Abstract
The sequencing of the genomes of key organisms and the subsequent identification of genes merely leads us to the next real challenge in modern biology—revealing the precise functions of these genes. Further, detailed knowledge of how the products of these genes behave in space and time is required, including their interactions with other molecules. In order to tackle these considerable tasks, a large and continuously expanding toolbox is required to probe the functions of proteins on a cellular level. Here, the currently available tools are described and future developments are projected. There is no doubt that only the close interplay between the life science disciplines in addition to advances in engineering will be able to meet the challenge.
The major goal of molecule-oriented biology is to accumulate knowledge about all molecules in the organelle, cell, or organism of interest. Even in organisms where the genome is unraveled, so far only a fraction of the molecules have a function assigned to. When investigating the limited set of known proteins thoroughly, there is initially a need for quantitative data, the most basic of which is counting molecules. The resulting information would, even if all proteins were known, not suffice to describe a complex entity like the living cell. We would also need to know about the mobility of each molecule and its location in the cell or organism at a particular point in time. Further, we need to know when a molecule species is appearing or disappearing. Is this enough? The answer is “no.” Molecules might be abundant but inactive or the same molecule might exhibit different activities in different areas or time courses. The function of a large part of the genome, namely enzymatic activity, is usually strongly regulated by posttranslational modifications. It is therefore essential to know in what form of modification a protein of interest is and, more important, how active it is. The required spatial and temporal information is not limited to proteins. Other important molecules include the different types of RNA, DNA in its various forms of condensation and nuclear distribution, and, of course, ions and small molecules. The latter are nucleosides and nucleotides, carbohydrates, lipids, peptides, and different kinds of co-factors. Of these, lipids constitute the most diverse, the least investigated, and therefore the most difficult group of small molecules to study. Accordingly, we would like to monitor several thousand proteins at various stages of modification, plus several thousand RNAs and an estimated thousand small molecules. In addition, unknown or undetectable parameters need to be accounted for by modeling. Combining all this could be the basis for performing systems biology. The data set will need to be comparable with respect to organisms and external conditions and can then be fed into modeling programs designed to deal with complex systems.
About 80–90% of all genes in an organism such as Drosophila change expression levels over its lifespan (Arbeitman et al., 2002). What seems a humongous data acquisition task when extended to quantitative data acquisition could be eased a bit by looking only at those factors and events that significantly change during the time course of an experiment. For this interval, it can be speculated that the majority of molecules will remain in a steady state with respect to gene expression and protein location after cells are stimulated, unless major events like cell division or pathogen entry are happening. But even during the onset of house-keeping signaling the parameters of many hundreds of molecules will be altered (Iyer et al., 1999).
Ideally, we would like to monitor these alterations on a protein level simultaneously and within the intact cell. Measuring in living cells requires predominantly light microscopy approaches, in particular fluorescent methods that permit following molecules in realtime with good spatial resolution (Yuste, 2005). By using time-lapse microscopy of fluorescent reporters in living cells we can to date measure intracellular enzyme activities, the location of proteins or sometimes protein complexes, the formation of larger entities like microtubuli, and the synthesis or breakdown of a limited number of small molecules. In time-lapse fluorescence microscopy, so far most approaches are of qualitative or semi-quantitative nature. However, more quantitative approaches have been introduced. In particular, fluorescence lifetime imaging microscopy (FLIM) as a way to measure fluorescence resonance energy transfer (FRET) opens the possibility to distinguish populations of molecules within a defined volume element. In this form of global analysis, bound and unbound protein populations can be determined, provided that certain prerequisites, such as priori knowledge of the fluorescence decay kinetics, are fulfilled. (Verveer et al., 2000a,b). Other microscopy methods that deliver quantitative data are fluorescence correlation spectroscopy (FCS) (Haustein and Schwille, 2007) and single molecule microscopy. FCS allows observing a defined small volume within the cell and therefore provides information about the concentration as well as the size of the molecule (or the complex of molecules) of interest. In combination with evanescence illumination, single molecule microscopy was successfully used to image the internalization of single green fluorescent protein (GFP)-labeled growth factor receptors (Webb et al., 2007). FLIM, FCS, and single molecule microscopy are probably the most promising imaging techniques to generate quantitative data in living cells. The problem is that these techniques are not generally available, with FCS being the most common one. Both FLIM and single molecule microscopy require special instrumentation and significant skills on the user side.
To a limited extend the simultaneous acquisition of different data sets is possible with the above mentioned techniques. In any of the fluorescent imaging techniques, we are only able to measure a few intracellular parameters at the same time, allowing us to get a glimpse of the close timing that is involved in the intertwined biochemical processes of the cell. However, we are far from mastering the cell’s complexity simply because for spectral or frequency reasons we are unable to use more than four to six fluorescent sensors at the same time. In addition, the number of available fluorescent sensors is still very small.
How can complex events, such as signaling cascades, be tackled in intact cells in a quantitative manner? Classic biochemistry has produced a wealth of information on signaling networks and molecule turnover. From this literature we should be able to make intelligent guesses of where biochemical processes are linked sufficiently close that they are likely to form a module (Meyer and Teruel, 2003). Imaging a single module with a selected set of fluorescent sensors could produce a concise readout that reduces the number of relevant and therefore representative sensors for each module to a manageable minimum (Teruel and Meyer, 2002). Measuring several of these markers simultaneously might provide us with an idea of how modules are connected (Schultz et al., 2005; Teruel and Meyer, 2002). A second set of approaches would start again with the comparison of stimulated versus unstimulated cells. But instead of activating an entire signaling cascade or a physiological process by treating cells with a hormone, a neurotransmitter, a toxin, a drug, or a physical or electrical stimulus, tools are used that activate (or deactivate) a very limited subset of intracellular events. Instead of a global onset of the signaling machinery, ideally a small dissected set of immediate parameters downstream of the physiological stimulus, e.g., an activated receptor, will be visible. In the past, the most common approaches in this respect involved the use of pharmaceuticals, such as rapamycin or membrane-permeant cyclic nucleotide derivatives, just to name a few examples. Most of these activators or inhibitors, however, result in a response pattern that does not resemble a physiologically relevant pattern, at least on the time scale of seconds. A main reason for this is that the kinetics of small molecule-mediated triggers reflects predominantly cell entry of the small molecule, rather than target enzyme kinetics. Therefore it would be particularly important to use and develop methods that permit an initial onset of activity as well as the generation of entire activity patterns thereby reflecting the timing of processes in the intact cell. On the small molecule level, a limited number of photoactivatable compounds fulfill this task. Compared to the vast demand for such tools, very little has been achieved on the level of photoactivatable proteins, although “caged” enzymes would definitely be highly desirable, for instance to switch on subsets of signaling cascades. An alternative approach to photoactivation would be translocation-mediated signal switching. The different technical possibilities to rapidly dissect signal transduction pathways will be discussed in the main text. Some important tools and techniques, such as in vivo labeling (Gronemeyer et al., 2005; Chen and Ting, 2005; Miller and Cornish, 2005), the use of quantum dots (Jamieson et al., 2007) or small interfering RNAs (Hannon, 2002), have been reviewed before and are omitted due to space limitations. In addition, I would like to draw the attention to a few very recently published reviews in the field (Giepmans et al., 2006; Johnsson and Johnsson, 2007).
Firstly, current and future optical approaches to determine molecule numbers in cells and to measure intracellular events will be detailed.
SENSORS AND REPORTERS
Counting molecules
Counting molecules in living cells is a difficult task. In fact, if you will ask a biologist about copy numbers of his or her protein of interest per cell, you will often receive not more than a shrug. However, in order to eventually produce bottom-up models for cellular events we will need to know how many molecules we are dealing with. The largest problem results from the fact that we would like to avoid using cell ensembles and rather look into single cells. This practically excludes quantitative Western blotting and standard proteomic approaches, although even the determination of molecule numbers in batches of cells is not regularly performed. Up to now the focus was on global analysis of all proteins and it is certainly useful to establish an overall picture of expression levels under certain conditions (Ghaemmaghami et al., 2003; Huh et al., 2003). However, the required techniques (global homologous recombination plus Tap tagging or global GFP-tagging) are probably too cumbersome to be used for the average cell biology problem. Recently, though, single cell analysis of protein numbers has become available (Huang et al., 2007). Single cells are lysed using a microfluidic device which also includes separation of molecules by capillary electrophoresis. Proteins are then detected by fluorescent antibodies. Unfortunately, all these techniques require the destruction of the cell. The question arises whether it is possible to quantify protein levels inside living cells. The obvious way would be again the use of fluorescent fusion proteins. In a pioneering study, the Ellenberg lab determined the copy numbers of nuclear pore complex components in order to develop a thorough model on nuclear pore dynamics (Rabut et al., 2004). Each GFP-tagged pore protein was expressed in a stable cell line. Calibration was achieved by comparing the fluorescence intensity of each nuclear pore to single virus-like particles known to contain 120 GFP molecules each. The work required the stable expression of many different proteins as GFP fusions.
Are there ways to avoid these elaborate procedures or to refrain from tagging proteins in general? In the future, the constant improvements in cryoelectron tomography might eventually allow us to determine the precise number of large protein aggregates or even single large proteins in the frozen cell (Nickell et al., 2006). Another option might be provided by Raman microscopy because this technique does not require the introduction of labeled molecules to cells, but rather can detect any molecule, that provides a specific Raman band (van Manen et al., 2005). Vibrational spectroscopic methods, such as IR or Raman spectroscopy, are based on measuring the intrinsic atom-to-atom movement of endogenous molecules.
Although counting molecules is absolutely indispensable for bottom-up systems biology in the future, we are far from successfully tackling this crucial problem on a routine basis. Considering all the above-mentioned problems and technical demand it might be advisable to mainly use methods where relative changes in molecule levels are monitored, however, with the background information of total numbers in the resting cell or after thorough calibration. After all, even the sophisticated method introduced by Huang et al. (2007) does not permit analysis in a time-resolved manner, let alone information regarding the spatial distribution and activity of proteins within one cell.
Fluorescent sensors for time-lapse microscopy
Relative changes in concentration or activity of molecules in a living cell are mostly monitored by fluorescent probes, if possible by using a ratiometric approach. Ratiometric probes were initially developed by the Tsien lab for measuring changes in ion concentrations (Grynkiewicz et al., 1985; Tsien, 1981). Binding of an ion to the sensor changes either the excitation spectra as in, e.g., the Fura series of calcium sensors. Others change the emission properties and permit emission ratio analysis (e.g., Indo-1) (Grynkiewicz et al., 1985). The advantage of many of these probes is that the necessary calibration that yields concentration readouts has been thoroughly established.
Reporters that recognize molecules larger than ions hardly exist, mainly because the recognition structure needs to be excessively big, expanding to the size level of proteins. Accordingly, some sensors have been introduced where the binding of a small molecule, for instance a nucleotide, gives rise to conformational change of a respective binding protein. The first FRET probe of that type was FlCRhR, a sensor for adenosine 3′,5′-cyclic monophosphate (cAMP) levels based on its major binding protein, the cAMP-dependent protein kinase (PKA). Attachment of fluorescein to the catalytic and rhodamine to the regulatory subunit (hence the name) gave FRET as long as the holoenzyme was intact. Upon binding of four cAMP molecules the subunits separated and FRET became impossible (Adams et al., 1991). For experiments in living cells, FlCRhR needed to be microinjected. Following the development of the colorful array of fluorescent proteins (Ai et al., 2007; Chudakov et al., 2005; Shaner et al., 2004, 2005; Shu et al., 2006), there are now numerous FRET sensors that are genetically encoded (Zhang et al., 2002). Usually, the sensor unit is sandwiched between a fluorescent protein serving as a donor and another serving as the FRET acceptor. Despite the relative technical ease of preparing such a sensor, the outcome is hard to predict. Accordingly, the number of reporters that are able to monitor changes in molecule counts is very limited, but reporters for cAMP (Adams et al., 1991; Nikolaev et al., 2004; Zaccolo et al., 2000), cyclic guanosine 3′,5′-monophosphate (cGMP) (Honda et al., 2001), H2O2 (Belousov et al., 2006), myo-inositol 1,4,5-trisphosphate (Morii et al., 2002), glucose (Fehr et al., 2003), maltose (Fehr et al., 2002), ribose (Lager et al., 2003), phosphate (Gu et al., 2006), glutamate (Okumoto et al., 2005), and lipopolysaccharide (LPS) (Voss et al., 2007) have been introduced. For quantification in cells, in vitro derived binding constants could be used. In special cases, a priori knowledge can be used. For example, glucose concentrations in liver cells are known to be equilibrated by reversible GLUT-mediated plasma membrane transport (Fehr et al., 2005). Accordingly, intracellular glucose levels equal those in the extracellular space.
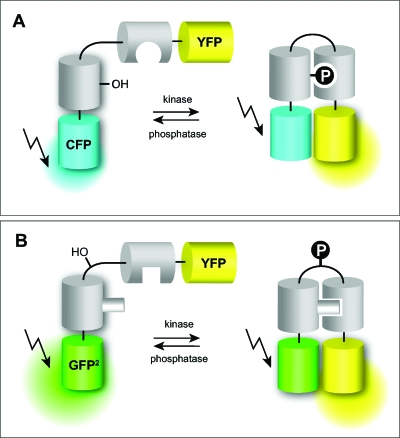
More common are genetically encoded sensors that report enzyme activities (Zhang et al., 2002). Especially phosphorylation events are readily picked up by conjugates constructed of a substrate sequence connected to a binding domain that recognizes one of the states of the substrate, for instance the phosphorylated peptide moiety but not the dephosphorylated one (Kunkel et al., 2005; Lin and Ting, 2004; Rothman et al., 2005; Schleifenbaum et al., 2004; Ting et al., 2001; Violin et al., 2003; Zhang et al., 2001). Recognition leads to a conformational change resulting in an alteration of the distance or the dipole moment of the two fluorophores or both [Fig. 1A]. As FRET is extremely sensitive to both factors (Jares-Erijman and Jovin, 2003; 2006), the sensors respond with a change in the emission ratio of the donor and the acceptor thereby providing a ratiometric readout for enzyme activity. Apart from phosphorylation∕dephosphorylation events, FRET sensors for such diverse functions as peptide methylation (Lin et al., 2004), GTP/GDP exchange rates (Mochizuki et al., 2001), Rho-GTPase activity (Pertz et al., 2006), or G-protein interaction have been developed (Janetopoulos et al., 2001).
