Abstract
During adhesion and spreading, cells form micrometer-sized structures comprising transmembrane and intracellular protein clusters, giving rise to the formation of what is known as focal adhesions. Over the past two decades these structures have been extensively studied to elucidate their organization, assembly, and molecular composition, as well as to determine their functional role. Synthetic materials decorated with biological molecules, such as adhesive peptides, are widely used to induce specific cellular responses dependent on cell adhesion. Here, we focus on how surface patterning of such bioactive materials and organization at the nanoscale level has proven to be a useful strategy for mimicking both physical and chemical cues present in the extracellular space controlling cell adhesion and fate. This strategy for designing synthetic cellular environments makes use of the observation that most cell signaling events are initiated through recruitment and clustering of transmembrane receptors by extracellular-presented signaling molecules. These systems allow for studying protein clustering in cells and characterizing the signaling response induced by, e.g., integrin activation. We review the findings about the regulation of cell adhesion and focal adhesion assembly by micro- and nanopatterns and discuss the possible use of substrate stiffness and patterning in mimicking both physical and chemical cues of the extracellular space.
Most cells from solid tissues grow as an adherent monolayer and, unless they have been transformed and become anchorage-independent, need to attach and spread in order to perform cellular functions, such as proliferation and differentiation. An essential part in the formation of tissues, in which cells are assembled and bound together, is played by the extracellular matrix (ECM). This complex network of cell-secreted molecules can vary in composition and organization to generate a variety of different forms, depending on the functional requirements of the tissue. There are three major functions for the ECM. First, it provides structural support and tensile strength. Second, it represents a substrate for cell adhesion and cell migration. Third, it regulates cellular differentiation and metabolic function, for example, modulating cell growth by binding of growth factors. Interdisciplinary efforts from the field of material science, chemistry, physics, and biology aim at functionalizing materials to obtain specific cell surface interactions and to direct cell functions at the material interface. These materials are often coated with cell adhesive proteins (Arnold et al., 2004; Cavalcanti-Adam et al., 2006; Kaehler et al., 1989) or tissue specific growth factors (Liu et al., 2006).
A major role in regulating cell adhesion and function is played by specific motifs or sequences present in ECM molecules, to which cells bind via distinct molecular mechanisms. The arginine-glycine-aspartate (RGD) sequence is the cell attachment site of a large number of adhesive ECM, blood, and cell surface proteins (Ruoslahti, 1996). Proteins that contain the RGD motif, together with the integrins that serve as their cellular receptors, constitute a major recognition system for cell adhesion. Such short sequences, if in solution, inhibit the attachment of the cells to a fibronectin matrix. However, if coupled to a solid surface, they promote cell adhesion to it (Pierschbacher et al., 1994; Pierschbacher and Ruoslahti, 1987). The RGD sequence is by far the most often employed peptide sequence for stimulated cell adhesion on surfaces. As the integrin-mediated cell attachment influences and regulates cell migration, growth, differentiation, and apoptosis, RGD peptides can be used to probe integrin functions in various biological systems. One important aspect in surface functionalization with adhesive peptides is the way the peptide is linked to the surface; this is usually obtained by covalent bond formation between a surface carboxylic group and the amino-terminus of the peptide (Dee et al., 1998), though the use of thiol groups introduced in the peptide sequence provides control over the orientation and higher stability (Pfaff et al., 1994). Furthermore, the spatial arrangement of the ligand is also essential in determining the interactions with the receptor (Xiong et al., 2002). By using surface patterning methods at the micro- and nanometer length scale it is possible to combine both precision in the presentation of the ligand and control over the density and spacing of these adhesive sequence. These techniques allow for elucidating the role of spatial organization and geometry of the ECM during cell adhesion and spreading events.
