enzymes because their activity could be easily measured and it was assumed that proteins lacking any enzymatic activity are hardly targetable by small molecules. However, this assumption has to be revised because recently specific small molecule inhibitors of protein-protein interactions were identified (Kiessling et al., 2006; Oltersdorf et al., 2005; Vassilev et al., 2004). In the case of protein-based screens, the target is already known, making target identification obsolete. In analogy to traditional genetics, the screening against a known protein is termed “reverse” chemical genetics, while all other screening designs involving cell extracts or whole organisms are termed the “forward” strategy [Fig. 1B]. In both “traditional” and chemical genetics, the reverse approach starts with the manipulation of a known protein or its function, while the forward approach starts with random manipulation to obtain a defined phenotype and the responsible protein is identified in a second step. It is rather easy to miniaturize an in vitro assay and adapt it for high-throughput environments, which makes reverse screens more accessible than forward screens. During the last decade, however, automated microscopes and image analysis algorithms became available that allow the use of complex phenotypes as a read-out parameter for screens. This has led to rising interest in the forward approach, which will be taken into consideration by dedicating the last part of the article to this topic. The decision between a forward and a reverse approach can influence the choice of the library to be screened. A reverse chemical genetic screen with a known target might be more successful if a “biased” library is used, containing compounds selected for the structure of the protein or derived from inhibitors of other proteins of the same family, e.g., adenosine triphosphate (ATP) analogues to screen for inhibitors of protein kinases. However, such screens are more likely to identify compounds that inhibit more than one member of the protein family.
Historically, most compounds used in chemical genetics and drug development were natural products or derivatives thereof. In the 1990’s, however, progress in combinatorial chemistry changed the focus slightly from natural products to less complex molecules. After ten more years, initial expectations regarding the success of these giant synthetic libraries have been dampened a bit because of lower than expected hit rates. Although it is not entirely clear if the reasons for this are not rather technical than conceptual, interest in natural compounds has been increasing recently (Ortholand and Ganesan, 2004; Clardy and Walsh, 2004). This has led to the idea to synthesize new compounds inspired from backbones provided by nature, based on the notion that these structures have already been selected by evolution for biological activity. It is hoped that these synthetic molecules distinguish themselves from their natural counterparts by improved or even novel modes of action (Breinbauer et al., 2002; Noren-Muller et al., 2006; Piggott and Karuso, 2004). All in all, although the choice of the library is obviously very important for the outcome of a small molecule screen, there are still few objective rules to make this choice more rational. In real life, it is often determined by availability, infrastructure (only laboratories with a chemical background may be able to synthesize their own library) and cost of the library, since huge libraries are still rather expensive. In the last 20 years, though, tremendous progress has been made in high-throughput synthesis of small molecules (Rupasinghe and Spaller, 2006), which has reduced the cost and resources required for synthesis dramatically. It has now become more affordable to buy libraries from commercial suppliers and it is also possible for specialized laboratories to synthesize their own libraries in house at a reasonable cost (Hergenrother, 2006).
Irrespective of which screening strategy has been initially applied, the identified compounds have to give proof of their specificity. Since conclusions drawn from small molecule studies are only valid when the applied substances specifically modulate the activity of the assumed cellular target, the question of compound specificity has to be of highest priority. Ensuring specificity is one of the most difficult tasks in chemical genetics beside target identification. Typically, the first step to demonstrate specificity is to test in vitro the effect of the identified compounds on proteins related to the assumed target proteins. Compounds passing these studies can then be analyzed in vivo where they have to prove that they do not affect cellular processes unrelated to the function of the desired target. Additionally, it is a common strategy to confirm that the compound-induced phenotype mimics the one caused by RNAi-mediated depletion of the assumed target protein. However, as mentioned before, modulating the activity of a protein might result in a phenotype different from the one induced by removing the protein from the system (Knight and Shokat, 2007).
No matter how thoroughly these studies are performed, they only raise the level of confidence but do not provide the final ultimate proof of compound specificity. An example highlighting the importance of compound specificity is provided by Peter Cohen, who tested 42 molecules previously known as selective inhibitors of distinct protein kinases against a panel of more than 20 protein kinases in vitro (Davies et al., 2000; Bain et al., 2003). Surprisingly, most compounds were found to affect two different kinases or more, sometimes even more effectively than their presumed target. Less biased, proteome-wide target identification approaches often reveal even more affected proteins. Godl et al. (2003) tried such an approach for the compound SB203580, which had also been tested by Davies et al. (2000) and thought to be a relatively selective inhibitor of p38 kinase (Godl et al., 2003). Their affinity matrix based approach (chemical proteomics) identified many additional binding partners of SB203580 and one of them, the kinase RICK, was shown to be inhibited by SB203580 in cells, pointing out the importance of thorough specificity studies.
The final proof to demonstrate that the compound-induced phenotype is indeed mediated by affecting the function of the assumed target protein is to replace the wild-type endogenous protein with a mutant form resistant to the action of the small molecule. If these genetic manipulations suppress the phenotype mediated by the small molecule one can take it for granted that the assumed target is the relevant binding partner of the identified compound at the given concentration. Examples on how this approach can be used for target identification are presented below.