Figure 1. Design of some FRET-based reporters of phosphorylation levels.
(A) Recognition of a phosphorylated residue by a phosphoserine or tyrosine binding domain results in a more compact conformation with increased FRET. (B) Phosphorylation of the loop structure of a pleckstrin fragment leads to a conformational change of the adjacent domains and alteration of the FRET properties.
Similar approaches are based on proteins that are intrinsically responding to enzymatic modification with a conformational change. This type of sensor could in principle be the basis for a general phosphorylation sensor, provided that phosphorylation is only recognized by a change in the number of negative charges. Any specific substrate sequence should then lead to a new specific kinase∕phosphatase activity sensor. As an example, pleckstrin as the major substrate for protein kinase C in platelets was picked (Schleifenbaum et al., 2004). Following nuclear magnetic resonance (NMR) data, the N-terminal pleckstrin homology domain and the DEP domain flank an unstructured substrate loop (M. Sattler and co-workers, unpublished results). Upon phosphorylation of its three neighboring substrate sites the molecule undergoes a conformational change [Fig. 1B]. When fluorophores like GFP2 (FRET donor) and EYFP (FRET acceptor) are attached to the C- and N-termini, respectively, phosphorylation in vivo and in vitro leads to an increase in FRET efficiency which can easily be monitored by the ratio of donor and acceptor emission intensities (Schleifenbaum et al., 2004). This type of conformational changes, however, is very difficult to predict. For instance, if the last amino acids of the DEP domain are removed, the sensor responds with a decrease in emission ratio (Brumbaugh et al., 2006).
In addition, the fluorophores themselves seem to participate in the sensor dynamics, since non-dimerizing variants of the fluorescent proteins render the sensor non-functional (C. Jost and C. Schultz, unpublished results 2007). Only dynamic NMR analysis of the sensors during the phosphorylation procedure will provide us with a useful model for sensor performance in the future. Finally, the original idea of generating a general sensor platform, was so far unsuccessful: introduction of standard protein kinase A substrate peptides gave mostly inactive sensors (A. Schleifenbaum, unpublished results). However, the introduction of a PKA-sensitive loop at the C-terminus of KCP-1 gave a dual parameter sensor that is able to monitor both PKC and PKA with an increase and a decrease in FRET, respectively (Brumbaugh et al., 2006). A more general approach to prepare sensors that monitor intracellular enzyme activities should definitely be pursued, because there will be need for all possible kinase sensors in the future. In fact, it should be considered to prepare conformation sensors by sticking proteins between pairs of FRET-exhibiting fluorophores on a quasi-genome-wide basis, maybe excluding transmembrane proteins and proteins that are just too big for FRET approaches. This sensor array would provide information about protein conformational changes under any kind of conditions in living cells. This would greatly increase the number of available sensors.
All genetically encoded reporters only provide information of relative changes in phosphorylation levels etc. Accordingly, it is almost impossible to extract quantitative data from these measurements in cells. Nevertheless, these semiquantitative data will be indispensible for the analysis of signaling pathways or global cellular changes such as mitosis in the future.
An important feature of genetically encoded reporters should be that, as a true reporter, they should be inert to cell homeostasis. Ideally, the reporter is a molecule that does not occur in the cell of interest, for instance a bacterial protein expressed in mammalian cells. Most sensors are sufficiently artificial that little interference with enzymes is expected. Alternatively, some sensors are based on a very specialized protein from a particular cell type with little or no expression in the cell of interest. The above-mentioned pleckstrin, a protein predominantly expressed in platelets, is a good example. Sensors of small molecules might have the capacity to buffer the molecule of interest and therefore augment its biological effect. Therefore, sensors with a modest binding affinity just above the dissociation constant might be preferred over strong binders.
Translocation as readout
Instead of ratiometric probes, simple fusion constructs with one fluorophore may be used in cases where, as a response to an external stimulus, the protein of interest significantly changes locations within a cell (Teruel and Meyer, 2000). Many proteins recognize certain lipids in membranes via more or less selective lipid binding domains. The transient appearance of a lipid, for instance in the plasma membrane, will lead to a transient translocation of a lipid binding domain-bearing protein from the cytosol to the plasma membrane. This technique has been successfully used for measuring lipids such as diacylglycerol (Oancea and Meyer, 1998; Oancea et al., 1998) and phosphatidylinositol polyphosphates (Gray et al., 2003; Halet, 2005; Lee et al., 2005;Varnai et al., 1999). Alternatively, the breakdown of a lipid with a constantly high residual concentration in the plasma membrane, such as phosphatidylinositol 4,5-bisphosphate (PIP2), can be monitored after stimulating the cell with an agonist that leads to phospholipase C activation (van der Wal et al., 2001). In principle, there are many more lipids that could be monitored this way. However, the method has its limitations: for instance, the pleckstrin homology domain (PH domain) used to monitor PIP2 will recognize the lipid only in the plasma membrane, although the abundance of PIP2 in microsomal membranes and in the nucleus has been shown (Balla et al., 2000).
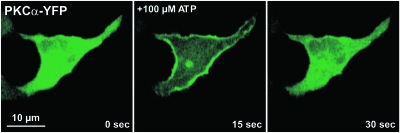
Another target for translocating sensors are calcium ions (Rizo and Sudhof, 1998), (Stahelin and Cho, 2001a). The C2 domain of protein kinase C isoforms and other proteins is known to translocate to the plasma membrane when binding calcium. Therefore a fusion of a C2 domain with a fluorescent protein is an easy to handle sensor for monitoring changes in intracellular calcium concentrations. Alternatively, fluorescent fusions of protein kinase C isoforms containing C2 domains (Fig. 2) may be used (Oancea and Meyer, 1998; Reither et al., 2006). Other translocating calcium binding proteins include phospholipases (Stahelin and Cho, 2001b; Stahelin et al., 2003), synaptotagmin (Shao et al., 1998), and annexins (Gerke et al., 2005; Piljic and Schultz, 2006).
Figure 2. Translocation as a readout of dynamic intracellular signaling.
Protein kinase C alpha (PKCα) transiently translocates to the plasma membrane indicative of an increase in calcium and diacylglycerol levels. In fact, the time course closely follows the calcium signal (Reither et al., 2006).
Translocation as a sensing tool and the use of lipid binding domains is still underdeveloped, especially considering the high number of proteins equipped with these domains. For instance, Meyer and co-workers recently showed that most of the 48 plasma membrane-associated small GTPases have polybasic domains permitting the molecule to interact with negatively charged phospholipids (Heo et al., 2006).
Translocation phenomena are not limited to lipid binding proteins. Other groups of proteins are transported through the Golgi and ER networks (Keller et al., 2001) or are shuffling between cytosol and nucleus. Transcription factors have to enter the nucleus to exhibit their function. Accordingly, fusion proteins of transcription factors such as NFκB subunits or STATs have been used to monitor nuclear entry (Birbach et al., 2004; Kretzschmar et al., 2004; Pranada et al., 2004). Another obvious group of proteins are those involved in nuclear import and export (Plafker and Macara, 2002). Finally, artificially induced translocation may become an important tool for switching protein function inside cells (see the following). Translocating probes are again not able to provide quantitative data, but the possibility to use many of these probes simultaneously is giving valuable insight in the timing of intracellular events.
Photoactivatable and photoswitchable fluorophores
Dynamic measurements often require spatial tracking of a molecule or an entire organelle within a cell. The tools of choice for these purposes are photoactivatable fluorescent proteins (PAFPs) (Lukyanov et al., 2005). PAFPs are structurally similar to GFP but are sensitive to photoinduced conversions, such that their optical properties are modified upon irradiation with light of a specific wavelength. Photoactivation of some PAFPs, such as PA-GFP (Patterson and Lippincott-Schwartz, 2002), enhances their quantum yield, which may give the appearance of switching on the fluorescence. The mechanism involves a decarboxylation of a glutamate residue in the fluorophore binding pocket, which leads to rearrangements and chromophore deprotonation (van Thor et al., 2002; Bell et al., 2003). In others, the emission wavelength of a bright fluorophore may be switched. For example, photoconversion of PS-CFP or PS-CFP2 with an initial emission around 470 nm yields a green fluorescent protein emitting at 510 nm. Other examples are Kaede and EOS-FP, with UV-induced green-to-red conversion (Ando et al., 2002; Wiedenmann et al., 2004). These may be used to locally mark a subpopulation of molecules in one particular region of the cell. Motility of the labeled molecules can thence be tracked with good spatial resolution over an extended period of time. This method is therefore often superior over fluorescence recovery after photobleaching (FRAP) experiments, which reports overall molecule motility but not the destiny of the mobile component. Even more sophisticated PAFPs are reversible in their photoswitching behavior. The tetrameric kindling fluorescent protein (KFP1) was developed from anthozoa-derived chromoproteins. KFP1 is switched from its dark form into a red fluorescent protein upon irradiation with intense green light. The underlying reaction is a trans–cis isomerization of the fluorophore itself (Andresen et al., 2005). In the dark this process is slowly reversed, unless very strong illumination induces an irreversible red fluorescent state. Irradiation with 450 nm switches the fluorescence off immediately (Chudakov et al., 2003, 2006). The green probe “Dronpa” can be reversibly bleached by excitation at 470 nm and fluorescence is restored after irradiation with 400 nm. This photoswitching can be repeated many times (Ando et al., 2007, 2004). In the future, improved probes with longer excitation and photoconversion-inducing wavelength are needed for applications such as the tracking of cells in a developing embryo and other long-term experiments. However, even longer wavelength light may be severely damaging when absorbed by a fluorophore, due to the formation of reactive radical species.
FRET sensors based on small molecules
These sensors have the advantage of being able to enter cells and therefore do not require transfection or other manipulations which are often very difficult in tissue or even fully differentiated cells. Almost all of these sensors are targeting hydrolytic enzymes that cleave the sensor molecule with the effect that the two attached FRET pair-forming fluorophores separate indefinitely. Although these sensors often exhibit huge changes in the emission ratio, an asset for screening applications, the performance of these reporter molecules is irreversible. This limits the use in long-term time lapse experiments where the reporter is eventually used up. However, this feature might also be an advantage when very little enzyme activity needs to be monitored. In some cases the accumulation of the product helps determining the activity of as little as 50 enzyme molecules in one cell (Zlokarnik et al., 1998). Another disadvantage is that these molecules need to be prepared by preparative organic chemistry, a notoriously slow and tedious process, which includes the attachment of two fluorophores and often the necessity to introduce bioactivatable protecting groups to permit cell entry (Schultz, 2003). Many FRET-based probes have been introduced, but so far intracellular applicable FRET probes based on small molecules were only developed for a few hydrolytic enzymes such as proteases, lipases, and β-lactamase.
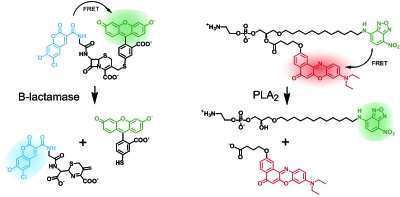
Due to its absence in eucaryote cells, β-lactamase is particularly suitable as a reporter enzyme. When placed under the control of the same promoter that regulates the expression of the protein of interest, visualization of β-lactamase activity directly reports co-expression. The assay requires a synthetic fluorogenic substrate. A membrane-permeant β-lactam derivative, equipped with FRET donor and acceptor was developed (Zlokarnik et al., 1998). Upon cleavage of the β-lactam the fluorescent acceptor is released from the substrate via a β-elimination step, leading to a total loss in FRET (Fig. 3). Emission ratio changes of this probe are enormous which makes the assay particularly suitable for super high-throughput applications. In addition, the ratio approach is well suited for quantification of enzyme activity and is therefore superior to a simple GFP expression assay. More recently, a set of FRET probes was developed that reports the activity of phospholipase A2 (Wichmann et al., 2007, 2006). Again, the phospholipid-based probes with donor and acceptor attached to the tips of the two fatty acids exhibited 20–30-fold changes in the emission ratio upon cleavage in cells. A particular advantage of these small molecule probes was their passive entry into cells and entire organisms made possible by the use of bioactivatable protecting groups such as acetoxymethyl and S-acetylthioethyl esters.
Figure 3. Small molecule FRET probe performance is based on substrate cleavage via β-elimination (left) or hydrolysis (right).
Shown is the reporter gene assay FRET substrate CCF and the PLA2 FRET sensor PENN. Both reporters may be used in living cells when equipped with bioactivatable protecting groups (see the text) (Wichmann et al., 2006; Zlokarnik et al., 1998).
Protease sensors have an obvious construction plan: a specific substrate peptide is flanked by two flurophores or a fluorophore and a quencher which serve as a FRET pair. Upon cleavage the fluorophores separated and FRET becomes impossible. This concept was successfully applied to measure matrix metalloproteases, caspases, cathepsins, as is summarized in several reviews (Baruch et al., 2004; Fonovic and Bogyo, 2007). Although many proteases act in extracellular space, it was in some cases possible to generate a membrane-penetrating sensor, for instance for caspase-3 and some cysteine proteases (Blum et al., 2005; Komoriya et al., 2000). For the future, it would be desirable to use amino acid derivatives carrying bioactivatable protecting groups (see the following) to generally allow peptides and peptide-based sensors to enter cells.
FRET sensors that report enzyme activity with spatial resolution
As discussed earlier, an enzyme might be distributed throughout the cell, but only exhibit its activity in one particular region of the cell. This can be due to local phosphorylation/dephosphorylation events, other posttranslational modifications, or the availability of certain co-factors, for instance lipids. It would therefore be very desirable to monitor not only the enzyme distribution but also enzyme activity with spatial resolution. Freely diffusing sensors, as they are discussed previously, are redistributing too fast to provide this type of information. A solution is a sensor that is strictly located to one area of the cell. This may be achieved by adding a lipidation or a nuclear location sequence (Violin et al., 2003). However, differential analysis would require a set of identical reporters each equipped with specific location sequences.