SURFACE-BOUND ADHESIVE PEPTIDES DIRECT CELLULAR ADHESION VIA INTEGRINS
Surface coatings with ECM adhesive proteins are widely used to elicit tissue specific cellular responses. For example, biomaterials designated to bone or skin regeneration are decorated with specific ECM proteins, such as collagen type I or laminin, to allow selective cell adhesion, modulate cell morphology and adhesion strength, and induce cell differentiation (Keselowsky et al., 2005; Kleinman et al., 1981; Yamamoto et al., 2000). Protein-surface interactions are highly dependent on surface properties such as charge, hydrophobicity and hydrophilicity (Bergkvist et al., 2003). These features directly determine the conformation of the adsorbed protein, which is vital for the maintenance of its biological activity (Yamamoto et al., 2007). While structural changes in ECM adhesive proteins during adsorption onto a substrate affect the molecular binding of these proteins to cell receptors and, in turn, mediate cell response (Garciaet al., 1999; Keselowsky et al., 2004), the number of available sites for the interaction with cell receptors cannot be controlled because of unpredictability of orientation and accessibility of the binding sites (Elbert et al., 2001).
ECM proteins contain many different cell recognition motives, leading to the initiation of several and undistinguished signaling cascades. On the other hand, small adhesive peptides represent only one single motif. Therefore, they can selectively address one particular type of cell adhesion receptors, such as a specific integrin receptor dimer (Meyer et al., 2006). The type of peptide anchorage to the surface and the conformation and sequence of the peptide itself are crucial aspects for surface biofunctionalization. Molecules can be either adsorbed or covalently linked via functional groups onto the surface; stable linking to a surface is essential to withstand the cells contractile forces (Katz et al., 2000) or prevent internalization (Memmo and McKeown-Longo, 1998). Smaller peptidic sequences show higher selectivity for a specific integrin type; for example, fragments consisting essentially of the 10th type III domain of fibronectin, bind better to a vitronectin receptor (αvβ3 integrin) than to a fibronectin receptor (α5β1) (Pytela et al., 1985). However, not all ligands functional minimal peptide sequences are available or identified. Therefore, the selective immobilization of entire proteins to surfaces is still necessary for many applications. When chemically or physically adsorbed to a surface, proteins undergo conformational changes leading to denaturation and loss of function. In order to preserve the activity of proteins on the surface site-directed immobilization via NTA∕his-tag or biotin∕streptavidin is the method of choice (Groll et al., 2005;Wolfram et al., 2007).
In many in vitro studies, investigators have bound peptides to surfaces to test their impact on cell behavior and to prove their applicability for biological use; these studies include investigation of cell attachment and spreading, cytoskeletal reorganization and formation of focal adhesions (FAs), as well as integrin-dependent signaling, such as proliferation and survival. In general, the bidirectionality of integrin-mediated cellular responses is exemplified by the fact that the extracellular binding activity of integrins is regulated from the inside of the cell (inside-out signaling), while the binding to the ECM elicits signals that are transmitted into the cell (outside-in signaling) (Hynes, 1992). The large variability in ligand recognition sequences and ligand structure results in a variety of different cellular responses, each dependent on integrin engagement with ECM-derived adhesive motifs (Giancotti, 2000; Giancotti and Ruoslahti, 1999). Laminin-based peptides, such as the adhesion ligand YIGSR, have been used to promote cell spreading and stress fiber formation when its conformation was constrained by covalent immobilization through the glycine residue at the N-terminus (Massia et al., 1993). Rezania and Healy (1999) showed that the response of cells on RGD-grafted surfaces is dependent on the specific peptide sequence. RGD peptides enhance the rate of cell spreading and adhesion strength (McFarland et al., 1999; Rezania et al., 1997). Cell binding to covalently linked RGD peptides causes increase in expression of integrins and FA molecules and promotes resistance to apoptotic stimuli (Cavalcanti-Adam et al., 2002; Grigoriou et al., 2005). Furthermore, the combination of RGD-containing adhesive peptides covalently linked to surfaces with soluble growth factors has been shown to affect cell proliferation and motility of endothelial cells (Dee et al., 1995). Finally, mixed peptide surfaces presenting either motifs of different ECM molecules, for example, ligands for different integrin receptors, or two distinct binding domains of the same ECM protein, such as cell-and heparin-binding domains of bone sialoprotein, further enhance the effects of surfaces as compared to those coated with single adhesive peptides (Healy et al., 1999) while decreasing the specificity of the adhesion-dependent signaling.