CONTRIBUTION OF CHEMICAL GENETICS TO ELUCIDATION OF THE FUNCTION OF GSK3
For a better illustration of how chemical genetics can generate new findings and complement other approaches, we would like to explain how our knowledge about glycogen synthase kinase-3 (GSK3) evolved during the last ten years. GSK3 is a serine∕threonine protein kinase involved in several biochemical pathways and was initially discovered in 1980 as a protein involved in glycogen metabolism (Cohen and Frame, 2001). In the 1990’s, additional functions of this protein were discovered unexpectedly. In a genetic screen in Drosophila melanogaster, the fly homologue of GSK3 was identified as contributing to the Wnt pathway. This pathway is conserved across many species and involved in embryogenesis and cancer pathogenesis. In 1995 it was shown that expression of a catalytically inactive mutant of GSK3 in Xenopus laevis resulted in duplication of the dorsal axis. A similar developmental defect had been shown to be induced by lithium ions in 1986. This led to the idea that lithium might be an inhibitor of GSK3, which was finally proven in 1996. Thus, the first pharmacological inhibitor of GSK3 was identified, which was followed by the discovery of a plethora of highly potent inhibitors with varying specificity profiles from both natural sources and synthetic libraries (Meijer et al., 2004). Notably, GSK3 is one of the few examples for which inhibitors were identified using both protein- and phenotype-based approaches. An example for a phenotype-based approach is provided by Ding et al., who screened for small molecules that induce neuronal differentiation in murine embryonic stem cells (Ding et al., 2003). Using a luciferase reporter construct under the control of a neuron-specific promoter they were able to detect neuronal differentiation in a highly reproducible manner in the 384-well format. Following structure-activity relationship studies, they identified one bioactive compound, named TWS 119, which was sufficiently potent to identify its binding partner via affinity chromatography (chemical proteomics). Mass spectroscopy analyses identified GSK3β s a TWS119-interacting protein. By now, it is well established by different experimental approaches that Wnt signaling is involved in the development of the nervous system (Ciani and Salinas, 2005). Thus, the different identified GSK3 inhibitors were key in many systems ranging from further biochemical characterization of the Wnt pathway in vitro (Zeng et al., 2005) to the revelation of a new role for GSK3 in spindle formation in mammalian cell lines (Wakefield et al., 2003) and to the study of the role of GSK3 in tissue protection against hypoxia mediated injury in rats (Gross et al., 2004). As mentioned previously, prerequisite for this success was the fact that small molecules can be effective across species and that they modulate protein activity on a fast time scale.
PHENOTYPE BASED SCREENS: EXAMPLES
Finally, we would like to focus on recent developments in forward chemical genetics starting from the simplest setup, performed in cell extracts, and finishing with projects using whole animals.
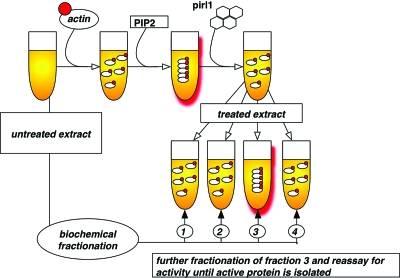
The recent discovery of a small molecule inhibitor of actin polymerization was performed in extract from Xenopus laevis oocytes (Peterson et al., 2006) which are a convenient and well established source for highly concentrated protein solution. Using a fluorescence-based assay, the authors succeeded in the identification of pirl1, a micromolar inhibitor of the phosphatidylinositol-bisphosphate (PIP2)-induced actin polymerization in Xenopus extract. Due to the low target affinity of pirl1 (IC50:3 μM), the authors decided to identify the binding partner of this compound via a biochemical complementation assay (Fig. 2). This target identification approach is based on the assumption that the inhibitory effect of the small molecule can be suppressed by adding the target protein in high concentrations. To this end, the authors added biochemical fractions of untreated extract to pirl1-treated samples and assayed for the restoration of PIP 2-mediated actin polymerization. One out of nine fractions counteracted the effect of pirl1. Importantly, this fraction was also able to enhance PIP2-induced actin polymerization in the absence of pirl1 suggesting that the observed complementation was specific and not mediated by an unspecific binding partner of pirl1. Further subfractionations followed by mass spectroscopy narrowed the search down to two possible target protein complexes of pirl1: Cdc42∕RhoGDI and Arp2∕3. Subsequent in vitro assays revealed that pirl1 inhibits CDC42∕RhoGDI but not Arp2∕3, which is acting downstream of CDC42∕RhoGDI. Thus, Peterson et al. provide an elegant example where the cellular target of a compound derived from a phenotype-based approach was identified by biochemical methods.
Figure 2. Biochemical suppression as a target identification strategy.
Scheme of the experiments performed by Peterson et al. to identify the target of the actin polymerization inhibitor pirl1 (Peterson et al., 2006). Actin polymerization in Xenopus laevis egg extract was monitored using a fluorescence-based assay. Biochemical fractions were assayed for their ability to restore actin polymerization in the presence of the small molecule pirl1. Active fraction (3) is subfractionated, re-assayed and relevant proteins identified via mass spectroscopy.
A successful example of chemical genetics applied in bacteria is provided by the group of Lucy Shapiro who was interested in the function of MreB, the prokaryotic homologue of actin, in the process of chromosome segregation (Gitai et al., 2005). While bacteria are easily amenable to genetic manipulation, the fast action of small molecules prompted the authors to focus on A22, a compound that had been previously identified in a phenotype based screen for generation of anucleate E. coli cells (Iwai et al., 2002). Although its target was not known, the phenotype of A22 was reminiscent of MreB mutants and some other target candidates had already been excluded. Indeed, by screening for compound-resistant bacteria, Gitai et al. identified MreB as the cellular target of A22. Sequence analyses revealed that in all cases a single point mutation within MreB accounted for the observed resistance of bacteria against A22. Using this newly established tool in combination with fluorescence-based live-cell microscopy, Gitai et al. were able to dissect the role of MreB in bacterial cell division. By adding A22 at different stages of the cell cycle the authors demonstrated that the activity of MreB is dispensable for DNA replication in Caulobacter crescentus but essential for correct segregation of the ori loci (origin of replication). These findings suggest a model reminiscent of the centromere-microtubule interaction in eukaryotic cells. While in higher organisms, these concepts are already well established [many of them using small molecules (Peterson and Mitchison, 2002)], in bacteria, the cytoskeleton has been discovered only recently (Gitai, 2005) and the systematic mechanistic analysis of bacterial cell division is just beginning.
