Abstract
Genetic recombination in eukaryotes requires the pairing of homologous chromosomes to allow precise molecular exchanges between chromosome pairs at intertwined structures called Holliday junctions, the formation of which requires the action of the RecA protein. The mechanism behind the precise pairing of structures as long as chromosomes remains mysterious. In yeast, during the initial phases of meiosis, chromosomes are paired at approximately 65 kilobase intervals via paranemic interactions that do not involve strand breakage nor the intervention of analogs of the RecA protein. It has been proposed that these paranemic interactions could occur between G-rich chromosomal regions, but putting in register stretches of homologous sequences hundreds of kb long remains challenging. Recent developments on the theory of the physicochemical properties of DNA in aqueous solutions, in presence of di- or multivalent counterions, leads to the prediction that molecules with the same sequence tend to pair spontaneously by paranemic interactions depending on the electrostatic properties of DNA. Experimental support for this prediction has now been provided in vitro with naked DNA. This newly discovered property of DNA duplexes may thus provide a clue to solve the puzzle of the premeiotic pairing.
Genetic recombination is essentially based on the complementary properties of the structure of the DNA duplex, via the introduction of double strand breaks (DSBs) in the involved homologues and the formation of the Holliday junction, the latter mediated by specific proteins like the product of the Escherichia coli RecA gene and its eukaryotic analogues. In other words, two homologous sequences can pair precisely thanks to the specificity of the Watson and Crick pairing rules: a single-stranded end produced in a molecule can invade a nearby homologous duplex and anneal to the complementary strand thanks to the catalytic mediation of the RecA protein or its analogs. But a very serious difficulty remains unexplained by this (otherwise well demonstrated) process: how do the two duplexes involved align closely enough to allow this molecular exchange to occur? The RecA-like proteins require an identity length of 8 bp to complete the process (Hsieh et al., 1992); can we imagine the two homologous duplexes exploring the whole length of the involved chromosomes looking for this level of homology? This is a particularly formidable task in the eukaryotic organisms, where recombination occurring during meiosis requires the formation of a close pairing of the whole homologous chromosomes in the synaptonemal complex, i.e., the precise pairing of DNA molecules of an average size, in the human genome, of 130 Mb, or 40 mm in length.
Two other considerations rule out the RecA-like proteins for assuring chromosome pairing preliminary to recombination. Since the length requirement for pairing via these proteins is only 8 bp, approximately 100,000 identities of this length are predictable in a genome like the human one, hence, the possibility of mistakes would be enormous. Furthermore, it is well demonstrated that chromosome pairing during meiosis occurs before, and in absence, of DSBs, an absolute requirement for RecA-mediated exchanges. How do living organisms achieve this precise pairing? A development based on theoretical considerations of the physicochemical properties of the DNA molecule and on recent fascinating and intriguing experiments sheds a possible light on this mysterious and key aspect of the recombination process. But let us review briefly the current ideas on this subject prior to the recent developments.
POSSIBLE MECHANISMS FOR LONG-RANGE HOMOLOG RECOGNITION AND PAIRING
The first hypothesis proposed for solving this conundrum was based on the known homology of centromeric and, particularly, of telomeric sequences and for the tendency of the latter to interact: thus, pairing of these structures could begin to align homologs (Loidl, 1990), whereas the subsequent intervention of DSBs and RecA-like proteins could do the rest. But this does not explain by itself the selection of homologs, since those repetitive structures are common to all chromosomes, hence, a close vicinity of the specific partner should be first assured (see also below); furthermore, even if the problem of pairing telomeres and centromeres exclusively of homologs were resolved, the lengths of these associated sequences would still be so high to be unable to meet the challenges exposed before for whole chromosomes.