Recently, a sensor termed “Rango” was introduced that monitored the spatial distribution of the Ran-importin system during mitosis. Rango sensed a RanGTP gradient driven by the guanine exchange factor RCC1. FLIM analysis in HeLa cells showed significant decrease in Rango-importin-β binding around chromatin. Further, the RanGTP gradient seems to be required for spindle assembly (Kaláb et al., 2006).
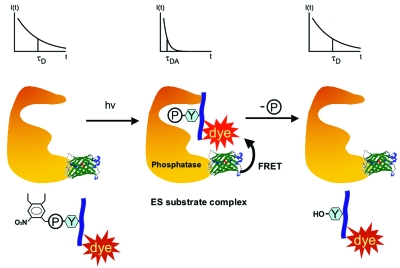
In a different approach, the lab of Philippe Bastiaens and our lab very recently developed a new FRET-based method that makes the detection of spatially resolved enzyme activity possible (Yudushkin et al., 2007). Using the example of phosphotyrosine phosphatase 1B (PTP1B), we constructed a probe that is able to monitor the formation of a PTP1B-substrate complex (Fig. 4). FRET between the two molecular entities was measured by FLIM. A GFP attached to the PTP1B served as the FRET donor and a rhodamine derivative linked to a small phosphorylated peptide constituted the FRET acceptor. The formation of the enzyme-substrate (ES) complex was induced by removing a photoactivatable group from the phosphate moiety. Subsequently, the lifetime of the donor decreased significantly. As the donor is exclusively attached to the enzyme of interest, a very specific readout for this enzyme is provided. This allows monitoring one enzyme in a large pool of competing enzymes. In the setup chosen, the peptide carrying the acceptor is not only dephosphorylated but also re-phosphorylated leading to a steady-state of ES complex levels. When normalized over the amount of donor in a region of interest, the fluorescent lifetime is a direct readout for the amount of ES complex. A large amount of ES complex then indicates regions with little enzyme activity and vice versa, giving us a crude map of where enzyme activity is high and where not. By this means it was confirmed that PTP1B activity is high near microsomal membranes and low at the plasma membrane. It remains to be determined whether this technique can be applied to many other enzymes.
Figure 4. The formation of enzyme-substrate (ES) complexes is monitored by the binding of a FRET acceptor-tagged substrate to a donor-labeled enzyme, here phosphotyrosine phosphatase 1B.
A photoactivatable group on the substrate phosphate prevents premature product formation. Upon photolysis changes in the fluorescence lifetime of the donor emission (upper panels) indicate ES complex formation (Yudushkin et al., 2007).
MOLECULES TO DISSECT INTRACELLULAR SIGNALING CASCADES
An early class of intracellular acting molecules used to influence signaling were derivatives of the few water-soluble small molecules involved in theses processes, such as cyclic nucleotides (Schwede et al., 2000) or inositol polyphosphates (Li et al., 1997; Potter and Lampe, 1995) as well as some lipids. In addition, a growing set of intracellular acting enzyme inhibitors accumulated over the decades. Many of these derivatives are natural compounds and have been identified by serendipity or through screens, e.g., for anticancer activity (Gough and Crews, 2007). In the last decade chemical genetics, the generation of “preferred structure” libraries, and sometimes computer-aided design have started to provide us with new tools to interfere with intracellular events (Breinbauer et al., 2007; Haggarty and Schreiber, 2007; Breinbauer et al., 2002; Schreiber, 1998). In the following paragraphs I will not dwell on small molecule modulators such as kinase inhibitors or effectors of the cytoskeleton. I will rather discuss molecules that have been equipped with functional alterations such as groups for photoactivation, artificially induced enzyme specificity or membrane-permeability as well as molecules useful in translocating and switching enzymes.
As discussed in the introduction, the action of small molecules interacting with intracellular targets is often depending on cell entry kinetics. Therefore, when fast intracellular processes are triggered, the output kinetics might predominantly reflect membrane penetration. On the other hand compounds like rapamycin seem to enter cells very rapidly, allowing the observation of changes inside cells within a few seconds. One way to make the compound of interest available within a fraction of a second is the use of photoactivatable (“caged”) compounds. In a few cases this concept has been expanded to larger molecules up to entire proteins. Alternatively, bioactivatable derivatives of the molecule of interest might be used that provide a sufficiently fast cellular uptake. The methods of how to attach and the choice of photoactivatable groups as well as their applications have recently been reviewed in great detail (Giordano et al., 2007; Goeldner and Givens, 2005).
Photoactivatable small molecules
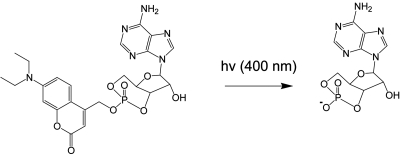
Photoactivatable groups are usually introduced chemically at a position of the molecule that is critical for its biological activity. For instance, for cAMP this would be the charged phosphate residue. Rapid release of the photoactivatable group from caged cAMP generates the biologically active cAMP within the fraction of a second (Fig. 5) (Hagen et al., 2003). The number of small photoactivatable molecules today is still very limited, although it is 20 years ago that the pioneering work on caged ATP has been published (Trentham et al., 1992; Walker et al., 1988). The most common signaling molecules such as the cyclic nucleotides (Hagen et al., 2001, 2002), including cyclic ADP ribose (Aarhus et al., 1995), neurotransmitters (Fedoryak et al., 2005; Furuta et al., 1999; Makings and Tsien, 1994), and a few inositol polyphosphates (Dinkel and Schultz, 2003;Li et al., 1998) have been prepared. In cases, where membrane-permeant versions were available, the tools were successfully used in cells (Furuta et al., 1999; Hagen et al., 2002; Li et al., 1998).
Figure 5. Photoactivation of caged cAMP by near UV light is possible within a fraction of a second ( Eckardt et al , 2002 ).
The reaction generates cAMP and 7-(diethylamino)coumarin-4-yl methanol as a by-product. Only one of the two possible cage-bearing diastereomers is shown.
More recently, the photoactivation of peptides became of interest, mainly because peptides are frequently very potent modulators of protein function. For instance, many kinases are controlled by inhibitor peptides. A successful caging strategy for peptides usually involves the masking of a crucial residue by a photoactivatable group. These are the amino acid side chains that are posttranslationally modified or those that are crucial for protein interaction. Peptides can now routinely be prepared by solid phase peptide synthesis which also permits the introduction of caged amino acids. An interesting aspect is the caging of phosphopeptides at the phosphate group. There are now several examples where caged phosphorylated peptide tools have successfully been employed in vitro (Vazquez et al., 2003) and in vivo (Nguyen et al., 2004; Yudushkin et al., 2007). As usual the problem is cell membrane penetration. Peptides require an extended stretch of basic residues in order to pass the cell membrane. Alternatively, special peptide sequences, usually also dominated by basic amino acids, permit cell entry. The most commonly used penetrating sequences are derived from the Drosophila antennapedia homeoprotein (Gratton et al., 2003) or the HIV Tat transactivator (Frankel and Pado, 1988; Green et al., 1988). The mechanism of entry is still not fully understood but endocytosis seems to play a crucial role (Fischer et al., 2005).
Photoactivatable proteins
Caged proteins are probably among the most desirable tools for cell biology. With a flash of light and therefore in a fraction of a second, the protein of interest is switched on or taken out of the game. This would dramatically facilitate the analysis of intracellular signaling pathways or the formation and interplay of protein complexes. The problems in realizing these tools are obvious: either the caged protein needs to be microinjected (Ghosh et al., 2004) or expressed in a photoactivatable form. For ex-vivo caging often multiresidue caging is performed. Alternatively, single cysteine mutants are employed to permit the site-specific introduction of the cage in aqueous environment, for instance by a nitrobenzyl bromide reagent (Chang et al., 1998, 1995). An alternative to cysteines are thiophosphorylated serines. This unnatural modification can be achieved by performing the in vitro phosphorylation with ATP(γ)S as the thiophosphate donor. The increased nucleophilicity of the thiophosphate then permits specific introduction of the cage (Zou et al., 2002, 2001).
The generation of caged enzymes in living cells to date requires the introduction of unnatural, photosensitive amino acids via site-directed unnatural amino acid mutagenesis. This technique is based on the expansion of the natural genetic code (Xie and Schultz, 2006). So far caged aspartates (Mendel et al., 1991; Short et al., 1999), serines (Cook et al., 1995), tyrosines (Miller et al., 1998; Philipson et al., 2001), cysteines (Philipson et al., 2001), and lysines (Endo et al., 2004b) were used to effectively incorporate caged amino acids into proteins. Mostly nitrobenzyl groups were used as photoactivatable groups to liberate a crucial residue after photolysis. Sometimes it would be useful to photolytically cleave the peptide backbone, for instance to mimick protease activity. This can be achieved by using o-nitro-phenylglycine as an artificial amino acid (Endo et al., 2004a). Alternatively, other photolinkers can be employed as was demonstrated for Smad2 (Hahn and Muir, 2004; Pellois et al., 2004). However, so far these applications were only successfully performed in vitro. For the future, one could imagine that the use of membrane-permeant photocleavable peptides combined with expressed chemical ligation might permit the in vivo preparation of photocleavable proteins.
The down-regulation of protein activity via light is already possible in another way. In fact, photodestruction can be employed to switch off molecules: the red fluorescent protein variant “Killer-Red” generates sufficient reactive oxygen species in the vicinity of the fluorophore that proteins residing nearby are damaged. This can be used to selectively eliminate protein activity within a very small sub-compartment of the cell (Lukyanov et al., 2005).
Photoaffinity labeling
Another group of photosensitive tools are photoactivatable ligands. These compounds are used to provide a covalent link between a small molecule and its protein target. In many cases this method is instrumental in identifying targets of small molecules in cells or cell extracts. A photosensitive group is attached to the molecule of interest. Illumination with short wavelength light usually generates a sufficiently reactive intermediate, such as a nitren, a carben, or a stabilized radical intermediate. These readily react with residues in their immediate environment thereby forming a covalent bond to the binding protein. Reaction mechanisms and application examples have been thoroughly summarized previously (Dorman and Prestwich, 2000). Often the preparation of a successful photoaffinity probe is fairly difficult and labeling specificities vary due to the high reactivity of the intermediates. Very recently, solid phase-based synthetic approaches have been introduced that allow preparing an array of probes with different photosensitive groups, linkers, and protein-binding ligands (Kan et al., 2007). This might vastly improve the feasibility of future photoaffinity applications.
Translocation as a switching tool
In cells translocation is an important phenomenon. First, it is used to transport cargo from one compartment to another. For instance, importins and exportins are part of a shuttle system to exchange molecules between the cytosol and the nucleus. In addition, many proteins such as transcription factors translocate from the cytosol to one of the many cellular membranes or vice versa. Second, translocation is often used as a switch, through which an enzyme may get into the vicinity of its substrate. For instance, PI 3-kinase is required to translocate to the plasma membrane to find PIP2 which is subsequently phosphorylated to the second messenger PIP3 (Cantley, 2002). In turn, this lipid serves as an anchor for the two kinases, PDK1 and Akt. Both bind PIP3 and its metabolite PI(3,4)P2 via pleckstrin homology domains. PDK1 phosphorylates Akt and the required interaction is only possible at the two-dimensionality of the membrane. Especially when rare molecules are involved, the concentration effect at membranes over the three-dimensional cytosol or nucleosol is critical. Assuming that two reaction partners have a binding constant of 100 nM, but are only abundant in the cytosol in 10 nM concentrations, a successful reaction will be unlikely. This changes dramatically when both partners translocate to the same membrane, where the apparent concentration will be increased by at least one order of magnitude. When artificially induced at a small fraction of, e.g., early endosomes or centrosomes, this concentration effect could be used to trigger signaling events with a minimal number of molecules, just by concentrating them within a very limited area.
In order to induce translocation artificially, lipid binding domains may be used. For instance, the proteins of interest could be equipped with PI(3)P-binding FYVE domains. With the help of a photoactivatable PI(3)P derivative, translocation could be specifically induced in single endosomes within a few seconds leading to a rapid onset of protein-protein interaction. If this kind of approach was realized, it would provide photoactivation without the need to install a photosensitive group into a protein. Unfortunately, this form of photoactivation via an PI(3)P anchor will likely lead to longlasting target activation, due to the slow metabolism of PI(3)P in endosomal membranes. It would be of great interest, however, to generate activity patterns, i.e., single short ones or a number of repetitive activity spikes. For this purpose, a much more transient anchoring molecule is required. This would allow shuttling to and from membranes. Different activation patterns are expected to result in a diverse set of unique response patterns, as it has been shown in calcium signaling (Li et al., 1998). It needs to be determined whether these artificially created patterns will match endogenous signaling sequences. This brings us back to the importance of analyzing signaling events with good time resolution.
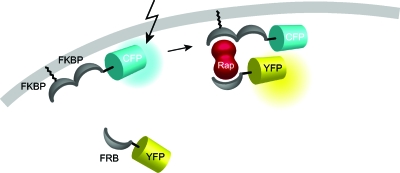
An elegant solution to rapid changes in the absence of photoactivatable groups are methods like the “chemical dimerizer strategy.” Here, a heterodimerization of protein domains that depend on the immunosuppressant rapamycin or one of its analogs is employed (Choi et al., 1996; Inoue et al., 2005). Specifically, the protein of interest, e.g., Rac1 or Cdc42, was tagged with the rapamycin-binding domain of mTor (FRB) and the membrane anchor, a transmembrane receptor, was conjugated to a FK506-binding protein (Fig. 6). Addition of rapamycin brought the GTPases to the membrane of interest (Castellano et al., 1999; Inoue et al., 2005). By a similar approach, addition of the drug translocated an enzyme connected to the cytosolic portion of the complex to the plasma membrane, where the enzyme found its substrate. In the cases published so far, enzymes that dephosphorylate phosphoinositides like PIP2 or PIP3 were as successful used as those that biosynthesize these second messenger lipids. With this method, it was shown for the first time that the opening of KCNQ ion channels is solely regulated by PIP2 levels and independent from other messengers such as calcium, diacylglycerol, Ins(1,4,5)P3, or PIP3 (Suh et al., 2006). In addition, the requirement of concerted binding of proteins with polybasic clusters to PIP2 and PIP3 could be demonstrated (Heo et al., 2006). In the future, it would be of tremendous interest to switch more than one parameter at the same time. For that purpose, more chemical dimerizers able to act independently are needed.