Yeast is one of the most important organisms used for genetic studies, mainly because its genetic information can be easily modified and because of its robustness. Leland Hartwell and Paul Nurse, for example, identified the first cyclin dependent kinases as key regulators of cell growth in yeast, which led to the universal concept of “cell cycle” (Hartwell, 2002; Nurse, 2002). Given its long tradition as a genetic model organism, yeast has also proven to be a powerful system for the identification of small molecule targets. These genetic approaches are based on one of the following principles: (1) Increasing the copy number of the target protein causes resistance of the yeast mutants against the small molecule, a so-called high-copy number suppression screen. A proof-of-concept study using genetic suppression was published recently by Luesch et al. (2005) who identified an inhibitor for the Pkc1 protein kinase in yeast. (2) Decreasing the copy number of the target protein renders yeast strains hypersensitive toward the small molecule, a so-called haplo-insufficiency screen. Using an unbiased genome-wide haplo-insufficiency screen Baetz et al. could demonstrate that dihydromotuporamine C, a compound in preclinical studies that inhibits angiogenesis and metastasis by an unknown mechanism, targets sphingolipid metabolism (Baetz et al., 2004). (3) Mutagenizing the binding site of the small molecule on its target protein confers resistance against the compound.
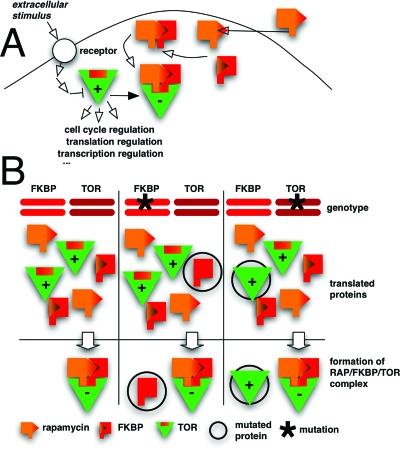
For the latter approach, one of the most prominent examples is target identification of rapamycin, a natural product derived from the bacteria Streptomyces hygroscopicus. While rapamycin was originally identified as an antibiotic with antifungal activity (Vezina et al., 1975), its mode of action became of interest when its immunosuppressive activity was recognized. In vitro assays revealed that rapamycin binds to and inhibits the peptidyl-prolyl cis-trans isomerase FKBP12 (FK506 binding protein of 12 kDa), which was initially identified through a chemical proteomics approach as the binding partner of FK506, a drug structurally related to rapamycin [Bierer et al., 1990; Harding et al., 1989, Fig. 3A]. However, the observation that rapamycin derivatives inhibit FKBP12 but do not immunosuppress argued against the idea that the inhibition of FKBP12 accounts for the immunosuppressive effect of rapamycin. This puzzle was finally solved by the groups of Michael Hall and George Livi who harnessed the power of yeast genetics to identify the targets of rapamycin in budding yeast (Heitman et al., 1991; Kunz et al., 1993; Cafferkey et al., 1993). By screening for spontaneous yeast mutants resistant to the antifungal effect of rapamycin they identified TOR1∕DRR1 and TOR2∕DRR2 (target of rapamycin∕dominant rapamycin resistant) as the cellular targets of rapamycin. Further analyses revealed that TOR∕DRR proteins are evolutionarily conserved protein kinases that control cell growth in response to nutrient availability (Raught et al., 2001; Crespo and Hall, 2002). In contrast to yeast, higher eukaryotes seem to possess only one TOR∕DRR (mTOR) which was discovered based on its ability to interact in vitro with the rapamycin-FKBP12 complex or by a two-hybrid screen (Chen et al., 1994; Sabers et al., 1995; Sabatini et al., 1994; Brown et al., 1994; Chiu et al., 1994). The potential of genetics to identify the target of small molecules becomes apparent when we consider how much information about the mode of action of rapamycin was provided by the original yeast studies (Fig. 3). In these initial screens, all recessive mutations were mapped to FKBP12 and all dominant mutations were mapped to TOR∕DRR. Rapamycin and FKBP12 form a complex that binds to TOR∕DRR and thus inhibits its activity. That means that rapamycin is effective as long as cells express at least a minimal amount of wildtype FKBP12 (corresponding to a heterozygous FKBP12 mutation), while it is not effective if there is a minimal amount of TOR∕DRR, which cannot bind to the rapamycin-FKBP12 complex and thus stays active (corresponding to a heterozygous TOR∕DRR mutation).
Figure 3. Mechanism of action of rapamycin and discovery of TOR proteins.
Rapamycin is a membrane-permeable small molecule that binds to and inhibits FKBP12. The rapamycin-FKBP12 complex inactivates its target TOR, a protein kinase family regulating cell cycle progression in response to nutrient availability. B. Effect of heterozygous mutations in the FKBP12 and TOR genes, respectively. A heterozygous mutation in one copy of the FKBP12 alleles (marked with an asterisk) impairs the interaction between FKBP12 and rapamycin. The remaining wildtype FKBP12 is sufficient to inhibit the TOR protein in complex with rapamycin. Thus, a homozygous mutation of FKBP12 is required for rapamycin resistance. Mutant TOR protein unable to bind rapamycin-FKBP12, however, is dominant in conferring resistance against rapamycin.