The problem was later investigated in detail in Saccharomyces cerevisiae, particularly by the group of Nancy Kleckner in Harvard (Weiner and Kleckner, 1994; Burgess et al., 1999); by fluorescence in situ hybridization analysis these investigators observed the pairing of homologous chromosomes before meiosis, without compaction of the chromosomes, in absence of double-strand breaks and without the intervention of RecA-like proteins. Also, the pairing disappears during meiotic DNA duplication and is restored immediately afterwards. The pairing occurs via multiple interstitial interactions, one every ∼65 kb and these interactions probably correspond to the subsequent recombination events. The same type of pairing occurs also in vegetatively cycling cells, in G1 and G2, disappearing in S. Thus, these authors conclude that both premeiotic and somatic pairings occur via paranemic interactions of homologous stretches of the chromosomes, at sites spaced approximately 65 kb from each other, prior to meiotic or vegetative DNA duplication; pairing is dissolved by duplication, but it is reestablished immediately afterwards in the same paranemic mode; during meiosis, only later the DSBs required by recombination and the action of the RecA-like proteins occur in the already aligned stretches, leading to the plectonemic interactions in the context of the synaptonemal complex.
This elegant process begins to dispel some of the mystery of homologous chromosome pairing, but shifts the puzzle to another level: what is the molecular nature of this paranemic interaction? Is it purely determined by DNA sequence (at some level, it has to be) or is it mediated by specific proteins (that must in any way be able to distinguish the multiple pairing sites)? The authors have no hypothesis on the molecular basis of this periodic specific paranemic interaction, but emphasize the prerequisite of topographical vicinity of the homologous chromosomes for this finer interaction to occur, a prerequisite in line with the mounting evidence for the presence of specific compartments for the different chromosomes in the nucleus.
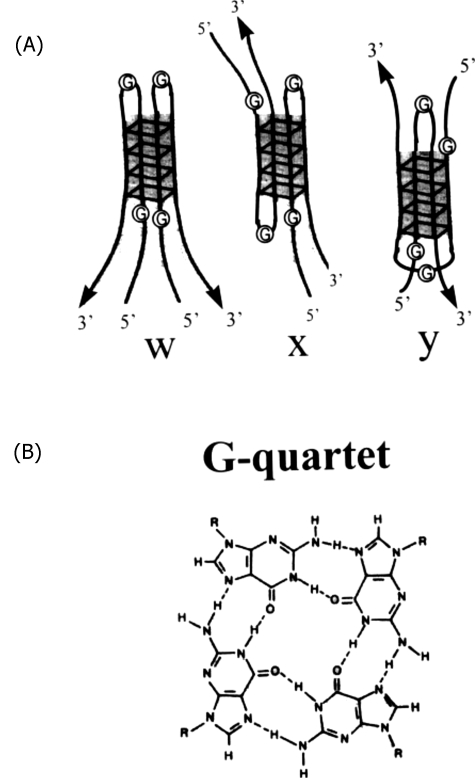
What kind of sequence-specific paranemic interactions can we envisage to explain these data? A possible candidate is given by the unusual properties of stretches of G residues; these are known to be able to form structures called G-quartets held together by Hoogsten hydrogen bonds (see Fig. 1) in which four strands can be linked together, either with two parallel and two antiparallel strands, or with four parallel strands (Marco-Haviv et al., 1999). Arrays of G are interspersed throughout the chromosomes and one may envisage that they could (helped possibly by specific proteins) bulge out as hairpins with a certain periodicity and form four-stranded structures with a nearby homolog, without the need of introducing breaks. These interactions can be relatively dynamic and it could be predicted that they adapt to the least hindered and most stable overall structure, when the two homologous sequences are in register.
Figure 1. (A) Three possible structures formed by bulged-out G -hairpins in nearby molecules.
(B) Structure of the hydrogen-bonded G-quartets. Reprinted from Marco-Haviv et al. (1999). DNA molecules can drive the assembly of other DNA molecules into specific four-stranded structures. (Reprinted from J. Mol. Biol.286, 45–56, with permission from the American Chemistry Society.)
It is tempting to possibly identify these G-mediated interactions with the sites of pairing identified by the work of Kleckner and her collaborators, but this will require specific investigation. A tantalizing hint in this sense comes from the observation of a strong correlation between G-rich isochors and presence of recombinational “hotspots” in yeast chromosomes (Baudat and Nicolas, 1997).