Figure 6. Translocation to the plasma membrane is induced by addition of the heterodimeric ligand rapamycin which ties the molecule of interest to a membrane-bound anchor.
In this example the cargo is a fluorescent protein. See Inoue et al. (2005).
Ligand-switched proteins
As discussed earlier, the use of photoactivatable compounds is particularly beneficial for fast signaling events. However, many processes in cells are sufficiently slow that cell entry of ligands targeting the protein of interest is not rate limiting. The most obvious compounds would be natural ligands such as second messengers. Unfortunately, these molecules often bind to a large number of binding proteins and hence a multitude of responses may be expected. Therefore, artificial molecules designed to bind specifically to a target proteins are preferred. Such molecules are exceedingly identified by chemical genetics procedures or from selected compound libraries, although to date many of the valuable tools still originate from natural sources. For a recent example of effective PI 3-kinase ligand discovery that is based on “principle component analysis” (see Knight et al., 2006; Knight and Shokat, 2005).
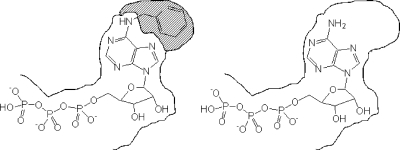
Protein specificity can also be achieved by artificially altering the protein in a way that a designed compound is necessary to induce or inhibit activity (Fig. 7). One of the first examples for engineering a protein by site-directed mutagenesis was the GTPase elongation factor Tu (EF-Tu), an enzyme responsible for loading amino-acyl tRNAs onto ribosomes (Hwang and Miller, 1987). One of the key amino acids to recognize the guanine base is 138Asp. After altering Asp to Asn, GTP was unable to bind to the enzyme, but xanthine triphosphate (XTP) was. Therefore EF-Tu could now be selectively triggered by XTP, a nucleotide with negligible affinity for common nucleotide binding sites. Intriguingly, this procedure can be used for any single GTPase as the guanine ring binding motif NKXD is conserved for all known GTPases (Bishop et al., 2000). This technique, now called orthogonal pair design or orthogonal chemical genetic approach, has since been expanded to receptors, kinases, kinesins, ion channels, and antibodies (Bishop et al., 2000; Simon and Shokat, 2007). The method is often referred to as the “hole and bump” strategy (Fig. 7).
Figure 7. In order to create a one ligand-one target situation in cells, the ligand binding pocket of an enzyme, here the ATP binding site of a kinase, is altered in a way that it preferentially accommodates a modified ligand such as a 6-substituted ATP derivative.
Due to its increased spatial demand, the latter is unable to occupy endogenous ATP binding sites (left). On the other hand, unmodified ATP has a largely reduced affinity for the widened binding site (right).
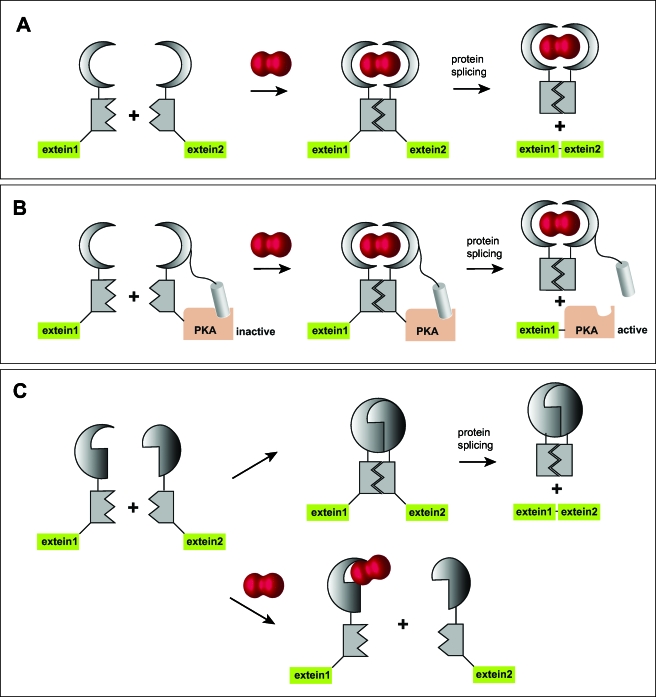
Another prominent example of small molecules being used as designed switching devices is the above-mentioned immunosuppressant rapamycin, a structurally related compound to FK506. The compound was shown to bind to the FK506-binding protein as well as another protein complex. The latter was named target of rapamycin, which is today giving name to an entire signaling cascade (Crespo and Hall, 2002). Apart from inducing translocation of the protein of interest, rapamycin-induced dimerization may also be used to switch enzyme activity. In this innovative approach, called conditional protein splicing, protein splicing via artificial inteins is used to generate a functional protein in living cells. The intein is instrumental in fusing the two halves of the protein of interest into one peptide chain. The intein itself has been artificially split into N- and C-terminal halves and will only be allowed to unite to an active intein when rapamycin is binding to both of its interaction partners, FKBP and FRB [Fig. 8A] (Mootz and Muir, 2002; Schwartz et al., 2005). Very recently, this conditional splicing system was used to generate active luciferase in Drosophila melanogaster (Schwartz et al., 2007). A similar procedure was employed to target proteins such as kinases, i.e., protein kinase A, which might not easily fold into an active conformation after protein splicing. In this approach an intrinsically attached inhibitor peptide was lost in the splicing process, leaving the kinase in the active form after addition of rapamycin [Fig. 8B]. In other words, the authors presented a triggerable kinase that could be activated with a small molecule instead of being inhibited (Mootz et al., 2004). In the future, it would be desirable to stimulate this process in the course of seconds rather than minutes. Even more intriguing would be the possibility to rapidly switch off kinase activity after the initial trigger. This would allow generating signaling patterns that are otherwise very hard to initiate and follow. The possibility to inhibit protein formation by a rapamycin-induced intein splicing event has indeed been demonstrated: a mutant of the FKBP (F36M) leads to FKBP dimers. If both parts of the split intein were tagged with this mutant, the intermolecular dimerization was constantly producing an intact intein and hence protein formation [Fig. 8C]. The presence of rapamycin disrupted dimer formation and therefore inhibited the splicing to the protein of interest (Brenzel and Mootz, 2005). Other attempts to provide controllable intein formation in living cells include temperature and small molecule-sensitive split inteins (Skretas and Wood, 2005; Zeidler et al., 2004).
Figure 8. Rapamcyin-induced protein splicing.
By using a split intein with each half fused to one of the rapamycin-binding proteins FKBP and FRB, respectively, a functional intein is generated as soon as rapamycin is present, leading to the generation of the protein of interest (a). Alternatively, an inhibitor peptide may be fused to half of the intein carrying the protein of interest, here protein kinase A. The kinase is activated as soon as rapamycin is permitting protein splicing, resulting in the separation of the inhibitor peptide and the now active kinase (b). Finally, the procedure can be reversed by using dimerizing mutants of rapamycin-binding proteins that constantly produce the protein of interest. Rapamycin addition will in this case inhibit protein synthesis (c).
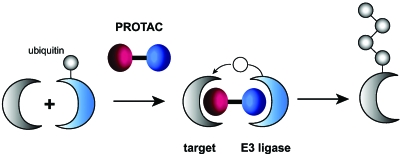
A combination of the hole and bump strategy and heterobifunctional molecules is known as PROteolysis Targeting Chimeric moleculeS (PROTACS). In this approach, the gene of interest is eliminated by small molecule-induced proteolysis (Fig. 9). Targeted proteolytic degradation is complementary to genetic knockouts and RNAi knockdowns on the protein level. The necessary ubiquitination of the target protein is induced targeting a modified E3 ubiquitin ligase and the E2 ubiquitin transfer enzyme by a small molecule to form a complex. The modification at the E3 ligase permits binding to one part of the bifunctional small molecule ligand, whereas the other part is recognized by the target protein. After initial in vitro studies for proof of principle (Sakamoto et al., 2001, 2003), cell permeable PROTACs were developed (Schneekloth et al., 2004). The target (EGFP) was fused to the F36V mutation of FK506 binding protein (FKBP12). This mutation creates the “hole” which is recognized by the artificial ligand AP21998. This ligand was then equipped with a seven amino acid sequence, including a hydroxylatable proline, to be accepted by the E3 recognition domain. Finally, a short polyarginine sequence was added to permit passive cell entry. Proteolysis could be monitored in the presence of 25 μM heterodimeric ligand by a loss of fluorescence in living cells. Due to the high specificity of the approach, it is likely that several degradation processes may be performed simultaneously or that certain patterns of protein (dis)appearance could be generated.
Figure 9. The principle of the PROTAC approach.
Targeted degradation of proteins is achieved by attracting an E3 ubiquitin ligase through a heterobifunctional molecule, the so-called PROteolysis Targeting Chimeric molecule (PROTAC)
Finally, in the past decade a large number of approaches have been developed that interfere with gene expression on the mRNA level. Targeted mRNA destruction or silencing can be achieved by using a variety of complementary oligonucleotides such as antisense oligodeoxynucleotides, chemically modified oligonucleotides, ribozymes, DNAzymes, or short interfering RNAs (siRNAs). Especially the latter has become extremely popular due to its simple design and preparation. An additional class of oligonucleotide molecules targeting genes on the protein level are aptamers. Aptamers are short single-stranded oligonucleotides that fold into a distinct three-dimensional structure. They bind their targets with high affinity by complementary shape interactions (Famulok and Mayer, 2005; Famulok et al., 2000). The high specificity is generated by using combinatorial sequence libraries and a method called SELEX (systematic evolution of ligands by exponential enrichment), an in vitro selection process. Any molecular structure of interest might be a target, even entire parasites (Goringer et al., 2003). When combined with gold nanoparticles, very effective sensors for small or large molecules can be formed, with the aptamer being the sensing unit (Liu and Lu, 2006; Liu et al., 2006).
Although originally aptamers were used to target extracellular proteins, more recently, major focus is given to aptamers that inhibit intracellular targets, so-called intramers. Intramers can be introduced to cells by transfection methods or by particular intramer expression systems. In fact, once inside cells intramers are fairly stable against endogenous nucleases (Thesis et al., 2004). Among other applications, they have been employed to downregulate proteins crucial to virus development and the elucidation of signaling pathways (Famulok and Mayer, 2005). As such they represent an alternative approach to siRNAs. In addition, aptamer-target complexes are used as part of screening assays to identify small molecule binders to the target (Hartig and Famulok, 2002; Hartig et al., 2002; Mayer and Famulok, 2006).
Membrane permeability
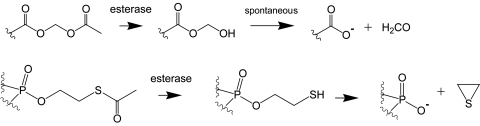
It is obvious that both sensors and modulators are only able to exhibit the desired activity when they can reach the intracellular target sites. Usually this demands that the import happens via diffusion rather than endocytosis, because endocytosed molecules often stay in endosomal structures and are not released to the cytosol. The ideal intracellularly working compound is lipophilic enough to pass the plasma membrane. However, these compounds are usually able to leave cells as quickly as they enter. Functional groups that prevent cell penetration most efficiently are negatively charged groups. Due to the negative potential of the plasma membrane maintained by most cells (from −50 to −70 mV), negatively charged compounds are literately repelled from crossing membranes, while partially positively charged molecules are more likely to penetrate. Medicinal chemists therefore developed bioactivatable protecting groups such as alkyoxymethyl esters and S-alkylthioethyl esters, just to name a couple (Friis and Bundgaard, 1996; Peyrottes et al., 2004; Schultz, 2003). The resulting protected molecules are usually referred to as prodrugs or “Trojan horse” compounds. The protecting groups mask charged residues such as carboxylic acid groups or phosphates in a way that the compound of interest is neutral and hence able to pass membranes. Once inside cells, endogenous enzymes, mostly esterases and lipases, remove the groups thereby restoring biological activity. Since the hydrophilic groups are now deprotected, the compounds are impermeant again and are trapped in cells. Therefore these prodrugs potentially lead to an accumulation of the biologically active species. This of course depends largely on the metabolism of the modulator or sensor. The most commonly used groups for negatively charged moieties are alkyloxymethyl esters, especially acetoxymethyl and pivaloxymethyl esters, and S-alkyloxythioethyl esters. Upon enzymatic cleavage they release carboxylic acid or phosphate and the spacer to the enzyme-sensitive protected group. In case of the alkyloxymethyl esters this is formaldehyde, for S-alkyloxythioethyl esters it is thiirane (Fig. 10). Most work so far has been done on ion chelators (Grynkiewicz et al., 1985; Tsien, 1981), nucleotides (Friis and Bundgaard, 1996; Peyrottes et al., 2004; Schultz, 2003; Schultz et al., 1994, 1993), inositol phosphates (Li et al., 1998; Rudolf et al., 2003; Vajanaphanich et al., 1994), phosphoinositides (Dinkel et al., 2001; Jiang et al., 1998), and other lipid derivatives (Dinkel et al., 2003). In addition, some small molecule sensor molecules (Wichmann et al., 2006; Zlokarnik et al., 1998) but also some commercial drugs rely on this technique (Krapcho et al., 1988; Starret et al., 1994; von Daehne et al., 1970).
Figure 10. Bioactivatable protecting groups release negatively charged residues upon enzymatic hydrolysis.
Both, acetoxymethyl (AM) esters and S-acetylthioethyl (SATE) esters are suitable for masking carboxylic acids and phosphates.