Small molecule studies have been and are still key for our understanding of mitosis and the function of the mitotic cytoskeleton in eukaryotes. Examples are the use of the microtubule poison benomyl to identify the checkpoint proteins in yeast (Hoyt et al., 1991; Li and Murray, 1991) or the discovery of tubulin as the target of colchicine (Borisy and Taylor, 1967a; Borisy and Taylor, 1967b). However, most of the known antimitotic compounds target the same protein, namely tubulin. Thus, in order to identify compounds that induce a mitotic arrest in mammalian cells by a different mode of action, Mayer et al. applied a combination of two phenotype-based screens and in vitro assays to select for antimitotic molecules that do not target tubulin (Mayer et al., 1999). One compound identified in these screens induced the collapse of the bipolar spindle and the formation of a monoaster like spindle, a phenotype that named the compound monastrol. This monoaster phenotype was reminiscent of cells lacking the activity of Eg5, a mitotic kinesin essential for spindle bipolarity in eukaryotes. In vitro assays demonstrated that monastrol was indeed a reversible inhibitor of Eg5. To gain insights into the mechanism of action, Lawrence Kuo and colleagues determined the structure of the motor-domain of Eg5 in complex with Mg2+ ADP and monastrol (Raught et al., 2001). These studies revealed that monastrol binds to an induced-fit, allosteric site of Eg5, about 12 Å away from the nucleotide binding site. By now, monastrol is a well established tool in cell biology that provided insights into the process of spindle formation, chromosome capture, and the activation of the spindle assembly checkpoint, which would have been hardly if at all amenable to traditional techniques (Kapoor et al., 2000; Kapoor and Mitchison, 2001; Khodjakov et al., 2003).
Recently, chemical genetics has also started to embrace multicellular organisms as a screening system especially in the field of developmental biology. The main obstacle in screening whole organisms is to adapt the assay for high throughput experiments. Obviously, only small organisms can be kept and are able to survive in multi-well plates. Although the application of small molecule screens to multicellular organisms is a rather novel approach, recent publications already indicate the impact chemical genetics will have on this aspect of biology in the future. Caenorhabditis elegans has been shown to be a powerful tool for target identification already two decades ago, e.g., with the identification of tubulin as a target of nematocides (Driscoll et al., 1989) and was also useful for the characterization of the target proteins of drug candidates (Fitzgerald et al., 2006) and drugs like Fluoxetine (Prozac®) (Choy and Thomas, 1999). A recent example of the powerful combination of phenotypic screen and genetics possible in this system is provided by the group of Peter Roy who set up a phenotype-based screen to identify calcium channel inhibitors in the nematode Caenorhabditis elegans (Kwok et al., 2006). The authors screened about 18.000 compounds on 24-well plates for generation of phenotypic defects in worms, which were classified by visual examination. Of the 308 identified bioactive compounds they focused on one compound, named Nemadipine-A. This compound caused three different defects: a population growth defect called “Gro,” dramatic morphological defects called Vab (for Variable Abnormal) and Egg laying defects called an Egl phenotype. For target identification, similarly to strategies described above, they performed a genetic suppressor screen, selecting for random mutations conferring resistance to Nemadipine-A. By correlating the segregation pattern of Nemadipine-A resistance with the segregation of single nucleotide polymorphisms (Wicks et al., 2001) they discovered that Nemadipine-A resistance was always linked to mutations within the egl-19 gene locus, which encodes the only L-type calcium channel α1-subunit in C. elegans. All suppressor mutants carried missense mutations in the highly conserved region of egl-19 that form the pore of the channel. While the authors did not directly prove that Nemadipine-A affects the activity of EGL-19, the following line of evidence suggests that Kwok et al. identified the relevant binding partner of Nemadipine-A: (1) Overexpression of EGL-19 in C. elegans leads to resistance against Nemadipine-A. (2) Lowering the expression of EGL-19 phenocopies Nemadipine-A treatment. (3) Approved drugs that inhibit the α-subunit of L-type calcium channels in humans and are used for treatment of hypertension are structurally related to Nemadipine-A. In a further step, using Nemadipine-A, Kwok et al. were able to dissect the role of EGL-19 in egg laying and show a redundant positive effect of EGL-19 and the other two voltage-gated calcium channels in worms on egg laying. Since, in contrast to treatment with low doses of Nemadipine-A, the weakest egl-19 hypomorph is already egg-laying defective, genetic methods alone would not have revealed this finding.
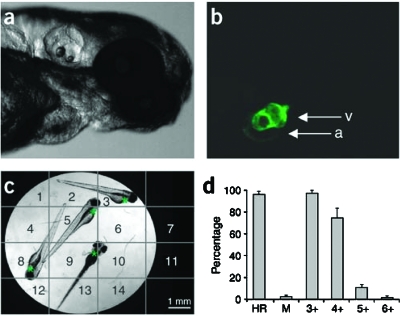
Finally, we would like to mention that chemical genetic screens have already been performed in vertebrates. One very useful model organism here is the zebrafish (Danio rerio), which has a long tradition in developmental biology. Zebrafish embryos can be grown for a few days even in 384-well plates and, exploiting their transparent appearance, sophisticated microscope-based methods have been developed to automatically monitor physiologic functions. For example, an assay that uses zebrafish embryos expressing green fluorescent protein (GFP) in the myocardium has been established to automatically image and extract heartbeat rates (Burns et al., 2005, Fig. 4). Since appropriate mutants that mimic human disease already exist, this could be used to identify new drug candidates for treatment of arrhythmia. Another study tried to identify compounds able to rescue the phenotype of the “gridlock” mutation (Peterson et al., 2004). This mutation was originally discovered by the same group (Weinstein et al., 1995) and induces a vascular defect that impedes blood flow to the tail. This phenotype is highly reminiscent of a developmental defect observed in humans called aortic coarctation. They found a compound able to restore the normal blood flow in the gridlock mutation background. Although Peterson et al. were not able to identify the target, they could demonstrate that zebrafish embryos treated with the identified compound have elevated levels of the angiogenic cytokine VEGF. Notably, injection of VEGF cDNA—similarly to the application of the identified substance—is sufficient to rescue the gridlock phenotype, suggesting that VEGF is mediating the effect of the compound. For the future, it will be important to elucidate how the compound increases VEGF levels in D. rerio. While these examples highlight the potential of chemical genetics to address biological questions in whole organisms, they concurrently point out that the complexity of the system poses new challenges for the identification of the compound target and specificity studies. Target identification strategies which at least partially worked in zebrafish include the use of compound libraries already containing a linker molecule for immobilization on a solid matrix, which greatly facilitates chemical proteomics approaches (Khersonsky et al., 2003), and the educated guess approach (Anderson et al., 2007). Although this work is promising, functional tests to prove relevance of the identified targets are missing.