THE NANOSCALE ARRANGEMENT OF ECM ADHESIVE PEPTIDES CONTROLS CELLULAR RESPONSES
In vitro strategies for mimicking the extracellular space aim at addressing the variability of ECM architecture encountered by cells in vivo. In order to reproduce ECM topographies, like meshwork or fibrillar structures, systematic variation of collagen fibril densities in three-dimensional (3D) ECM microenvironments affects fibroblast size, morphology, and contractile force generation (Pizzo et al., 2005). The surface distribution of adhesive molecules has a strong influence on cell behavior; however, most of the studies on surface coating are based on the assumption that there is an equal distribution of these molecules on the surfaces. To better control the spatial arrangement of ECM molecules and peptides on cell-adhesive surfaces, several methods have been described over the past 10 years for patterning the immobilization of proteins or peptides. These methods combine a mean of imposing a pattern on the substrate with a mean of modifying surface properties so that one region of the substrate promotes cell attachment while the other region prevents adhesion.
Several methods have been developed for patterning surfaces at nanometer resolution to control the position of peptide clusters or of single adhesive ligands. At first, mere surface topography effects on cell adhesion, morphology, and gene expression in cells adhering to islands as low as 13 nm in height indicate that cells sense and react to such small surface features (Biggs et al., 2007; Dalby et al., 2004; 2002). Nanopits, ranging in diameter from 35 to 200 nm and arranged in well-ordered orthogonal or hexagonal patterns, have also been shown to affect cell responses (Curtis et al., 2004). Being nanometer topographies in the range of size of single biomacromolecules, the targeted functionalization of such platforms with adhesive peptides or small fragments of ECM molecules is gaining consensus in the field of cell adhesion. In particular, as indicated by crystallography, the size of single integrin receptors ranges from 9 to 12 nm (Xiong et al., 2001; 2002); patterning methods at the micrometer level do not allow exploring the relationships of integrin-mediated adhesion site size and distribution, with the control of integrin receptor clustering, while substrates patterned with ligands at the nanoscale level are suitable for addressing this aspect of cell-ECM interactions (Fig. 1).
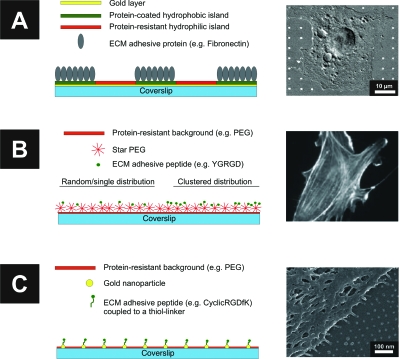
Figure 1. Surface patterning for studying cell adhesion: from micro-to nanometer-sized adhesive islands.
(A) Microcontact printing to generate fibronectin coated islands in the range of 0.3–3 μm2 and separated by 1–30 μm. The protein adsorbs only to the hydrophobic areas while the remaining regions are blocked with a protein resistant hydrophylic layer. Bottom, a cell adhering to a micropattern is shown by overlay of fluorescence and DIC microscopy [from Lehnert (2004), J. Cell. Sci.117, 41–52. Reproduced with permission of the Company of Biologists.]. (B) RGD peptides are presented on star polymers to vary the total average concentration and the spatial distribution. The actin cytoskeleton of a cell adhering to 9-RGD star polymer substrate is imaged by fluorescence microscopy [from Maheshwari (2001), J. Cell. Sci.113, 1677–1686. Reproduced with permission of the Company of Biologists.]. (C) RGD nanopatterns arranged in a hexagonal lattice over a nonadhesive background to study the effect of single ligand spacing on cell adhesion. Scanning electron microscopy (SEM) image of a cell adhering to a 58 nm interparticle distance between RGD-functionalized gold nanoparticles (adapted from Arnold, 2004).