An alternative to masking each negatively charged residue is the use of peptides with multiple positive charges, also called polybasic peptides. These are known to induce their penetration into cells in a receptor-independent way and can do so also when cargo is added (Joliot and Prochiantz, 2004). The most commonly used cell-permeant peptides are the HIV Tat transactivator and the third α-helix of the homeodomain of the Drosophila melanogaster transcription factor Antennapedia (Antp). The mechanism of penetration is usually not fully understood, but recently a study showed that apart from a minimum number of positive charges to interact with the phospholipid surface, certain aromatic residues, in particular tryptophans, are essential for interaction with the hydrophobic core of the membrane (Zhang and Smith, 2005). Another study stresses the presence of heparin sulfate as a crucial docking point on the cell surface (Ziegler et al., 2005). In some cases, for instance for the delivery of adenovirus to COS-7 cells, a covalent connection of cell-permeant peptide and cargo is not required. This means that the peptides work essentially as a transfection agent, which requires polybasic peptide concentrations above 100 μM. In general, the addition of a stretch of seven or eight positively charged amino acids suffices to generate a penetrating entity. This has been very smartly used in substrates for matrix metalloproteases (MMPs), where a substrate loop for these enzymes links a polybasic and a corresponding acidic stretch of amino acids. Upon MMP cleavage the polybasic part is able to enter cells. By attaching a cytotoxic compound to this sequence, targeted toxicity against cells secreting MMPs, i.e., cancer cells, can be achieved (Jiang et al., 2004). In other cases, it is necessary to synthesize a polybasic conjugate. This is now achievable by reacting a cysteine-bearing cargo with a mercapto-pyridine-activated cell-penetrating peptide forming a disulfide bridge. The latter is usually fairly stable outside cells but is readily opened in the reducing environment of the cell’s interior thereby releasing the cargo to the cytosol (Nguyen et al., 2004; Yudushkin et al., 2007).
CONCLUSIONS
Despite the enormous progress possible after unraveling several genomes and the introduction of green fluorescent protein, we are still witnessing the initial phase of intracellular network analysis. There is an urgent need for more quantitative data. The prerequisite for this is that microscopy techniques such as FLIM and FCS will become standard tools in the biology lab. These will only be available with more tools and more sophisticated functionally tailored molecules. Genome-wide analysis of protein function in living cells, for instance via high throughput RNAi screens (Neumann et al., 2006; Pepperkok and Ellenberg, 2006), will significantly increase the assignment of protein function in the near future. As a consequence, the need for suitable reporters and modulators will dramatically increase. In fact, these tools are likely to be sufficiently essential for systems biology, that their preparation will become a bottleneck for the progress in the field. Another bottleneck is data handling and storage, but it is more likely that these needs will be met sooner than later. It is therefore foreseeable that high throughput platforms for both, reporters and modulators, will be essential. High throughput screening (HTS) of small molecule libraries of course provides inhibitors and sometimes activators of proteins. Unlike in pharmaceutical research, the resulting compounds usually do not need to act in the lower nanomolar range. Therefore, today HTS already meets the needs of bottom-up systems biology to a certain degree. As discussed above, however, the ideal modulator needs to act within a fraction of a second. Again chemistry will be needed to render the modulators rapidly membrane-permeant and∕or photoactivatable. On the reporter molecule level, we are far from high throughput preparations. Therefore, additional efforts are needed, both on the molecular biology and the chemistry level. Progress will strongly depend on the instrumentation available. The data quality, for instance in imaging experiments, will determine the success of any large-scale experiment. New developments from the physics departments in microscopy and automation are indispensable for further advancement. Finally, we will see data handling and bioinformatics taking center stage in the near future. Amongst these will be modeling approaches that will hopefully produce useful approximations for describing cellular processes in time and space.
ACKNOWLEDGMENTS
The author would like to thank Petra Riedinger for drawing most of the figures. The author received research funding from the EMBL, the EU (LSHG-CT-2003-503259), the Volkswagenstiftung, the Human Frontiers Science Programme, and SB Cancer, a member of the Systems Biology Initiative of the Helmholtz Association, Germany. Key disciplines relevant to this article are organic chemistry, biophysics, cell biology, chemical biology, molecular biology, and molecular medicine.
References
- Aarhus, R, Gee, K, and Lee, H C (1995). “Caged cyclic ADP-ribose—synthesis and use.” J. Biol. Chem. 10.1074/jbc.270.13.7745 270, 7745–7749. [DOI] [PubMed] [Google Scholar]
- Adams, S R, Harootunian, A T, Buechler, Y J, Taylor, S S, and Tsien, R Y (1991). “Fluorescence ratio imaging of cyclic-amp in single cells.” Nature (London) 10.1038/349694a0 349, 694–697. [DOI] [PubMed] [Google Scholar]
- Ai, H W, Shaner, N C, Cheng, Z H, Tsien, R Y, and Campbell, R E (2007). “Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins.” Biochemistry 10.1021/bi700199g 46, 5904–5910. [DOI] [PubMed] [Google Scholar]
- Ando, R, Flors, C, Mizuno, H, Hofkens, J, and Miyawaki, A (2007). “Highlighted generation of fluorescence signals using simultaneous two-color irradiation on Dronpa mutants.” Biophys. J. 10.1529/biophysj.107.105882 92, L97–L99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, R, Hama, H, Yamamoto-Hino, M, Mizuno, H, and Miyawaki, A (2002). “An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.202320599 99, 12651–12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, R, Mizuno, H, and Miyawaki, A (2004). “Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting.” Science 10.1126/science.1102506 306, 1370–1373. [DOI] [PubMed] [Google Scholar]
- Andresen, M et al. (2005). “Structure and mechanism of the reversible photoswitch of a fluorescent protein.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0502772102 102, 13070–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman, M N, Furlong, E EM, Imam, F, Johnson, E, Null, B H, Baker, B S, Krasnow, M A, Scott, M P, Davis, R W, and White, K P (2002). “Gene expression during the life cycle of Drosophila melanogaster.” Science 10.1126/science.1072152 297, 2270–2275. [DOI] [PubMed] [Google Scholar]
- Balla, T, Bondeva, T, and Varnai, P (2000). “How accurately can we image inositol lipids in living cells?” Trends Pharmacol. Sci. 10.1016/S0165-6147(00)01500-5 21, 238–241. [DOI] [PubMed] [Google Scholar]
- Baruch, A, Jeffery, D A, and Bogyo, M (2004). “Enzyme activity—it’s all about image.” Trends Cell Biol. 10.1016/j.tcb.2003.11.002 14, 29–35. [DOI] [PubMed] [Google Scholar]
- Bell, A F, Stoner-Ma, D, Wachter, R M, and Tonge, P J (2003). “Light-driven decarboxylation of wild-type green fluorescent protein.” J. Am. Chem. Soc. 10.1021/ja034588w 125, 6919–6926. [DOI] [PubMed] [Google Scholar]
- Belousov, V V, Fradkov, A F, Lukyanov, K A, Staroverov, D B, Shakhbazov, K S, Terskikh, A V, and Lukyanov, S (2006). “Genetically encoded fluorescent indicator for intracellular hydrogen peroxide.” Nat. Methods 3, 281–286. [DOI] [PubMed] [Google Scholar]
- Birbach, A, Bailey, S T, Ghosh, S, and Schmid, J A (2004). “Cytosolic, nuclear and nucleolar localization signals determine subcellular distribution and activity of the NF-kappa B inducing kinase NIK.” J. Cell. Sci. 10.1242/jcs.01224 117, 3615–3624. [DOI] [PubMed] [Google Scholar]
- Bishop, A et al. (2000). “Unnatural ligands for engineered proteins: new tools for chemical genetics.” Annu. Rev. Biophys. Biomol. Struct. 10.1146/annurev.biophys.29.1.577 29, 577–606. [DOI] [PubMed] [Google Scholar]
- Blum, G, Mullins, S R, Keren, K, Fonovic, M, Jedeszko, C, Rice, M J, Sloane, B F, and Bogyo, M (2005). “Dynamic imaging of protease activity with fluorescently quenched activity-based probes.” Nat. Chem. Biol. 10.1038/nchembio728 1, 203–209. [DOI] [PubMed] [Google Scholar]
- Breinbauer, R, Hillisch, A, and Waldmann, H (2007). “Reverse chemical genetics—an important strategy for the study of protein function in chemical biology and drug discovery.” In Chemical Biology, Schreiber, S L, Kapoor, T, and Wess, G (eds), vol. 1, pp 355–384, Wiley-VCH, Weinheim. [Google Scholar]
- Breinbauer, R, Vetter, I R, and Waldmann, H (2002). “From protein domains to drug candidates—natural products as guiding principles in the design and synthesis of compound libraries.” Angew. Chem., Int. Ed. 41, 2879–2890. [DOI] [PubMed] [Google Scholar]
- Brenzel, S, and Mootz, H D (2005). “Design of an intein that can be inhibited with a small molecule ligand.” J. Am. Chem. Soc. 10.1021/ja043501j 127, 4176–4177. [DOI] [PubMed] [Google Scholar]
- Brumbaugh, J, Schleifenbaum, A, Gasch, A, Sattler, M, and Schultz, C (2006). “A dual parameter FRET probe for measuring PKC and PKA activity in living cells.” J. Am. Chem. Soc. 10.1021/ja0562200 128, 24–25. [DOI] [PubMed] [Google Scholar]
- Cantley, L C (2002). “The phosphoinositide 3-kinase pathway.” Science 10.1126/science.296.5573.1655 296, 1655–1657. [DOI] [PubMed] [Google Scholar]
- Castellano, F, Montcourrier, P, Guillemot, J C, Gouin, E, Machesky, L, Cossart, P, and Chavrier, P (1999). “Inducible recruitment of Cdc42 or WASP to a cell-surface receptor triggers actin polymerization and filopodium formation.” Curr. Biol. 10.1016/S0960-9822(99)80161-4 9, 351–360. [DOI] [PubMed] [Google Scholar]
- Chang, C Y, Fernandez, T, Panchal, R, and Bayley, H (1998). “Caged catalytic subunit of cAMP-dependent protein kinase.” J. Am. Chem. Soc. 10.1021/ja981649v 120, 7661–7662. [DOI] [Google Scholar]
- Chang, C Y, Niblack, B, Walker, B, and Bayley, H (1995). “A photogenerated pore-forming protein.” Chem. Biol. 10.1016/1074-5521(95)90220-1 2, 391–400. [DOI] [PubMed] [Google Scholar]
- Chen, I, and Ting, A Y (2005). “Site-specific labeling of proteins with small molecules in living cells.” Curr. Opin. Biotechnol. 10.1016/j.copbio.2004.12.003 16, 35–40. [DOI] [PubMed] [Google Scholar]
- Choi, J W, Chen, J, Schreiber, S L, and Clardy, J (1996). “Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP.” Science 10.1126/science.273.5272.239 273, 239–242. [DOI] [PubMed] [Google Scholar]
- Chudakov, D M, Belousov, V V, Zaraisky, A G, Novoselov, V, Staroverov, D B, Zorov, D B, Lukyanov, S, and Lukyanov, K A (2003). “Kindling fluorescent proteins for precise in vivo photolabeling (vol 21, pg 191, 2003).” Nat. Biotechnol. 10.1038/nbt778 21, 452–452. [DOI] [PubMed] [Google Scholar]
- Chudakov, D M, Chepurnykh, T V, Belousov, V V, Lukyanov, S, and Lukyanov, K A (2006). “Fast and precise protein tracking using repeated reversible photoactivation.” Traffic (Oxford, U. K.) 10.1111/j.1600-0854.2006.00468.x 7, 1304–1310. [DOI] [PubMed] [Google Scholar]
- Chudakov, D M, Lukyanov, S, and Lukyanov, K A (2005). “Fluorescent proteins as a toolkit for in vivo imaging.” Trends Biotechnol. 10.1016/j.tibtech.2005.10.00523, 605–613. [DOI] [PubMed] [Google Scholar]
- Cook, S N, Jack, W E, Xiong, X, Danley, L E, Ellman, J A, Schultz, P G, and Noren, C J (1995). “Photochemically initiated protein splicing.” Angew. Chem., Int. Ed. Engl. 10.1002/anie.199516291 34, 1629–1630. [DOI] [Google Scholar]
- Crespo, J L, and Hall, M N (2002). “Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae.” Microbiol. Mol. Biol. Rev. 10.1128/MMBR.66.4.579-591.2002 66, 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel, C, and Schultz, C (2003). “Synthesis of caged myo-inositol 1,3,4,5-tetrakisphosphate.” Tetrahedron Lett. 10.1016/S0040-4039(02)02834-4 44, 1157–1159. [DOI] [Google Scholar]
- Dinkel, C, Wichmann, O, and Schultz, C (2003). “Versatile reagents to introduce caged phosphates.” Tetrahedron Lett. 10.1016/S0040-4039(02)02832-0 44, 1153–1155. [DOI] [Google Scholar]