Figure 4. Screening setup to automatically assess heart rate in zebrafish on 96-well plates.
A. B. Transmitted and fluorescent light photographs of a zebrafish embryo expressing GFP in the myocardium. a: atrium, v: ventricle. C. Transmitted light photograph of a single well, showing the boundaries of virtual subsites used to identify fluorescent hearts. Asterisks mark the localization of embryo hearts. Four embryos per well were used for the screening. D. First-pass efficiency of the assay. The graph shows the mean percentages of scored heart rates (HR), moving embryos (M) which could therefore not be scored for heart rate, and wells in which three to six or more heart rates were scored (for four embryos) by software-based analysis. More than four heart rates per well are detected if a single heart is localized in two adjacent subsites and is thus scored twice. From (Burns et al., 2005).
CONCLUSIONS
Chemical genetics is still an emerging field. It has, however, already become too vast to cover all of its aspects adequately in one review article and we apologize to our colleagues whose work could not be cited due to space constraints. Still, we hope that this review gives a broad readership, from different research areas, an understanding of the potential of chemical genetics to solve fundamental questions in modern biology. To make this clear, we decided to put a focus on two special aspects.
First, we wanted to show that chemical genetics is a truly interdisciplinary approach with a long history and most of its fundamental ideas have been developed in different fields. There have been many examples of elegant chemical genetic solutions to biological problems in the past, long before this term even existed. What is new, however, is the coordinated effort of contributors from many disciplines like cell biology, structural biology, biochemistry, and chemistry to identify or develop new small molecules for the study of biological questions. In the past, this collaboration and also the exchange of information were very weak, and it often took decades until it was realized that a compound discovered by a chemist could be helpful for biologists and it was thus “rediscovered.” We think that this is also one of the main challenges in chemical genetics as an interdisciplinary approach: few laboratories exist that have enough experience in all the disciplines contributing to chemical genetics. Therefore, efficient collaborations are often mandatory for a project to be successful. We think that setting up core facilities providing libraries and screening infrastructure, which is happening in many institutions now, will be very helpful in this context. Our intention was to also show that chemical genetics is most powerful when used in combination with the more traditional tools of the field. While it is a very versatile tool that effectively complements the toolbox of biologists, chemical genetics is not a universal solution for any biological problem and success strongly depends on the right choice of the biological question to be answered.
Our second focus was on the rapidly expanding field of forward chemical genetics. We tried to present a few interesting examples, thus covering both the different strategies for target identification as well as the broad spectra of screening systems that have reached an astonishing complexity. Increasing complexity comes at a cost, though. While screening in complex organisms enables us to study problems that were out of reach using cell lines or unicellular organisms, target identification can often be very difficult. Although many strategies have been suggested and some of them have been shown to work, successful projects beyond proof-of-concept work are still scarce. Often, it is useful to “turn back” to a simpler system like yeast or cell lines for target identification. This can, however, only be successful if these organisms express homologues of the target protein and show a distinct phenotype upon compound treatment. It remains to be seen if the existing obstacles can be overcome, which would make these complex systems very useful for basic research. Screening in multicellular organisms is already of high importance for drug discovery, because this strategy helps to minimize the probability of failure of a drug candidate at later stages in drug development as compared to protein-based screens as a starting point.
The mentioned problems, however, should not leave the impression that chemical genetics requires alot of resources and is successful only in exceptional cases. A way to minimize the effort required to apply chemical genetics to biological problems is to use small molecules with identified targets. While this approach does not strictly require a profound expertise in chemical genetics it allows researchers from different areas to discover potentially novel and unexpected functions of proteins.
Finally, we want to emphasize that the ultimate goal of chemical genetics is to provide biologists with a toolbox of specific small molecules for every protein in a cell. Clearly, this is a challenge for both chemists and biologists and requires the discovery of sophisticated methods to synthesize complex molecules, to perform elaborate small molecule screens and finally to demonstrate specificity of the identified bioactive compounds. While this will be a long way to go, there is no doubt that chemistry already has and will continue to change our view on biology.