Yet, putting in register DNA homologs with an average periodicity of 65 kb would still pose serious problems for the close alignment necessary to allow RecA-mediated exploration for fine homology. The more so, since data obtained in several systems, both prokaryotic and eukaryotic, indicate the need for a 100–200 bp homology for achieving efficient homologous recombination (Watt et al., 1985; Rubnitz and Subramani, 1984). Can we identify other forms of paranemic, sequence-specific interactions, intrinsic to the DNA structure?
INTRINSIC SEQUENCE HOMOLOGY RECOGNITION BY DNA DUPLEXES
In the last decade, the study of the physical and physicochemical properties of DNA duplexes has addressed, in particular, the a priori unexpected tendency of DNA duplexes to associate in aqueous solutions, at least in certain conditions, like the presence of divalent or multivalent positive counterions, compensating the overall negative charge of this “acid” given by the phosphate residues. Experimentally, Parsegian and collaborators, at NIH (Gelbart et al., 2000) observed that, in the presence of these cationic counterions, DNA molecules display a surprising tendency to closely associate, a property for which the quantum-mechanically elicited Casimir force was invoked (or, more prosaically, old fashioned van der Waals interactions). At the same time, Leikin, also at NIH, and Kornyshev, now at Imperial College, by a combination of theory, computer simulations, and model experimental approaches, identified the importance of short range electrostatic interactions in favoring this association, a force not alternative to the van der Waals interactions, but possibly more important (Kornyshev and Leikin, 2001).
These authors brought forward this initial intuition by first developing an accurate a priori theory of the interactions of DNA duplexes in presence of divalent or multivalent counterions (Cherstvy et al., 2004; Kornyshev et al., 2007); the essential elements of this theory predict, in the first place, that, as, concentration rises, the molecules will tend to align along the major axis; not surprising, since this property is what allows the formation of the essentially one-dimensional crystals (or fibers) that allowed Franklin to obtain the crucial diffraction radiographs that brought to the discovery of DNA structure.
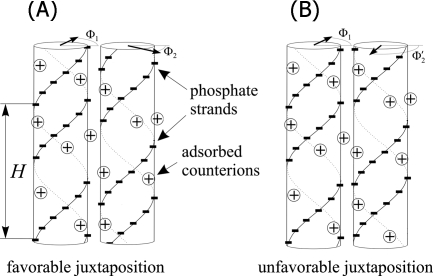
On top of this, the most surprising prediction issuing from the theoretical treatment was that the sequence of base pairs would influence the orientation of counterions spontaneously concentrated in the major and minor grooves, with accompanying slight local alterations of the base tilt and helical pitch that reflect the base pair sequence. But this was not all: this specific helical distribution of the counterions, with its sequence-dependent vagaries, would cause difficulty (thermodynamic barriers) in achieving the longitudinal associations to which the duplexes tend; difficulties, that is, unless the sequences are identical: in this case, the longitudinal interactions become optimal and the lowest energy state for association is achieved. In other words, optimal, thermodynamically favored association is achieved only between molecules of identical sequence. Actually, according to the theory, the optimal association is achieved between two duplexes, one longitudinally and azimuthally displaced from the other by the distance and azimuthal angle necessary to bring the phosphate residue of one base pair in front of the counterion present in the major groove of the corresponding (identical) base pair of the partner (see Fig. 2).
Figure 2. (a) If two molecules with identical sequences are exactly aligned with a reciprocal azimuthal orientation (Φ1 andΦ2) that puts the counterions bound in the grooves in juxtaposition with the phosphate chain, the two molecules specifically attract each other, whereas, if they are aligned in azimuthal orientations such that the phosphate chains face each other, the repulsion is predominant.
The base sequence determines small variations in helical pitch whereby the most thermodynamically favored situation occur when the sequences of the two duplexes are identical. Reprinted from Baldwin et al. (2008). Double helices recognize mutual sequence homology in a protein free environment. (Reprinted from J. Phys. Chem. B112, 1060–1064, with permission from Elsevier.)