Integrin-mediated cell adhesion depends not only on receptor occupancy but also on receptor clustering. In fact, binding of ligands to integrin receptors induces changes in conformation resulting in clustering of the receptors, an event which provides binding sites for the proximal intracellular components of FAs. This triggers rearrangement of the cytoskeleton, phosphorylation of cytoskeletal proteins, and activation of kinases and genomic responses. Being these events regulated by the ECM network, RGD-functionalized biomaterials have proven to stimulate different cellular responses, not only by different average peptide surface concentrations, but also by presenting such peptides in a way that enables or even triggers integrin aggregation. Systematic investigations on how the spatial organization and lateral distances of adhesive peptides affect cell adhesion and behavior are still very few. Maheshwari et al. (2000) functionalized star-shaped polymers with RGD-containing peptapeptide over a nonadhesive background to achieve a controlled independent and systematic variation of surface density and local spatial distribution of the peptide. When the RGD peptide was presented in clusters of at least five peptides per star, but not in case of random single RGD peptide per star, cells developed well-formed actin stress fibers and mature FAs.
To study the effects of ligand density and clustering on fibroblast adhesion and to relate adhesion forces to these parameters, substrates with comb-shaped copolymers presenting GRGDSPK in an ordered or clustered array were used (Koo et al., 2002). At higher ligand clusters and higher ligand densities adhesion reinforcement in response to force application indicated that ligand spacing and ECM rigidity are cues playing a central role in the control of cell responses to the environment. Dip-pen nanolithography has been proposed as method to directly pattern monolayer into nanometer scaled islands in the range of 30 nm; as such, higher flexibility in nonperiodic patterns can be achieved (Lee et al., 2002). Comisar et al. (2006) investigated the effects of RGD nanopatterns, which are independent of bulk density, on cell adhesion and differentiation. While the number of RGD peptides per alginate island was significant for cell spreading and differentiation, the patterns presenting more closely spaced islands favor cell spreading, but only in case of highly spaced islands cell differentiation was observed. These studies lead to the hypothesis that cell spreading might be dependent on critical densities of submicron integrin clusters to then begin the recruitment of FA and cytoskeleton proteins. Such a local integrin density could be critical for the initiation of mature and stable FA assembly. To address these open questions regarding the role of integrin local densities and lateral clustering, a peptide-patterning method using block copolymer nanolithography (BCN) has been developed, recently. This technique is based on the self-assembly driven deposition of spherical PS-b-PVP micelles on solid substrates. The micellar core can be loaded with a metal precursor salt like HAuCl4, which upon treatment with a reactive gas plasma form metal particles in the predefined hexagonal pattern. By using diblock copolymers of different molecular weight the separation distance between nanoparticles can be tuned (Glass et al., 2003). Such extended and regularly spaced platforms of nanometer-size particles represent a useful tool to study the dependence of ECM-cell interactions on ligand spacing. To selectively study integrin-mediated cell adhesion and function, the gold particles, having a size of <8 nm which allows the binding of a single integrin heterodimer, have been rendered bioactive by linking cyclic RGD peptides. The area between the particles was passivated by the deposition of a protein repellant poly(ethylene glycol) layer (Blümmel et al., 2007). These types of RGD-nanopatterns have been shown to promote cell adhesion via integrin αvβ3 receptors (Cavalcanti-Adam et al., 2006). The hypothesis that the nanometer lateral spacing between single integrin ligands regulates cell spreading and adhesion sites stability has been supported by several observations. Fibroblast initial spreading is delayed and, even though lamellipodia and spike-like structures are formed, increased ruffling of the cell membrane occurs if RGD peptides are too highly spaced, i.e., above a threshold of 73 nm. Furthermore, cells lose the stability of their contacts to the surface and undergo major changes in shape and polarity. These observations indicate that the ability of fibroblasts to form lamellipodia is not influenced by the distance between ECM ligands, but the formation of stable contacts, and the maintenance of cell shape (Cavalcanti-Adam et al., 2007) and cell adhesion force (Selhuber-Unkel et al., 2008) is dependent on such distance. When plated for 24 h on RGD nanopatterns these effects were still evident, indicating that less cells managed to adhere to patterns of RGD-peptide spacing of 73 nm and above and that the adhering cells were in a quiescent state (Arnold et al., 2004) (Fig. 2).