References
- Anderson, C, Bartlett, S J, Gansner, J M, Wilson, D, He, L, Gitlin, J D, Kelsh, R N, and Dowden, J (2007). “Chemical genetics suggests a critical role for lysyl oxidase in zebrafish notochord morphogenesis.” Mol. Biosyst. 3, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz, K, McHardy, L, Gable, K, Tarling, T, Reberioux, D, Bryan, J, Andersen, R J, Dunn, T, Hieter, P, and Roberge, M (2004). “Yeast genome-wide drug-induced haplo-insufficiency screen to determine drug mode of action.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0307122101 101, 4525–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain, J, McLauchlan, H, Elliott, M, and Cohen, P (2003). “The specificities of protein kinase inhibitors: an update.” Biochem. J. 371, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierer, B E, Mattila, P S, Standaert, R F, Herzenberg, L A, Burakoff, S J, Crabtree, G, and Schreiber, S L (1990). “Two distinct signal transmission pathways in T lymphocytes are inhibited by complexes formed between an immunophilin and either FK506 or rapamycin.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.87.23.9231 87, 9231–9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisy, G G, and Taylor, E W (1967a). “The mechanism of action of colchicine. Binding of colchicine-3H to cellular protein.” J. Cell Biol. 34, 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisy, G G, and Taylor, E W (1967b). “The mechanism of action of colchicine. Colchicine binding to sea urchin eggs and the mitotic apparatus.” J. Cell Biol. 34, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breinbauer, R, Vetter, I R, and Waldmann, H (2002). “From protein domains to drug candidates—natural products as guiding principles in the design and synthesis of compound libraries.” Angew. Chem., Int. Ed. 41, 2879–2890. [DOI] [PubMed] [Google Scholar]
- Brown, E J, Albers, M W, Shin, T B, Ichikawa, K, Keith, C T, Lane, W S, and Schreiber, S L (1994). “A mammalian protein targeted by G1-arresting rapamycin-receptor complex.” Nature (London) 10.1038/369756a0 369, 756–758. [DOI] [PubMed] [Google Scholar]
- Burdine, L, and Kodadek, T (2004). “Target identification in chemical genetics: the (often) missing link.” Chem. Biol. 10.1016/j.chembiol.2004.05.001 11, 593–597. [DOI] [PubMed] [Google Scholar]
- Burns, C G, Milan, D J, Grande, E J, Rottbauer, W, MacRae, C A, and Fishman, M C (2005). “High-throughput assay for small molecules that modulate zebrafish embryonic heart rate.” Nat. Chem. Biol. 10.1038/nchembio732 1, 263–264. [DOI] [PubMed] [Google Scholar]
- Cafferkey, R, Young, P R, McLaughlin, M M, Bergsma, D J, Koltin, Y, Sathe, G M, Faucette, L, Eng, W K, Johnson, R K, and Livi, G P (1993). “Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity.” Mol. Cell. Biol. 13, 6012–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y, Chen, H, Rhoad, A E, Warner, L, Caggiano, T J, Failli, A, Zhang, H, Hsiao, C L, Nakanishi, K, and Molnar-Kimber, K L (1994). “A putative sirolimus (rapamycin) effector protein.” Biochem. Biophys. Res. Commun. 10.1006/bbrc.1994.2140 203, 1–7. [DOI] [PubMed] [Google Scholar]
- Chiu, M I, Katz, H, and Berlin, V (1994). “RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12∕rapamycin complex.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.91.26.12574 91, 12574–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy, R K, and Thomas, J H (1999). “Fluoxetine-resistant mutants in C. elegans define a novel family of transmembrane proteins.” Mol. Cell 10.1016/S1097-2765(00)80362-7 4, 143–152. [DOI] [PubMed] [Google Scholar]
- Ciani, L, and Salinas, P C (2005). “WNTs in the vertebrate nervous system: From patterning to neuronal connectivity.” Nat. Rev. Neurosci. 10.1038/nrn1665 6, 351–362. [DOI] [PubMed] [Google Scholar]
- Clardy, J, and Walsh, C (2004). “Lessons from natural molecules.” Nature (London) 10.1038/nature03194 432, 829–837. [DOI] [PubMed] [Google Scholar]
- Cohen, P, and Frame, S (2001). “The renaissance of GSK3.” Nat. Rev. Mol. Cell Biol. 2, 769–776. [DOI] [PubMed] [Google Scholar]
- Crespo, J L, and Hall, M N (2002). “Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae.” Microbiol. Mol. Biol. Rev. 10.1128/MMBR.66.4.579-591.2002 66, 579–591, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, S P, Reddy, H, Caivano, M, and Cohen, P (2000). “Specificity and mechanism of action of some commonly used protein kinase inhibitors.” Biochem. J. 10.1042/0264-6021:3510095 351, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S, Wu, T Y, Brinker, A, Peters, E C, Hur, W, Gray, N S, and Schultz, P G (2003). “Synthetic small molecules that control stem cell fate.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0732087100 100, 7632–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll, M, Dean, E, Reilly, E, Bergholz, E, and Chalfie, M (1989). “Genetic and molecular analysis of a Caenorhabditis elegans beta-tubulin that conveys benzimidazole sensitivity.” J. Cell Biol. 10.1083/jcb.109.6.2993 109, 2993–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, P (1967). “Partial cell functions.” Nobel lecture, December 11, 1908. In Nobel Lectures, Physiology or Medicine 1901-1921, pp. 304–320, Elsevier, Amsterdam. [Google Scholar]