- Dinkel, C J, Moody, M, Traynor-Kaplan, A E, and Schultz, C (2001). “Membrane-permeant 3-OH-phosphorylated phosphoinositide derivatives.” Angew. Chem., Int. Ed. 40, 3004–3008. [DOI] [PubMed] [Google Scholar]
- Dorman, G, and Prestwich, G D (2000). “Using photolabile ligands in drug discovery and development.” Trends Biotechnol. 10.1016/S0167-7799(99)01402-X 18, 64–77. [DOI] [PubMed] [Google Scholar]
- Eckardt, T, Hagen, V, Schade, B, Schmidt, R, Schweitzer, C, and Bendig, J (2002). “Deactivation behavior and excited-state properties of (coumarin-4-yl)methyl derivatives. 2. photocleavage of selected (coumarin-4-yl)methyl-caged adenosine cyclic 3′,5′-monophosphates with fluorescence enhancement.” J. Org. Chem. 10.1021/jo010692p 67, 703–710. [DOI] [PubMed] [Google Scholar]
- Endo, M, Nakayama, K, Kaida, Y, and Majima, T (2004a). “Design and synthesis of photochemically controllable caspase-3.” Angew. Chem., Int. Ed. 10.1002/anie.200460889 43, 5643–5645. [DOI] [PubMed] [Google Scholar]
- Endo, M, Nakayama, K, and Majima, T (2004b). “Design and synthesis of photochemically controllable restriction endonuclease BamHI by manipulating the salt-bridge network in the dimer interface.” J. Org. Chem. 10.1021/jo035774n 69, 4292–4298. [DOI] [PubMed] [Google Scholar]
- Famulok, M, and Mayer, G (2005). “Intramers and aptamers: applications in protein-function analyses and potential for drug screening.” ChemBioChem 10.1002/cbic.200400299 6, 19–26. [DOI] [PubMed] [Google Scholar]
- Famulok, M, Mayer, G, and Blind, M (2000). “Nucleic acid aptamers—from selection in vitro to applications in vivo.” Acc. Chem. Res. 10.1021/ar960167q 33, 591–599. [DOI] [PubMed] [Google Scholar]
- Fedoryak, O D, Sul, J Y, Haydon, P G, and Ellis-Davies, G CR (2005). “Synthesis of a caged glutamate for efficient one- and two-photon photorelease on living cells.” Chem. Commun. (Cambridge) , 3664–3666. [DOI] [PubMed] [Google Scholar]
- Fehr, M, Frommer, W B, and Lalonde, S (2002). “Visualization of maltose uptake in living yeast cells by fluorescent nanosensors.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.142089199 99, 9846–9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, M, Lalonde, S, Lager, I, Wolff, M W, and Frommer, W B (2003). “In vivo imaging of the dynamics of glucose uptake in the cytosol of COS-7 cells by fluorescent nanosensors.” J. Biol. Chem. 10.1074/jbc.M301333200 278, 19127–19133. [DOI] [PubMed] [Google Scholar]
- Fehr, M, Takanaga, H, Ehrhardt, D W, and Frommer, W B (2005). “Evidence for high-capacity bidirectional glucose transport across the emdoplasmatic reticulum membrane by genetically encoded fluorescence resonance energy transfer nanosensors.” Mol. Cell. Biol. 10.1128/MCB.25.24.11102-11112.2005 25, 11102–11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, R, Fotin-Mleczek, M, Hufnagel, H, and Brock, R (2005). “Break on through to the other side—Biophysics and cell biology shed light on cell-penetrating peptides.” ChemBioChem 10.1002/cbic.200500044 6, 2126–2142. [DOI] [PubMed] [Google Scholar]
- Fonovic, M, and Bogyo, M (2007). “Activity based probes for proteases: applications to biomarker discovery, molecular imaging and drug screening.” Curr. Pharmaceut. Design 13, 253–261. [DOI] [PubMed] [Google Scholar]
- Frankel, A D, and Pabo, C O (1988). “Fingering too many proteins.” Cell 10.1016/0092-8674(88)90083-9 53, 675–675. [DOI] [PubMed] [Google Scholar]
- Friis, G J, and Bundgaard, H (1996). “Prodrugs of phosphates and phosphonates: novel lipophilic alpha-acyloxyalkyl ester derivatives of phosphate- or phosphonate containing drugs masking the negative charges of these groups.” Eur. J. Pharm. Sci. 4, 49–59. [Google Scholar]
- Furuta, T, Wang, S SH, Dantzker, J L, Dore, T M, Bybee, W J, Callaway, E M, Denk, W, and Tsien, R Y (1999). “Brominated 7-hydroxycoumarin-4-ylmethyls: photolabile protecting groups with biologically useful cross-sections for two photon photolysis.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.96.4.1193 96, 1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke, V, Creutz, C E, and Moss, S E (2005). “Annexins: linking Ca2+ signalling to membrane dynamics.” Nat. Rev. Mol. Cell Biol. 6, 449–461. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami, S, Huh, W, Bower, K, Howson, R W, Belle, A, Dephoure, N, O’Shea, E K, and Weissman, J S (2003). “Global analysis of protein expression in yeast.” Nature (London) 10.1038/nature02046 425, 737–741. [DOI] [PubMed] [Google Scholar]
- Ghosh, M, Song, X, Mouneimne, G, Sidani, M, Lawrence, D S, and Condeelis, J S (2004). “Cofilin promotes actin polymerization and defines the direction of cell motility.” Science 10.1126/science.1094561 304, 743–746. [DOI] [PubMed] [Google Scholar]
- Giepmans, B NG, Adams, S R, Ellisman, M H, and Tsien, R Y (2006). “The fluorescent toolbox for assessing protein location and function.” Science 10.1126/science.1124618 312, 217–224. [DOI] [PubMed] [Google Scholar]
- Giordano, A, Zarbakhsh, S, and Schultz, C (2007). “Controlling protein function by caged compounds.” In Chemical Biology, Schreiber, S L, Kapoor, T and Wess, G (eds), vol. 1, pp 140–173, VCH, Weinheim. [Google Scholar]
- Goeldner, M, and Givens, R SE (2005). Dynamic Studies in Biology, Wiley-VCH, Weinheim. [Google Scholar]
- Goringer, H U, Homann, M, and Lorger, M (2003). “In vitro selection of high-affinity nucleic acid ligands to parasite target molecules.” Int. J. Parasitol. 33, 1309–1317. [DOI] [PubMed] [Google Scholar]
- Gough, J D, and Crews, C M (2007). “Using natural products to unravel cell biology.” In Chemical Biology, Schreiber, S L, Kapoor, T, and Wess, G (eds), vol. 1, pp 95-114, Wiley-VCH, Weinheim. [Google Scholar]
- Gratton, J P, Yu, J, Griffith, J W, Babbitt, R W, Scotland, R S, Hickey, R, Giordano, F J, and Sessa, W C (2003). “Cell-permeable peptides improve cellular uptake and therapeutic gene delivery of replication-deficient viruses in cells and in vivo.” Nat. Med. 9, 357–362. [DOI] [PubMed] [Google Scholar]
- Gray, A, Olsson, H, Batty, I H, Priganica, L, and Downes, P C (2003). “Nonradioactive methods for the assay of phosphoinositide 3-kinases and phosphoinositide phosphatases and selective detection of signaling lipids in cell and tissue extracts.” Anal. Biochem. 10.1016/S0003-2697(02)00607-3 313, 234–245. [DOI] [PubMed] [Google Scholar]
- Green, M, Loewenstein, P M, Pusztai, R, and Symington, J S (1988). “An adenovirus E1a-protein domain activates transcription in vivo and in vitro in the absence of protein synthesis.” Cell 10.1016/S0092-8674(88)90429-1 53, 921–926. [DOI] [PubMed] [Google Scholar]
- Gronemeyer, T, Godin, G, and Johnsson, K (2005). “Added value to fusion proteins through covalent labelling.” Curr. Opin. Biotechnol. 10.1016/j.copbio.2005.06.001 16, 453–458. [DOI] [PubMed] [Google Scholar]
- Grynkiewicz, G, Poenie, M, and Tsien, R Y (1985). “A new generation of Ca2+ indicators with greatly improved fluorescence properties.” J. Biol. Chem. 260, 3440–3450. [PubMed] [Google Scholar]
- Gu, H, Lalonde, S, Okumoto, S, Looger, L L, Scharff-Poulsen, A M, Grossman, A R, Kossmann, J, Jakobsen, I, and Frommer, W B (2006). “A novel analytical method for in vivo phosphate tracking.” FEBS Lett. 10.1016/j.febslet.2006.09.048 580, 5885–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, V, Bendig, J, Frings, S, Eckardt, T, Helm, S, Reuter, D, and Kaupp, U B (2001). “Highly efficient and ultrafast phototriggers for cAMP and cGMP by using long-wavelength UV/vis-activation.” Angew. Chem., Int. Ed. 40, 1045–1048. [DOI] [PubMed] [Google Scholar]
- Hagen, V, Frings, S, Bendig, J, Lorenz, D, Wiesner, B, and Kaupp, U B (2002). “Fluorescence spectroscopic quantification of the release of cyclic nucleotides from photocleavable [bis(carboxymethoxy)coumarin-4-yl]methyl esters inside cells.” Angew. Chem., Int. Ed. 41, 3625–3628. [DOI] [PubMed] [Google Scholar]
- Hagen, V, Frings, S, Wiesner, B, Helm, S, Kaupp, U B, and Bendig, J (2003). “[7-(dialkylamino)coumarin-4-yl]methyl-caged compounds as ultrafast and effective long-wavelength phototriggers of 8-bromo-substituted cyclic nucleotides.” ChemBioChem 10.1002/cbic.200300561 4, 434–442. [DOI] [PubMed] [Google Scholar]
- Haggarty, S J, and Schreiber, S L (2007). “Forward chemical genetics.” In Chemical Biology, Schreiber, S L, Kapoor, T, and Wess, G (eds), vol. 1, pp 299–354, Wiley-VCH, Weinheim. [Google Scholar]
- Hahn, M E, and Muir, T W (2004). “Photocontrol of Smad2, a multiphosphorylated cell-signaling protein, through caging of activating phosphoserines.” Angew. Chem., Int. Ed. 10.1002/anie.200461141 43, 5800–5803. [DOI] [PubMed] [Google Scholar]
- Halet, G (2005). “Imaging phosphoinositide dynamics using GFP-tagged protein domains.” Biol. Cell 97, 501–518. [DOI] [PubMed] [Google Scholar]
- Hannon, G J (2002). “RNA interference.” Nature (London) 10.1038/418244a 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Hartig, J S, and Famulok, M (2002). “Reporter ribozymes for real-time analysis of domain-specific interactions in biomolecules: HIV-1 reverse transcriptase and the primer-template complex.” Angew. Chem., Int. Ed. 41, 4263–4266. [DOI] [PubMed] [Google Scholar]
- Hartig, J S, Najafi-Shoushtari, S H, Grune, I, Yan, A, Ellington, A D, and Famulok, M (2002). “Protein-dependent ribozymes report molecular interactions in real time.” Nat. Biotechnol. 10.1038/nbt0702-717 20, 717–722. [DOI] [PubMed] [Google Scholar]
- Haustein, E, and Schwille, P (2007) “Fluorescence correlation spectroscopy: novel variations of an established technique.” Annu. Rev. Biophys. Biomol. Struct. 10.1146/annurev.biophys.36.040306.132612 36, 151–169. [DOI] [PubMed] [Google Scholar]
- Heo, W D, Inoue, T, Park, W S, Kim, M L, Park, B O, Wandless, T J, and Meyer, T (2006). “PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane.” Science 10.1126/science.1134389 314, 1458–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, A, Adams, S R, Sawyer, C L, Lev-Ram, V, Tsien, R Y, and Dostmann, W RG (2001). “Spatiotemporal dynamics of guanosine 3′,5′-cyclic monophosphate revealed by a genetically encoded, fluorescent indicator.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.051631298 98, 2437–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B, Wu, H K, Bhaya, D, Grossman, A, Granier, S, Kobilka, B K, and Zare, R N (2007). “Counting low-copy number proteins in a single cell.” Science 10.1126/science.1133992 315, 81–84. [DOI] [PubMed] [Google Scholar]
- Huh, W K, Falvo, J V, Gerke, L C, Carroll, A S, Howson, R W, Weissman, J S, and O’Shea, E K (2003). “Global analysis of protein localization in budding yeast.” Nature (London) 10.1038/nature02026 425, 686–691. [DOI] [PubMed] [Google Scholar]
- Hwang, Y W and Miller, D L (1987). “A mutation that alters the nucleotide specificity of elongation factor-Tu, a Gtp regulatory protein.” J. Biol. Chem. 262, 13081–13085. [PubMed] [Google Scholar]
- Inoue, T, Do Heo, W, Grimley, J S, Wandless, T J, and Meyer, T (2005). “An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways.” Nat. Methods 2, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, V R et al. (1999). “The transcriptional program in the response of human fibroblasts to serum.” Science 10.1126/science.283.5398.83 283, 83–87. [DOI] [PubMed] [Google Scholar]
- Jamieson, T, Bakhshi, R, Petrova, D, Pocock, R, Imani, M, Seifalian, A M (2007). “Biological applications of quantum dots.” Biomaterials 10.1016/j.biomaterials.2007.07.014 28, 4717–4732. [DOI] [PubMed] [Google Scholar]
- Janetopoulos, C, Jin, T, and Devreotes, P (2001). “Receptor-mediated activation of heterotrimeric G-proteins in living cells.” Science 10.1126/science.1055835 291, 2408–2411. [DOI] [PubMed] [Google Scholar]
- Jares-Erijman, E A, and Jovin, T M (2003). “FRET imaging.” Nat. Biotechnol. 10.1038/nbt896 21, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Jares-Erijman, E A, and Jovin, T M (2006). “Imaging molecular interactions in living cells by FRET microscopy.” Curr. Opin. Chem. Biol. 10.1016/j.cbpa.2006.08.021 10, 409–416. [DOI] [PubMed] [Google Scholar]
- Jiang, T, Olson, E, Nguyen, Q, Roy, M, Jennings, P A, and Tsien, R (2004). “Tumor imaging by means of proteolytic activation of cell-penetrating peptides.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0408191101 101, 17867–17872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, T, Sweetney, G, Rudolf, M T, Klip, A, Traynor-Kaplan, A, and Tsien, R Y (1998). “Membrane-permeant esters of phosphatidylinositol 3,4,5-trisphosphate.” J. Biol. Chem. 18, 11017–11024. [DOI] [PubMed] [Google Scholar]