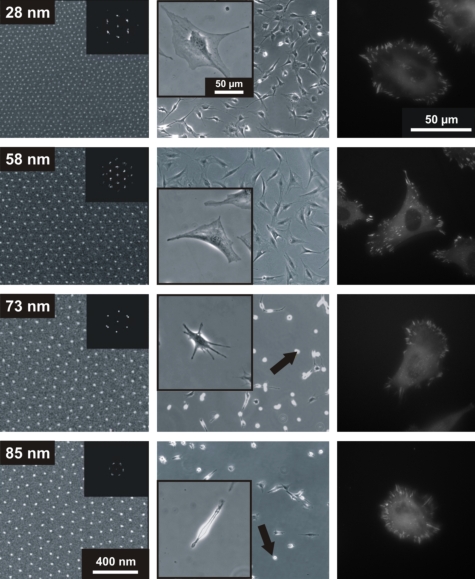
Figure 2. The spacing between hexagonal arrays of gold nanoparticles functionalized with RGD affects cell spreading and focal adhesion formation.
First column: SE micrographs of nanopatterns of gold dots separated by 28, 58, 73, and 85 nm, respectively. Second column: phase contrast microscopy images of cells adhering to the nanopatterned surfaces presenting gold dots functionalized with cRGDfK peptides. The arrows indicate nonadherent cells [from Arnold et al. (2004), ChemPhysChem5(3), 383–388. Copyright © Wiley-VCH Verlag GmbH & Co. KGaA.]. Third column: fibroblasts stably transfected with YFP-paxillin (a focal adhesion molecule) adhering to RGD nanopatterns imaged by fluorescence microscopy.
UNCOUPLING LOCAL FROM GLOBAL LIGAND DENSITY BY MICRONANOPATTERN
Micropatterning methods allow the spatial control of adhesion of entire cells onto discrete regions of the substrate by combining both topographical and chemical cues of the ECM (Jung et al., 2001). Chen et al. (1997) showed that the restriction in cell spreading and shape by adhesive islands of decreasing size controls cell fate, regardless of the type of ECM coating used. This study pointed out for the first time the role of geometry on cell shape and survival, a mechanism, which might participate to the regulatory function of cell microenvironment during morphogenesis or malignant transformation. The geometric limits of ECM binding sites for fibronectin molecules necessary for cell attachment and spreading have been investigated by using regular micrometer-sized island produced by microcontact printing (Lehnert et al., 2004). Three different thresholds, which affect cell behavior, could be identified: the molecule density per island, the maximal distance between the islands and, lastly, the surface coverage with the ECM molecules. These parameters proved to be relevant for the regulation of the amount of adhesion sites in cells and in turn for the support early cell spreading.
The number of attached cells is clearly related to the peptide density on the surface: a critical minimum average (global) density for cell response sufficient for cell spreading and formation of FAs and stress fibers was measured (Massia and Hubbell, 1991). Based on ligand density, an average distance between the ligands could be estimated, remaining, however, still difficult since it is not possible to control and determine the clustering of ligands in the matrix and such local ligand density.