- Fenteany, G, Standaert, R F, Lane, W S, Choi, S, Corey, E J, and Schreiber, S L (1995). “Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin.” Science 10.1126/science.7732382 268, 726–731. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, K, et al. (2006). “Chemical genetics reveals an RGS∕G-protein role in the action of a compound.” PLoS Genet. 2, e57 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai, Z (2005). “The new bacterial cell biology: moving parts and subcellular architecture.” Cell 10.1016/j.cell.2005.02.026 120, 577–586. [DOI] [PubMed] [Google Scholar]
- Gitai, Z, Dye, N A, Reisenauer, A, Wachi, M, and Shapiro, L (2005). “MreB actin-mediated segregation of a specific region of a bacterial chromosome.” Cell 10.1016/j.cell.2005.01.007 120, 329–341. [DOI] [PubMed] [Google Scholar]
- Godl, K, et al. (2003). “An efficient proteomics method to identify the cellular targets of protein kinase inhibitors.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.2535024100 100, 15434–15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, E R, Hsu, A K, and Gross, G J (2004). “Opioid-induced cardioprotection occurs via glycogen synthase kinase beta inhibition during reperfusion in intact rat hearts.” Circ. Res. 10.1161/01.RES.0000122392.33172.09 94, 960–966. [DOI] [PubMed] [Google Scholar]
- Harding, M W, Galat, A, Uehling, D E, and Schreiber, S L (1989). “A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase.” Nature (London) 10.1038/341758a0 341, 758–760. [DOI] [PubMed] [Google Scholar]
- Hartwell, L H (2002). “Yeast and cancer.” Nobel lecture, December 9, 2001. In Les Prix Nobel. The Nobel Prizes 2001, Frängsmyr, T (ed), pp. 246–265. Nobel Foundation, Almqvist & Wiskell International, Stockholm. [Google Scholar]
- Heitman, J, Movva, N R, and Hall, M N (1991). “Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast.” Science 10.1126/science.1715094 253, 905–909. [DOI] [PubMed] [Google Scholar]
- Hemler, M, Lands, W E, and Smith, W L (1976). “Purification of the cyclooxygenase that forms prostaglandins. Demonstration of two forms of iron in the holoenzyme.” J. Biol. Chem. 251, 5575–5579. [PubMed] [Google Scholar]
- Hergenrother, P J (2006). “Obtaining and screening compound collections: a user’s guide and a call to chemists.” Curr. Opin. Chem. Biol. 10.1016/j.cbpa.2006.04.005 10, 213–218. [DOI] [PubMed] [Google Scholar]
- Hoyt, M A, Totis, L, and Roberts, B T (1991). “S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function.” Cell 10.1016/0092-8674(81)90014-3 66, 507–517. [DOI] [PubMed] [Google Scholar]
- Iwai, N, Nagai, K, and Wachi, M (2002). “Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2.” Biosci., Biotechnol., Biochem. 66, 2658–2662. [DOI] [PubMed] [Google Scholar]
- Kapoor, T M, Mayer, T U, Coughlin, M L, and Mitchison, T J (2000). “Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5.” J. Cell Biol. 10.1083/jcb.150.5.975 150, 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T M, and Mitchison, T J (2001). “Eg5 is static in bipolar spindles relative to tubulin: Evidence for a static spindle matrix.” J. Cell Biol. 10.1083/jcb.200106011 154, 1125–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky, S M, Jung, D W, Kang, T W, Walsh, D P, Moon, H S, Jo, H, Jacobson, E M, Shetty, V, Neubert, T A, and Chang, Y T (2003). “Facilitated forward chemical genetics using a tagged triazine library and zebrafish embryo screening.” J. Am. Chem. Soc. 10.1021/ja035334d 125, 11804–11805. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A, Copenagle, L, Gordon, M B, Compton, D A, and Kapoor, T M (2003). “Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis.” J. Cell Biol. 10.1083/jcb.200208143 160, 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling, A, Sperl, B, Hollis, A, Eick, D, and Berg, T (2006). “Selective inhibition of c-Myc∕Max dimerization and DNA binding by small molecules.” Chem. Biol. 13, 745–751. [DOI] [PubMed] [Google Scholar]
- Knight, Z A, and Shokat, K M (2007). “Chemical genetics: where genetics and pharmacology meet.” Cell 10.1016/j.cell.2007.01.021 128, 425–430. [DOI] [PubMed] [Google Scholar]
- Kunz, J, Henriquez, R, Schneider, U, Deuter-Reinhard, M, Movva, N R, and Hall, M N (1993). “Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression.” Cell 10.1016/0092-8674(93)90144-F 73, 585–596. [DOI] [PubMed] [Google Scholar]
- Kwok, T C, Ricker, N, Fraser, R, Chan, A W, Burns, A, Stanley, E F, McCourt, P, Cutler, S R, and Roy, P J (2006). “A small-molecule screen in C. elegans yields a new calcium channel antagonist.” Nature (London) 10.1038/nature04657 441, 91–95. [DOI] [PubMed] [Google Scholar]
- Li, R, and Murray, A W (1991). “Feedback control of mitosis in budding yeast.” Cell 10.1016/0092-8674(81)90015-5 66, 519–531. [DOI] [PubMed] [Google Scholar]
- Luesch, H, Wu, T Y, Ren, P, Gray, N S, Schultz, P G, and Supek, F (2005). “A genome-wide overexpression screen in yeast for small-molecule target identification.” Chem. Biol. 10.1016/j.chembiol.2004.10.015 12, 55–63. [DOI] [PubMed] [Google Scholar]
- Mayer, T U, Kapoor, T M, Haggarty, S J, King, R W, Schreiber, S L, and Mitchison, T J (1999). “Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen.” Science 286, 971–974. [DOI] [PubMed] [Google Scholar]
- Meijer, L, Flajolet, M, and Greengard, P (2004). “Pharmacological inhibitors of glycogen synthase kinase 3.” Trends Pharmacol. Sci. 25, 471–480. [DOI] [PubMed] [Google Scholar]
- Mitchison, T J (1994). “Towards a pharmacological genetics.” Chem. Biol. 10.1016/1074-5521(94)90034-5 1, 3–6. [DOI] [PubMed] [Google Scholar]