This exciting prediction has recently received experimental confirmation; in the first place, the group of Ohyama at Waseda University studied in vitro association of DNA fragments of ∼200 bp by electrophoresis and atomic force microscopy and they detected a preferential association of like over nonlike molecules at 10 nM concentrations and in presence of Mg ions (Inoue et al., 2007). They interpreted this association as mediated by local formation of bubbles and base flipping with subsequent association of the fleetingly opened stretches, an interpretation that begs the question of how specific can the interaction be if you cannot anneal for a certain length the flipped out bases.
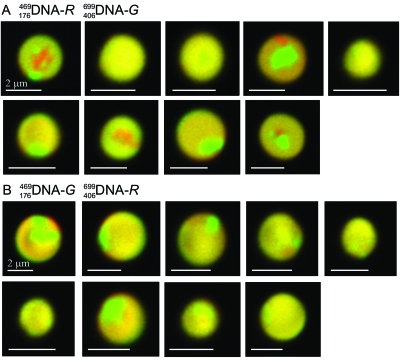
This interpretation is not shared by Kornyshev, Leikin, and their collaborators who performed very ingenuous experiments in which they concentrated, with poly-ethylene glycol, mixtures of two 294 bp fragments of the same base composition but different sequence, one labeled with a green-fluorescent and the other with a red-fluorescent tag (Baldwin et al., 2008). The condensed droplets, or “spherulites,” were kept at room temperatures for two weeks and showed eventually an unambiguous spontaneous separation of the two colors in the concentrated solutions, demonstrating that the fragments with the same sequence preferentially associated to each other (see Fig. 3). These authors note that, from previous data, the associated fragments are separated by layers of water molecules between 1 and 3 nm thick, making it unlikely that they associate by base flipping.
Figure 3. Droplets (spherulites) containing concentrated mixtures of two 294 bp DNA fragments with the same base composition but different sequences, one (A) labeled in red (DNA- R ) and the other in green (DNA- G ), or the reverse (B), after two weeks at room temperature show a clear separation of colors at confocal microscopy; each molecule appears to preferentially associate with molecules of the same sequence.
Reprinted from Baldwin et al. (2008). Double helices recognize mutual sequence homology in a protein free environment. (Reprinted from J. Phys. Chem. B112, 1060–1064, with permission from Elsevier.)
More work will certainly be necessary to demonstrate to general satisfaction that the association of like molecules is mediated by the electrostatic interactions described by Leikin and Kornyshev, as there are obvious limitations to this work, considering that the experiments are performed with naked DNA (as opposed to chromatin, which constitutes the actual chromosomes), and that the incubation required to observe the spontaneous pairing is very long. Nevertheless, the hypothesis proposed by Leikin and Kornyshev appears particularly attractive for its apparent general applicability, for having been derived from a thorough theoretical treatment of the matter from first principles (Kornyshev et al., 2007) and from the elegant experimental confirmation of a theory (Baldwin et al., 2008). Another element in favor of this interpretation is that the theory predicts that, for the electrostatic interactions between two identical duplexes to be most thermodynamically favored, fragment lengths of at least 100–200 bp are necessary; this agrees well with the above-mentioned requirements of a minimal length of 100–200 bp homology for efficient homologous recombination.
CONCLUSION
McClintock stated in 1933 that “there is a tendency for chromosomes to associate 2-by-2 in the prophase of meiosis.” With all the data collected subsequently on the process and the novel observations summarized here, we can dare to indicate a possible five-step flow of events in meiotic recombination. (1) In the first place, chromosome pairs acquire a topographical closeness within a nuclear compartment, via specific interactions concerning nuclear functional architecture that have yet to be understood. (2) Telomeres and centromeres may now realize an initial chromosome pairing not disturbed by the presence of other chromosomes. (3) G-arrays distributed along the chromosomes may begin to interact and find a more stable paired structure when the sequences are in register (do the 65 kb-spaced interstitial pairing interactions described in yeast correspond to the G-arrays?). (4) At this point, the electrostatically determined pairing interactions described above bring the duplexes in close paranemic, sequence-specific contact. (5) Breaks of the DNA sequence provide the substrates for the RecA-mediated reciprocal exploration of the two aligned duplexes for precise Watson-and-Crick determined exchanges, eventually leading to plectonemic interactions in the context of the synaptonemal complex.