As described above, block-copolymer micelle nanolithography is able to spatially control ligand receptor interaction at the nanometer scale. However, by changing the interparticle distance the density of particles on the surface is concomitantly altered. Substrates where micrometer-sized nanostructured patches are surrounded by bare substratum overcome this constraint. These micronanostructured surfaces can help to differentiate whether a signal triggered by an array of biofunctionalized gold nanoparticles is resulting from the induced proximity of the ligand-coupled receptor clusters or simply depending on the amount of bound receptor. As shown by (Arnold et al., 2004), not the total number of RGD functionalized nanoparticles, but rather the spatial confinement of the integrin αvβ3 was crucial for proper cell attachment and spreading. Here, cells on surfaces with an interparticle spacing of 73 nm could not adhere and no FAs were formed, but FAs were perfectly established on surfaces with 58 nm spaced nanoparticles in micropatches even though the average (global) concentration of particles on the surface was lower than in the latter case. The fabrication process involves the selective irradiation of a micellar monolayer with ultraviolet light (Gorzolnik et al., 2006), focused ions (Mela et al., 2007) or electrons (Glass et al., 2003) and removal of nonirradiated parts. The different methods exhibit advantages and limitations with regard to structured surface area, feature size, and necessary equipment. Micellar electron-beam lithography has shown to be well suitable to meet the demands of cell adhesion studies. Arbitrary patterns can be written with feature sizes in the sub 100 nm regime—the distance of the micelles themselves—with surface areas sufficient for microscopy studies (Fig. 3).
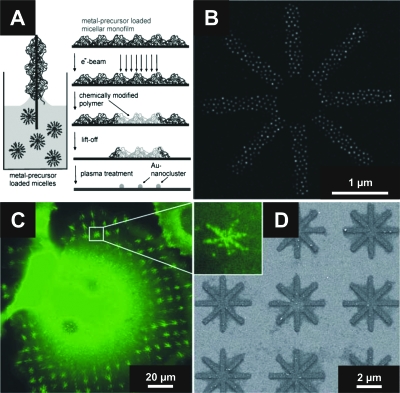
Figure 3. Hexagonally ordered gold nanoparticle arrays can be arbitrarily patterned by means of micellar electron beam lithography.
(A) The fabrication process involves formation of micellar monolayer, irradiation with electrons to pin micelles to the substrate, removal of nonirradiated parts, and a plasma process to generate the particles [from Glass et al. (2003), Adv. Funct. Mat.13(7), 567–575. Copyright © Wiley-VCH Verlag GmbH & Co. KGaA.]. (B) shows a SE micrograph of a star-shaped micronanostructure [from Glass et al. (2003), Nanotechnology14(10), 1153–1160. Copyright © IOP Publishing Ltd.]. (C) REF52-YFP-paxillin cells were seeded onto an array (D) of cRGDfK-functionalized star-shaped micronanostructures and imaged by fluorescence microscopy. Focal contact formation visualized by accumulation of YFP fluorescence exclusively occurred on micronanostructures and adapted the star-like shape as illustrated in the inset.
DEFINED GRADIENTS OF ECM PEPTIDES DIRECT CELL POLARIZATION AND MIGRATION
Cell polarization and directed cell migration play a crucial role in many physiological processes, such as embryonic development, immune response and angiogenesis, as well as in pathological processes, like inflammation and cancer metastasis (Lauffenburger and Horwitz, 1996). The biased migration of eukaryotic cells towards a gradient of soluble chemoattractant molecules, defined as chemotaxis, has been extensively investigated in leukocytes and Dictyostelium [recently reviewed in Kay et al. (2008)]. The orientation of cells in these chemotactic gradients is highly dependent on the gradient strength: while steep gradients, particularly if cells are not polarized, induce polarization and movement towards the source of chemoattractant (Zhelev et al., 2004), weaker gradients cause cell steering and gradual reorientation due to random splitting of pseudopodes and favoring of the most accurate structures formed along the gradient (Andrew and Insall, 2007).