- Johnsson, N, and Johnsson, K (2007). “Chemical tools for biomolecular imaging.” ACS Chem. Biol. 2, 31–38. [DOI] [PubMed] [Google Scholar]
- Joliot, A, and Prochiantz, A (2004). “Transduction peptides: from technology to physiology.” Nat. Cell Biol. 10.1038/ncb0304-189 6, 189–196. [DOI] [PubMed] [Google Scholar]
- Kaláb, P, Pralle, A, Isacoff, E Y, Heald, R, and Weis, K (2006). “Analysis of a RanGTP-regulated gradient in mitotic somatic cells.” Nature (London) 10.1038/nature04589 440, 697–701. [DOI] [PubMed] [Google Scholar]
- Kan, T, Kita, Y, Morohashi, Y, Tominari, Y, Hosoda, S, Tomita, T, Natsugari, H, Iwatsubo, T, and Fukuyama, T (2007). “Convenient synthesis of photoaffinity probes and evaluation of their labeling abilities.” Org. Lett. 10.1021/ol070376i 9, 2055–2058. [DOI] [PubMed] [Google Scholar]
- Keller, P, Toomre, D, Diaz, E, White, J, and Simons, K (2001). “Multicolour imaging of post-Golgi sorting and trafficking in live cells.” Nat. Cell Biol. 10.1038/35055042 3, 140–149. [DOI] [PubMed] [Google Scholar]
- Knight, Z A et al. (2006). “A pharmacological map of the PI3-K family defines a role for p110 alpha in insulin signaling.” Cell 10.1016/j.cell.2006.09.018 127, 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, Z A, and Shokat, K M (2005). “Features of selective kinase inhibitors.” Chem. Biol. 10.1016/j.chembiol.2005.04.011 12, 621–637. [DOI] [PubMed] [Google Scholar]
- Komoriya, A, Packard, B Z, Brown, M J, Wu, M L, and Henkart, P A (2000). “Assessment of caspase activities in intact apoptotic thymocytes using cell-permeable fluorogenic caspase substrates.” J. Exp. Med. 191, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapcho, J et al. (1988). “Angiotensin-converting enzyme-inhibitors - mercaptan, carboxyalkyl dipeptide, and phosphinic acid inhibitors incorporating 4-substituted prolines.” J. Med. Chem. 31, 1148–1160. [DOI] [PubMed] [Google Scholar]
- Kretzschmar, A K, Dinger, M C, Henze, C, Brocke-Heidrich, K, and Horn, F (2004). “Analysis of Stat3 (signal transducer and activator of transcription 3) dimerization by fluorescence resonance energy transfer in living cells.” Biochem. J. 10.1042/BJ20030708 377, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, M T, Ni, Q, Tsien, R Y, Zhang, J, and Newton, A C (2005). “Spatio-temporal dynamics of protein kinase B∕Akt signaling revealed by a genetically encoded fluorescent reporter.” J. Chem. Biol. 280, 5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lager, I, Fehr, M, Frommer, W B, and Lalonde, S W (2003). “Development of a fluorescent nanosensor for ribose.” FEBS Lett. 553, 85–89. [DOI] [PubMed] [Google Scholar]
- Lee, J S, Kim, J H, Jang, I H, Kim, H S, Han, J M, Kazlauskas, A, Yagisawa, H, Suh, P G, and Ryu, S H (2005). “Phosphatidylinositol (3,4,5)-trisphosphate specifically interacts with the phox homology domain of phospholipase D1 and stimulates its activity.” J. Cell. Sci. 10.1242/jcs.02564 118, 4405–4413. [DOI] [PubMed] [Google Scholar]
- Li, W H, Llopis, J, Whitney, M, Zlokarnik, G, and Tsien, R Y. (1998). “Cell-permeant caged InsP(3) ester shows that Ca2+ spike frequency can optimize gene expression.” Nature (London) 10.1038/31965 392, 936–941. [DOI] [PubMed] [Google Scholar]
- Li, W H, Schultz, C, Llopis, J, and Tsien, R Y (1997). “Membrane-permeant esters of inositol polyphosphates, chemical syntheses and biological applications.” Tetrahedron 10.1016/S0040-4020(97)00714-X 53, 12017–12040. [DOI] [Google Scholar]
- Lin, C W, Jao, C Y, and Ting, A Y (2004). “Genetically encoded fluorescent reporters of histone methylation in living cells.” J. Am. Chem. Soc. 10.1021/ja038854h 126, 5982–5983. [DOI] [PubMed] [Google Scholar]
- Lin, C W, and Ting, A Y (2004). “A genetically encoded fluorescent reporter of histone phosphorylation in living cells.” Angew. Chem., Int. Ed. 10.1002/anie.200353375 43, 2940–2943. [DOI] [PubMed] [Google Scholar]
- Liu, J W, and Lu, Y (2006). “Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles.” Angew. Chem., Int. Ed. 10.1002/anie.200502589 45, 90–94. [DOI] [PubMed] [Google Scholar]
- Liu, J W, Mazumdar, D, and Lu, Y (2006). “A simple and sensitive “dipstick” test in serum based on lateral flow separation of aptamer-linked nanostructures.” Angew. Chem., Int. Ed. 10.1002/anie.200603106 45, 7955–7959. [DOI] [PubMed] [Google Scholar]
- Lukyanov, K A, Chudakov, D M, Lukyanov, S, and Verkhusha, V V (2005). “Photoactivatable fluorescent proteins.” Nat. Rev. Mol. Cell Biol. 6, 885–891. [DOI] [PubMed] [Google Scholar]
- Makings, L R, and Tsien, R Y (1994). “Caged nitric-oxide - stable organic-molecules from which nitric-oxide can be photoreleased.” J. Biol. Chem. 269, 6282–6285. [PubMed] [Google Scholar]
- Mayer, G, and Famulok, M (2006). “High-throughput-compatible assay for glmS riboswitch metabolite dependence.” ChemBioChem 10.1002/cbic.200500490 7, 602–604. [DOI] [PubMed] [Google Scholar]
- Mendel, D, Ellman, J A, and Schultz, P G (1991). “Construction of a light-activated protein by unnatural amino-acid mutagenesis.” J. Am. Chem. Soc. 10.1021/ja00007a063 113, 2758–2760. [DOI] [Google Scholar]
- Meyer, T, and Teruel, M N (2003). “Fluorescence imaging of signaling networks.” Trends Cell Biol. 10.1016/S0962-8924(02)00040-5 13, 101–106. [DOI] [PubMed] [Google Scholar]
- Miller, J C, Silverman, S K, England, P M, Dougherty, D A, and Lester, H A (1998) “Flash decaging of tyrosine sidechains in an ion channel.” Neuron 10.1016/S0896-6273(00)81001-6 20, 619–624. [DOI] [PubMed] [Google Scholar]
- Miller, L W, and Cornish, V W (2005). “Selective chemical labeling of proteins in living cells.” Curr. Opin. Chem. Biol. 10.1016/j.cbpa.2004.12.007 9, 56–61. [DOI] [PubMed] [Google Scholar]
- Mochizuki, N, Yamashita, S, Kurokawa, K, Ohba, Y, Nagai, T, Miyawaki, A, and Matsuda, M (2001). “Spatio-temporal images of growth-factor-induced activation of Ras and Rap1.” Nature (London) 10.1038/35082594 411, 1065–1068. [DOI] [PubMed] [Google Scholar]
- Mootz, H D, Blum, E S, and Muir, T W (2004). “Activation of an autoregulated protein kinase by conditional protein splicing.” Angew. Chem., Int. Ed. 10.1002/anie.200460941 43, 5189–5192. [DOI] [PubMed] [Google Scholar]
- Mootz, H D, and Muir, T W (2002). “Protein splicing triggered by a small molecule.” J. Am. Chem. Soc. 10.1021/ja026769o 124, 9044–9045. [DOI] [PubMed] [Google Scholar]
- Morii, T, Sugimoto, K, Makino, K, Otsuka, M, Imoto, K, and Mori, Y (2002.) “A new fluorescent biosensor for inositol trisphosphate.” J. Am. Chem. Soc. 10.1021/ja016824d 124, 1138–1139. [DOI] [PubMed] [Google Scholar]
- Neumann, B, Held, M, Liebel, U, Erfle, H, Rogers, P, Pepperkok, R, and Ellenberg, J (2006). “High-throughput RNAi screening by time-lapse imaging of live human cells.” Nat. Methods 3, 385–390. [DOI] [PubMed] [Google Scholar]
- Nguyen, A, Rothman, D M, Stehn, J, Imperiali, B, and Yaffe, M B (2004). “Caged phosphopeptides reveal a temporal role for 14-3-3 in G1 arrest and S-phase checkpoint function.” Nat. Biotechnol. 10.1038/nbt997 22, 993–1000. [DOI] [PubMed] [Google Scholar]
- Nickell, S, Kofler, C, Leis, A P, and Baumeister, W (2006). “A visual approach to proteomics.” Nat. Rev. Mol. Cell Biol. 7, 225–230. [DOI] [PubMed] [Google Scholar]
- Nikolaev, V O, Bunemann, M, Hein, L, Hannawacker, A, and Lohse, M J (2004). “Novel single chain cAMP sensors for receptor-induced signal propagation.” J. Biol. Chem. 10.1074/jbc.C400302200 279, 37215–37218. [DOI] [PubMed] [Google Scholar]
- Oancea, E, and Meyer, T (1998). “Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals.” Cell 10.1016/S0092-8674(00)81763-8 95, 307–318. [DOI] [PubMed] [Google Scholar]
- Oancea, E, Teruel, M N, Quest, A FG, and Meyer, T (1998). “Green fluorescent protein (GFP)-tagged cysteine-rich domains from protein kinase C as fluorescent indicators for diacylglycerol signaling in living cells.” J. Cell Biol. 10.1083/jcb.140.3.485 140, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto, S, Looger, L L, Micheva, K D, Reimer, R J, Smith, S J, and Frommer, W B (2005). “Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0503274102 102, 8740–8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G H, and Lippincott-Schwartz, J (2002). “A photoactivatable GFP for selective photolabeling of proteins and cells.” Science 10.1126/science.1074952297, 1873–1877. [DOI] [PubMed] [Google Scholar]
- Pellois, J P, Hahn, M E, and Muir, T W (2004). “Simultaneous triggering of protein activity, and fluorescence.” J. Am. Chem. Soc. 10.1021/ja0499142 126, 7170–7171. [DOI] [PubMed] [Google Scholar]
- Pepperkok, R, and Ellenberg, J (2006). “Innovation—high-throughput fluorescence microscopy for systems biology.” Nat. Rev. Mol. Cell Biol. 7, 690–696. [DOI] [PubMed] [Google Scholar]
- Pertz, O, Hodgson, L, Klemke, R L, and Hahn, K M (2006). “Spatiotemporal dynamics of RhoA activity in migrating cells.” Nature (London) 10.1038/nature04665 440, 1069–1072. [DOI] [PubMed] [Google Scholar]
- Peyrottes, S, Egron, D, Lefebvre, I, Gosselin, G, Imbach, J L, and Perigaud, C (2004). “SATE pronucleotide approaches: an overview.” Mini-Rev. Med. Chem. 4, 395–408. [DOI] [PubMed] [Google Scholar]
- Philipson, K D, Gallivan, J P, Brandt, G S, Dougherty, D A, and Lester, H A (2001). “Incorporation of caged cysteine and caged tyrosine into a transmembrane segment of the nicotinic ACh receptor.” Am. J. Physiol.: Cell Physiol. 281, C195–C206. [DOI] [PubMed] [Google Scholar]
- Piljic, A, and Schultz, C (2006). “Annexin A4 self-association modulates general membrane protein mobility in living cells.” Mol. Biol. Cell 17, 3318–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plafker, K, and Macara, I G (2002). “Fluorescence resonance energy transfer biosensors that detect ran conformational changes and a Ran center dot GDP-importin-beta-RanBP1 complex in vitro and in intact cells.” J. Biol. Chem. 10.1074/jbc.M203006200 277, 30121–30127. [DOI] [PubMed] [Google Scholar]
- Potter, B VL, and Lampe, D (1995). “Chemistry of inositol lipid-mediated cellular signaling.” Angew. Chem., Int. Ed. Engl. 10.1002/anie.199519331 34, 1933–1972. [DOI] [Google Scholar]
- Pranada, A L, Metz, S, Herrmann, A, Heinrich, P C, and Muller-Newen, G (2004). “Real time analysis of STAT3 nucleocytoplasmic shuttling.” J. Biol. Chem. 10.1074/jbc.M312530200 279, 15114–15123. [DOI] [PubMed] [Google Scholar]
- Rabut, G, Doye, V, and Ellenberg, J (2004). “Mapping the dynamic organization of the nuclear pore complex inside single living cells.” Nat. Cell Biol. 10.1038/ncb1184 6, 1114–U1127. [DOI] [PubMed] [Google Scholar]
- Reither, G, Schaefer, M, and Lipp, P (2006). “PKC alpha: a versatile key for decoding the cellular calcium toolkit.” J. Cell Biol. 10.1083/jcb.200604033 174, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo, J, and Sudhof, T C (1998). “C-2-domains, structure and function of a universal Ca2+-binding domain.” J. Biol. Chem. 10.1074/jbc.273.26.15879 273, 15879–15882. [DOI] [PubMed] [Google Scholar]
- Rothman, D M, Shults, M D, and Imperiali, B (2005). “Chemical approaches for investigating phosphorylation in signal transduction networks.” Trends Cell Biol. 10.1016/j.tcb.2005.07.003 15, 502–510. [DOI] [PubMed] [Google Scholar]
- Rudolf, M T, Dinkel, C, Traynor-Kaplan, A E, and Schultz, C (2003). “Antagonists of myo-inositol 3,4,5,6-tetrakisphosphate allow repeated epithelial chloride secretion.” Bioorg. Med. Chem. 10.1016/S0968-0896(03)00188-3 11, 3315–3329. [DOI] [PubMed] [Google Scholar]
- Sakamoto, K M, Kim, K B, Kumagai, A, Mercurio, F, Crews, C M, and Deshaies, R J (2001). “Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.141230798 98, 8554–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K M, Kim, K B, Verma, R, Ransick, A, Stein, B, Crews, C M, and Deshaies, R J (2003). “Development of protacs to target cancer-promoting proteins for ubiquitination and degradation.” Mol. Cell Proteomics 2, 1350–1358. [DOI] [PubMed] [Google Scholar]
- Schleifenbaum, A, Stier, G, Gasch, A, Sattler, M, and Schultz, C (2004). “Genetically encoded FRET probe for PKC activity based on pleckstrin.” J. Am. Chem. Soc. 10.1021/ja0460155 126, 11786–11787. [DOI] [PubMed] [Google Scholar]
- Schneekloth, J S, Fonseca, F N, Koldobskiy, M, Mandal, A, Deshaies, R, Sakamoto, K, and Crews, C M (2004). “Chemical genetic control of protein levels: selective in vivo targeted degradation.” J. Am. Chem. Soc. 10.1021/ja039025z 126, 3748–3754. [DOI] [PubMed] [Google Scholar]