Future work will show if this picture is realistic; in any case, molecular biologists working with the DNA molecule cannot afford to ignore or neglect this important, until now unsuspected, property of the double helix.
A final note: this brilliant passage from physical theory, based on first principles, to experimental validation and to contribution at understanding the mechanism and function of a crucial biological process, cannot fail to remind an outstanding precedent: I refer to the development from first principles of the theory of the x-ray diffraction pattern of helical structures (Cochran et al., 1952) that brought a brilliant physicist to collaborate with a bright biologist in discovering the structure of DNA.
References
- Baldwin, G S, Brook, N Y, Robson, R E, Wynween, A, Goldar, A, Leikin, S, Seddon, J M, and Kornyshev, A A (2008). “Double helices recognize mutual sequence homology in a protein free environment.” J. Phys. Chem. B 112, 1060–1064. [DOI] [PubMed] [Google Scholar]
- Baudat, F, and Nicolas, A (1997). “Clustering of meiotic double-strand breaks on yeast chromosome. III.” Proc. Natl. Acad. Sci. U.S.A. 94, 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, S M, Kleckner, N, and Weiner, B M (1999). “Somatic pairing of homologs in budding yeast: existence and modulation.” Genes Dev. 13, 1627–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherstvy, A G, Kornyshev, A A, and Leikin, S (2004). “Torsional deformation of double helix in interaction and aggregation of DNA.” J. Phys. Chem. 108, 6508–6518. [DOI] [PubMed] [Google Scholar]
- Cochran, W, Crick, F HC, and Vand, V (1952). “The structure of synthetic polypeptides. I. The transform of atoms on a helix.” Acta Crystallogr. 10.1107/S0365110X52001635 5, 581–586. [DOI] [Google Scholar]
- Gelbart, W M, Bruinsma, R F, Pincus, P A, and Parsegia, A (2000). “DNA-inspired electrostatics.” Phys. Today 53, 38–44. [Google Scholar]
- Hsieh, P, Camerini-Otero, C S, and Camerini-Otero, R D (1992). “The synapsis event in homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.89.14.6492 89, 6492–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, S, Sugiyama, S, Travers, A A, and Ohyama, T (2007). “Self-assembly of double-stranded DNA molecules at nanomolar concentrations.” Biochemistry 46, 164–171. [DOI] [PubMed] [Google Scholar]
- Kornyshev, A A, Lee, D J, Leikin, S, and Wynween, A (2007). “Structure and interactions of biological helices.” Rev. Mod. Phys. 10.1103/RevModPhys.79.943 79, 943–995. [DOI] [Google Scholar]
- Kornyshev, A A, and Leikin, S (2001). “Sequence recognition in the pairing of DNA duplexes.” Phys. Rev. Lett. 10.1103/PhysRevLett.86.3666 86, 3666–3669. [DOI] [PubMed] [Google Scholar]
- Loidl, J (1990). “The initiation of meiotic chromosome pairing: the cytological view.” Chromosoma 100, 221–228. [DOI] [PubMed] [Google Scholar]
- Marco-Haviv, Y, Baran, N, and Manor, H (1999). “DNA molecules can drive the assembly of other DNA molecules into specific four-stranded structures.” J. Mol. Biol. 286, 45–56. [DOI] [PubMed] [Google Scholar]
- Rubnitz, J, and Subramani, S (1984). “The minimum amount of homology required for homologous recombination in mammalian cells.” Mol. Cell. Biol. 4, 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt, V M, Ingles, C J, Urdea, M S, and Rutter, W J (1985). “Homology requirements for recombination in Escherichia coli.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.82.14.4768 82, 4768–4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner, B M, and Kleckner, N (1994). “Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast.” Cell 10.1016/0092-8674(94)90438-3 77, 977–991. [DOI] [PubMed] [Google Scholar]