If exposed to a gradient of cellular adhesion sites, or substrate-bound chemoattractants, cells acquire a directional motility, termed haptotaxis. Such gradients are presented by ECM components in different tissues of the body. The functional role of ECM gradients still remains to be elucidated, though their role in guiding the migration of mesenchymal stem cells appears to be crucial during tissue repair or regeneration. For example, fibronectin, collagen I and vitronectin, presented in both soluble and bound gradient, are able to induce cell motility (Thibault et al., 2007). Also proteoglycans play a role in the spatial and temporal control of cell movement imposing directionality and stop-go choice of migrating cells (Cattaruzza and Perris, 2005). To create in vitro adhesive haptotactic gradients, different techniques, based on either microfluidic systems or on spatially controlled placement of adhesive molecules, have been developed. A microfluid system has been fabricated to obtain laminin gradients and to control the orientation of axons of neuronal cells towards the denser laminin concentrations (Dertinger et al., 2002). Furthermore, fibronectin gradients generated by microfluidics induce fibroblast haptotaxis towards higher fibronectin concentration; however, the net movement directly correlates with the gradient slope, while the overall rate of migration does not (Rhoads and Guan, 2007). Herbert et al. (1997) introduced a method for using photoimmobilization of peptides on self-assembled monolayer, while (Wang et al., 2004) merged electrochemical potential gradients with electrosorption of organothiols. Smith et al. (2004) produced surface-bound gradients of fibronectin by the crossdiffusion of inert and reactive functionalized alkanethiols on gold. The formation of self-assembled gradients of COOH functional groups allows the covalent link to N-groups of fibronectin. On these substrates, endothelial cells show morphological polarization, which correlates with cellular drift speed; the drift speed and the discrete cellular motion proportionally increase with the gradient slope, but no effects are observed on the persistence time or on random speed (Smith et al., 2006). As such, this system introduced for the first time the role of different haptotactic gradient slope in the regulation of cell migration. Finally, fibroblast adhesion to RGD gradients, which are generated in a polymerization apparatus with a gradient pump, leads to cell alignment and higher eccentricity on the gradient axis (Kang et al., 2004).
To date, however, only with few methods it is possible to obtain a highly controlled presentation of surface-bound gradients. Furthermore, it still remains difficult to exclude local aggregation effects and achieve a precise spatial positioning of the surface-bound ligands. The latter, however, is crucial for studying cooperative effects of receptor clustering and spacing in signal transduction and for understanding the mechanism of gradient sensing at membrane protrusions (Maheshwari et al., 2000). Different cellular responses can be triggered not only by varying the average bound concentration of bioactive molecules on the surface, but also by controlling the mere relative positioning of such molecules (as previously discussed). To apply this concept to immobilized molecular gradients, a modified substrate dip-coating process of BCN for the nanopatterning of solid surfaces has been recently developed (Arnold et al., 2008). Here, a linear increase of the distance between RGD-functionalized gold nanoparticles, surrounded by inert PEG background, is achieved at the nanoscale level. As shown in Fig. 4, with this technique a surface-bound cyclicRGDfK peptide nanoscale gradient with increasing ligand spacing from 50 to 80 nm can be produced. As the interparticle distances increase, the number of attached cells as well as the projected cell area become significantly smaller; this is in line with the observation that cell adhesion area depends strongly on the strength of adhesion to the surrounding environment (DiMilla et al., 1993). After 24 h in culture, a significant number of cells align toward the direction of the gradient. The most striking finding is that cells can sense small, but consistent, differences in ligand spacing presented along the front and the back of their body; this difference, which is as little as 1 nm across the cell diameter, seems to affects cell polarization and directed migration. By coupling opposite FAs via actin filaments and myosin-based contraction, it is very likely that cells test the mechanical stability of spatially distributed FAs and therefore migrate towards smaller spaced peptide presentations. Taken all together, these results point out the relevance of nanometer spatial variations of ECM cues in directing integrin receptor clustering-based responses; the differential positional clustering of integrins could be then be interpreted as a cellular control mechanism for essentially screen matrices for their nanoscopic peptide presentation which directly regulates cellular functions, such as spreading and migration.