- Noren-Muller, A, et al. (2006). “Discovery of protein phosphatase inhibitor classes by biology-oriented synthesis.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0601490103 103, 10606–10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse, P (2002). “Cyclin dependent kinases and cell cycle control.” Nobel lecture, December 9, 2001. In Les Prix Nobel. The Nobel Prizes 2001, Frängsmyr, T (ed), pp. 308–320. Nobel Foundation, Almqvist & Wiskell International, Stockholm. [Google Scholar]
- Oltersdorf, T, et al. (2005). “An inhibitor of Bcl-2 family proteins induces regression of solid tumours.” Nature (London) 10.1038/nature03579 435, 677–681. [DOI] [PubMed] [Google Scholar]
- Ortholand, J Y, and Ganesan, A (2004). “Natural products and combinatorial chemistry: Back to the future.” Curr. Opin. Chem. Biol. 10.1016/j.cbpa.2004.04.011 8, 271–280. [DOI] [PubMed] [Google Scholar]
- Peterson, J R, Lebensohn, A M, Pelish, H E, and Kirschner, M W (2006). “Biochemical suppression of small-molecule inhibitors: a strategy to identify inhibitor targets and signaling pathway components.” Chem. Biol. 13, 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, J R, and Mitchison, T J (2002). “Small molecules, big impact: a history of chemical inhibitors and the cytoskeleton.” Chem. Biol. 10.1016/S1074-5521(02)00284-3 9, 1275–1285. [DOI] [PubMed] [Google Scholar]
- Peterson, R T, Shaw, S Y, Peterson, T A, Milan, D J, Zhong, T P, Schreiber, S L, MacRae, C A, and Fishman, M C (2004). “Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation.” Nat. Biotechnol. 10.1038/nbt963 22, 595–599. [DOI] [PubMed] [Google Scholar]
- Piggott, A M, and Karuso, P (2004). “Quality, not quantity: The role of natural products and chemical proteomics in modern drug discovery.” Comb. Chem. High Throughput Screening 7, 607–630. [DOI] [PubMed] [Google Scholar]
- Radeke, H S, and Snapper, M L (1998). “Photoaffinity study of the cellular interactions of ilimaquinone.” Bioorg. Med. Chem. 10.1016/S0968-0896(98)00100-X 6, 1227–1232. [DOI] [PubMed] [Google Scholar]
- Rang, H P (2006). “Drug discovery.” In Drug Discovery and Development: Technology in Transition, Rang, H P (ed), pp. 41–56, Churchill Livingstone∕Elsevier, Edinburgh. [Google Scholar]
- Raught, B, Gingras, A C, and Sonenberg, N (2001). “The target of rapamycin (TOR) proteins.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.121145898 98, 7037–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, G J, and Majerus, P W (1975). “The mechanism of the effect of aspirin on human platelets. I. Acetylation of a particulate fraction protein.” J. Clin. Invest. 56, 624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, G J, Stanford, N, and Majerus, P W (1975). “Acetylation of prostaglandin synthase by aspirin.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.72.8.3073 72, 3073–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupasinghe, C N, and Spaller, M R (2006). “The interplay between structure-based design and combinatorial chemistry.” Curr. Opin. Chem. Biol. 10.1016/j.cbpa.2006.03.014 10, 188–193. [DOI] [PubMed] [Google Scholar]
- Sabatini, D M, Erdjument-Bromage, H, Lui, M, Tempst, P, and Snyder, S H (1994). “RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs.” Cell 10.1016/0092-8674(94)90570-3 78, 35–43. [DOI] [PubMed] [Google Scholar]
- Sabers, C J, Martin, M M, Brunn, G J, Williams, J M, Dumont, F J, Wiederrecht, G, and Abraham, R T (1995). “Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells.” J. Biol. Chem. 10.1074/jbc.270.2.815 270, 815–822. [DOI] [PubMed] [Google Scholar]
- Schreiber, S L (1998). “Chemical genetics resulting from a passion for synthetic organic chemistry.” Bioorg. Med. Chem. 10.1016/S0968-0896(98)00126-6 6, 1127–1152. [DOI] [PubMed] [Google Scholar]
- Taunton, J, Hassig, C A, and Schreiber, S L (1996). “A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p.” Science 10.1126/science.272.5260.408 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Vane, J R (1971). “Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs.” Nature New Biol. 231, 232–235. [DOI] [PubMed] [Google Scholar]
- Vane, J R, and Botting, R M (2003). “The mechanism of action of aspirin.” Thromb. Res. 110, 255–258. [DOI] [PubMed] [Google Scholar]
- Vassilev, L T, et al. (2004). “In vivo activation of the p53 pathway by small-molecule antagonists of MDM2.” Science 10.1126/science.1092472 303, 844–848. [DOI] [PubMed] [Google Scholar]
- Vezina, C, Kudelski, A, and Sehgal, S N (1975). “Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle.” J. Antibiot (Tokyo) 28, 721–726. [DOI] [PubMed] [Google Scholar]
- Wakefield, J G, Stephens, D J, and Tavare, J M (2003). “A role for glycogen synthase kinase-3 in mitotic spindle dynamics and chromosome alignment.” J. Cell. Sci. 10.1242/jcs.00273 116, 637–646. [DOI] [PubMed] [Google Scholar]
- Weinstein, B M, Stemple, D L, Driever, W, and Fishman, M C (1995). “Gridlock, a localized heritable vascular patterning defect in the zebrafish.” Nat. Med. 1, 1143–1147. [DOI] [PubMed] [Google Scholar]
- Wicks, S R, Yeh, R T, Gish, W R, Waterston, R H, and Plasterk, R H (2001). “Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map.” Nat. Genet. 10.1038/88878 28, 160–164. [DOI] [PubMed] [Google Scholar]
- Zeng, X, Tamai, K, Doble, B, Li, S, Huang, H, Habas, R, Okamura, H, Woodgett, J, and He, X (2005). “A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation.” Nature (London) 10.1038/nature04185 438, 873–877. [DOI] [PMC free article] [PubMed] [Google Scholar]