- Schreiber, S L (1998). “Chemical genetics resulting from a passion for synthetic organic chemistry.” Bioorg. Med. Chem. 10.1016/S0968-0896(98)00126-6 6, 1127–1152. [DOI] [PubMed] [Google Scholar]
- Schultz, C (2003). “Prodrugs of biologically active phosphate esters.” Bioorg. Med. Chem. 10.1016/S0968-0896(02)00552-7 11, 885–898. [DOI] [PubMed] [Google Scholar]
- Schultz, C, Schleifenbaum, A, Goedhart, J, and Gadella, T W, Jr. (2005). “Multiparameter imaging for the analysis of intracellular signaling.” ChemBioChem 10.1002/cbic.200500012 6, 1323–1330. [DOI] [PubMed] [Google Scholar]
- Schultz, C, Vajanaphanich, M, Genieser, H G, Jastorff, B, Barrett, K E, and Tsien, R Y (1994). “Membrane-permeant derivatives of cyclic-amp optimized for high potency, prolonged activity, or rapid reversibility.” Mol. Pharmacol. 46, 702–708. [PubMed] [Google Scholar]
- Schultz, C, Vajanaphanich, M, Harootunian, A T, Sammak, P J, Barrett, K E, and Tsien, R Y (1993). “Acetoxymethyl esters of phosphates, enhancement of the permeability and potency of camp.” J. Biol. Chem. 268, 6316–6322. [PubMed] [Google Scholar]
- Schwartz, E C, Saez, L, Young, M W, and Muir, T W (2005). “Rapid in vivo control of enzyme function through conditional protein splicing.” Biopolymers 80, 544–544. [Google Scholar]
- Schwartz, E C, Saez, L, Young, M W, and Muir, T W (2007). “Post-translational enzyme activation in an animal via optimized conditional protein splicing.” Nat. Chem. Biol. 10.1038/nchembio832 3, 50–54. [DOI] [PubMed] [Google Scholar]
- Schwede, F, Maronde, E, Genieser, H G, and Jastorff, B (2000). “Cyclic nucleotide analogs as biochemical tools and prospective drugs.” Pharmacol. Ther. 87, 199–226. [DOI] [PubMed] [Google Scholar]
- Shaner, N, Campbell, R E, Steinbach, P A, GBN, P, Palmer, A E, and Tsien, R Y (2004). “Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein.” Nat. Biotechnol. 10.1038/nbt1037 22, 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shaner, N C, Steinbach, P A, and Tsien, R Y (2005). “A guide to choosing fluorescent proteins.” Nat. Methods 2, 905–909. [DOI] [PubMed] [Google Scholar]
- Shao, X G, Fernandez, I, Sudhof, T C, and Rizo, J (1998). “Solution structures of the Ca2+-free and Ca2+-bound C(2)A domain of synaptotagmin I: does Ca2+ induce a conformational change?” Biochemistry 10.1021/bi981789h 37, 16106–16115. [DOI] [PubMed] [Google Scholar]
- Short, G F, Lodder, M, Laikhter, A L, Arslan, T, and Hecht, S M (1999). “Caged HIV-1 protease: dimerization is independent of the ionization state of the active site aspartates.” J. Am. Chem. Soc. 10.1021/ja9838054 121, 478–479. [DOI] [Google Scholar]
- Shu, X K, Shaner, N C, Yarbrough, C A, Tsien, R Y, and Remington, S J (2006). “Novel chromophores and buried charges control color in mFruits.” Biochemistry 10.1021/bi060773l 45, 9639–9647. [DOI] [PubMed] [Google Scholar]
- Simon, M D, and Shokat, K M (2007). “Revealing biological specificity by engineering protein-ligand interactions.” In Chemical Biology. Schreiber, S L, Kapoor, T, and Wess, G (eds), Wiley-VCH, Weinheim, vol 1, pp 115–139. [Google Scholar]
- Skretas, G, and Wood, D W (2005). “Regulation of protein activity with small-molecule-controlled inteins.” Protein Sci. 10.1110/ps.04996905 14, 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin, R V, and Cho, W (2001a). “Roles of calcium ions in the membrane binding of C2 domains.” Biochem. J. 359, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin, R V, and Cho, W H (2001b). “Differential roles of ionic, aliphatic, and aromatic residues in membrane-protein interactions: a surface plasmon resonance study on phospholipases A(2).” Biochemistry 10.1021/bi0020325 40, 4672–4678. [DOI] [PubMed] [Google Scholar]
- Stahelin, R V, Rafter, J D, Das, S, and Cho, W (2003). “The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A(2).” J. Biol. Chem. 10.1074/jbc.M212864200 278, 12452–12460. [DOI] [PubMed] [Google Scholar]
- Starrett, J E, Tortolani, D R, Russell, J, Hitchcock, M JM, Whiterock, V, Martin, J C, and Mansuri, M M (1994). “Synthesis, oral bioavailability determination, and in-vitro evaluation of prodrugs of the antiviral agent 9-[2-(phosphonomethoxy) ethyl]adenine (PMEA).” J. Med. Chem. 10.1021/jm00038a015 37, 1857–1864. [DOI] [PubMed] [Google Scholar]
- Suh, B C, Inoue, T, Meyer, T, and Hille, B (2006). “Rapid chemically induced changes of Ptdlns(4,5)P-2 gate KCNQ ion channels.” Science 10.1126/science.1131163 314, 1454–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teruel, M N, and Meyer, T (2000). “Translocation and reversible localization of signaling proteins: a dynamic future for signal transduction.” Cell 10.1016/S0092-8674(00)00109-4 103, 181–184. [DOI] [PubMed] [Google Scholar]
- Teruel, M N, and Meyer, T (2002). “Parallel single-cell monitoring of receptor-triggered membrane translocation of a calcium-sensing protein module.” Science 10.1126/science.1065028 295, 1910–1912. [DOI] [PubMed] [Google Scholar]
- Theis, M G, Knorre, A, Kellersch, B, Moelleken, J, Wieland, F, Kolanus, W, and Famulok, M (2004). “Discriminatory aptamer reveals serum response element transcription regulated by cytohesin-2.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0402901101 101, 11221–11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, A Y, Kain, K H, Klemke, R L, and Tsien, R Y (2001). “Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.211564598 98, 15003–15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentham, D R, Corrie, J ET, and Reid, G P (1992). “A new caged atp with rapid photolysis kinetics.” FASEB J. 6, A295–A295. [Google Scholar]
- Tsien, R Y (1981). “A non-disruptive technique for loading calcium buffers and indicators into Cells.” Nature (London) 10.1038/290527a0 290, 527–528. [DOI] [PubMed] [Google Scholar]
- Vajanaphanich, M, Schultz, C, Rudolf, M T, Wasserman, M, Enyedi, P, Craxton, A, Shears, S B, Tsien, R Y, Barrett, K E, and Traynor-Kaplan, A (1994). “Long-term uncoupling of chloride secretion from intracellular calcium levels by Ins(3,4,5,6)P4.” Nature (London) 10.1038/371711a0 371, 711–714. [DOI] [PubMed] [Google Scholar]
- van der Wal, J, Habets, R, Varnai, P, Balla, T, and Jalink, K (2001). “Monitoring agonist-induced phospholipase C activation in live cells by fluorescence resonance energy transfer.” J. Biol. Chem. 10.1074/jbc.M007194200 276, 15337–15344. [DOI] [PubMed] [Google Scholar]
- van Manen, H J, Kraan, Y M, Roos, D, and Otto, C (2005). “Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0502746102 102, 10159–10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Thor, J J, Gensch, T, Hellingwerf, K J, and Johnson, L N (2002). “Phototransformation of green fluorescent protein with UV and visible light leads to decarboxylation of glutamate 222.” Nat. Struct. Biol. 10.1038/nsb739 9, 37–41. [DOI] [PubMed] [Google Scholar]
- Varnai, P, Rother, K I, and Balla, T (1999). “Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton’s tyrosine kinase pleckstrin homology domain visualized in single living cells.” J. Biol. Chem. 10.1074/jbc.274.16.10983 274, 10983–10989. [DOI] [PubMed] [Google Scholar]
- Vazquez, M E, Nitz, M, Stehn, J, Yaffe, M B, and Imperiali, B (2003). “Fluorescent caged phosphoserine peptides as probes to investigate phosphorylation-dependent protein associations.” J. Am. Chem. Soc. 10.1021/ja0351847 125, 10150–10151. [DOI] [PubMed] [Google Scholar]
- Verveer, P J, Squire, A, and Bastiaens, P IH (2000a). “Global analysis of fluorescence lifetime imaging microscopy data.” Biophys. J. 78, 2127–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verveer, P J, Wouters, F S, Reynolds, A R, and Bastiaens, P IH (2000b). “Quantitative imaging of lateral ErbB1 receptor signal propagation in the plasma membrane.” Science 10.1126/science.290.5496.1567 290, 1567–1570. [DOI] [PubMed] [Google Scholar]
- Violin, J D, Zhang, J, Tsien, R Y, and Newton, A C (2003). “A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C.” J. Cell Biol. 10.1083/jcb.200302125 161, 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Daehne, W, Frederiksen, E, Gundersen, E, Lund, F, Morch, P, Petersen, H J, Roholt, K, Tybring, L, and Godtfredsen, W O (1970). “Acyloxymethyl esters of ampicillin.” J. Med. Chem. 10.1021/jm00298a005 13, 607–612. [DOI] [PubMed] [Google Scholar]
- Voss, S, Fischer, R, Jung, G, Wiesmuller, K H, and Brock, R (2007). “A fluorescence-based synthetic LPS sensor.” J. Am. Chem. Soc. 10.1021/ja065016p 129, 554–561. [DOI] [PubMed] [Google Scholar]
- Walker, J W, Reid, G P, Mccray, J A, and Trentham, D R (1988). “Photolabile 1-(2-nitrophenyl)ethyl phosphate-esters of adenine-nucleotide analogs—synthesis and mechanism of photolysis.” J. Am. Chem. Soc. 10.1021/ja00229a036 110, 7170–7177. [DOI] [Google Scholar]
- Webb, S E, Roberts, S K, Needham, S R, Tynan, C J, Rolfe, D J, Winn, M D, Clarke, D T, Barraclough, R, and Martin-Fernandez, M (2007). “Single molecule imaging and FLIM show different structures for high and low-affinity EGFRs in A431 cells.” Biophys. J. [DOI] [PMC free article] [PubMed]
- Wichmann, O, Gelb, M, and Schultz, C (2007). “Probing phospholipase A2 with fluorescent phospholipid substrates.” ChemBioChem 10.1002/cbic.200600462 8, 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann, O, Wittbrodt, J, and Schultz, C (2006). “A small-molecule FRET probe to monitor phospholipase A(2) activity in cells and organisms.” Angew. Chem., Int. Ed. 10.1002/anie.200500751 45, 508–512. [DOI] [PubMed] [Google Scholar]
- Wiedenmann, J, Ivanchenko, S, Oswald, F, Schmitt, F, Rocker, C, Salih, A, Spindler, K D, and Nienhaus, G U (2004). “EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0403668101 101, 15905–15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J, and Schultz, P G (2006). “A chemical toolkit for proteins—an expanded genetic code.” Nat. Rev. Mol. Cell Biol. 7, 775–782. [DOI] [PubMed] [Google Scholar]
- Yudushkin, I A, Schleifenbaum, A, Kinkhabwala, A, Neel, B G, Schultz, C, and Bastiaens, P IH (2007). “Live-cell imaging of enzyme-substrate interaction reveals spatial regulation of PTP1B.” Science 10.1126/science.1134966 315, 115–119. [DOI] [PubMed] [Google Scholar]
- Yuste, R (2005). “Fluorescence microscopy today.” Nat. Methods 2, 902–904. [DOI] [PubMed] [Google Scholar]
- Zaccolo, M, De Giorgi, F, Cho, C Y, Feng, L X, Knapp, T, Negulescu, P A, Taylor, S S, Tsien, R Y, and Pozzan, T (2000). “A genetically encoded, fluorescent indicator for cyclic AMP in living cells.” Nat. Cell Biol. 10.1038/71345 2, 25–29. [DOI] [PubMed] [Google Scholar]
- Zeidler, M P, Tan, C, Bellaiche, Y, Cherry, S, Hader, S, Gayko, U, and Perrimon, N (2004). “Temperature-sensitive control of protein activity by conditionally splicing inteins.” Nat. Biotechnol. 10.1038/nbt979 22, 871–876. [DOI] [PubMed] [Google Scholar]
- Zhang, J, Campbell, R E, Ting, A Y, and Tsien, R Y (2002). “Creating new fluorescent probes for cell biology.” Nat. Rev. Mol. Cell Biol. 3, 906–918. [DOI] [PubMed] [Google Scholar]
- Zhang, J, Ma, Y L, Taylor, S S, and Tsien, R Y (2001). “Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.211566798 98, 14997–15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W Y, and Smith, S O (2005). “Mechanism of penetration of Antp(43-58) into membrane bilayers.” Biochemistry 10.1021/bi050341v 44, 10110–10118. [DOI] [PubMed] [Google Scholar]
- Ziegler, A, Nervi, P, Durrenberger, M, and Seelig, J (2005). “The cationic cell-penetrating peptide Cpp(TAT) derived from the HIV-1 protein TAT is rapidly transported into living fibroblasts: optical, biophysical, and metabolic evidence.” Biochemistry 10.1021/bi0491604 44, 138–148. [DOI] [PubMed] [Google Scholar]
- Zlokarnik, G, Negulescu, P A, Knapp, T E, Mere, L, Burres, N, Feng, L X, Whitney, M, Roemer, K, and Tsien, R Y (1998). “Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter.” Science 10.1126/science.279.5347.84 279, 84–88. [DOI] [PubMed] [Google Scholar]
- Zou, K Y, Cheley, S, Givens, R S, and Bayley, H (2002). “Catalytic subunit of protein kinase A caged at the activating phosphothreonine.” J. Am. Chem. Soc. 10.1021/ja020405e 124, 8220–8229. [DOI] [PubMed] [Google Scholar]
- Zou, K Y, Miller, W T, Givens, R S, and Bayley, H (2001). “Caged thiophosphotyrosine peptides.” Angew. Chem., Int. Ed. 40, 3049–3051. [DOI] [PubMed] [Google Scholar]