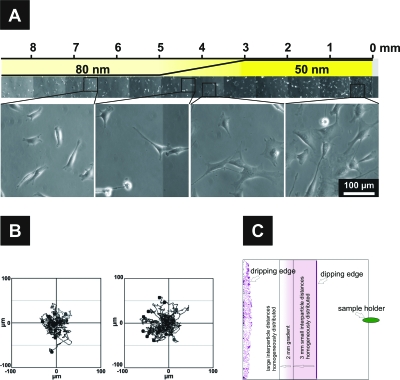
Figure 4. Biofunctionalized cRGDfK particle spacing gradient.
(A) Composite phase-contrast micrographs showing the adhesion of cells to the different areas of the substrate after 21 h and close-up on patch spacing, at substrate areas offering 50, 60, 70, and 80 nm patch spacing. (B) Migration pathways of fibroblasts after 13 h for 12 h with a time lapse of 10 min on a constant ligand patch spacing of 60 nm; and (left) a ligand patch gradient with a strength of Δ2 nm∕mm covering 60–110 nm spacing (right) [Reprinted with permission from the Americal Chemical Society, from Arnold et al. (2008), Nano Lett.8(7), 2063–2069. Copyright © 2008 American Chemical Society.]. (C) Schematic drawing of the dip-coated substrates illustrating the differently patterned areas, including the dripping edge, an area of uncontrolled nanoparticle aggregation forming upon solvent evaporation [from Hirschfeld–Warneken et al. (2008), Eur. J. Cell Biol.87(8–9), 743–750.].
EMERGING STRATEGIES FOR COMBINING PHYSICAL AND CHEMICAL PROPERTIES IN PATTERNED MATERIALS
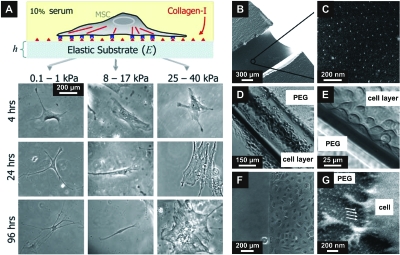
As discussed in the previous sections, the ECM displays adhesive ligands crucial for proper adhesion-dependent responses; however, the ECM exhibits also a number of relevant physical properties. In fact, the ability of the ECM to regulate cell fate has been recently investigated, particularly regarding the role of its composition, concentration, and stiffness in the regulation of key cellular processes, such as motility (Pelham and Wang, 1997), phagocytosis (Beningo and Wang, 2002), and differentiation (Cukierman et al., 2001). For example Engler et al. (2006) have mirrored tissue-level elasticity by using soft matrices to induce neurogenic differentiation, stiffer matrices for myogenic differentiation, and more rigid matrices for osteogenic differentiation (Fig. 5).
Figure 5. Substrate compliance influences cellular differentiation.
(A) Naïve mesenchymal stem cells on soft substrates differentiate into lineages with the respective native tissue elasticity. Collagen coated soft matrices induce neurogenic, stiffer matrices myogenic, and relatively rigid substrates osteogenic lineage commitment [from Engler et al. (2006), Cell126(4), 677–689. Copyright © 2006 by Cell Press.]. (B) and (C) Transfer lithography allows the nanopatterning of soft poly(ethylene glycol) hydrogels with elastic moduli similar to those used in (A). (D) and (E) Patterned tubes inside an inert hydrogel matrix overgrown with adherent cells mimic a blood vessel. (F) and (E) Cells adhere exclusively to patterned areas, while bare PEG hydrogels sustain their protein and cell repellent properties over days, as evidenced by the sharp line with nanostructures on the right side [Reprinted with permission from the American Chemical Society, from Graeter et al. (2007), Nano Lett.7(5), 1413–1418. Copyright © 2007 American Chemical Society.].
It remains still a challenge to determine how matrix stiffness couples with ligand density to modulate cellular responses; recently, it has been suggested that substrate compliance and ligand density are orthogonal determinants (Cukierman et al., 2001; Geiger, 2001) of similar importance in regulating, for example, migratory responses. In general, very soft substrates are perceived by a cell as inadequate for supporting anchorage-dependent events, while harder ones trigger acto-myosin contractility and sustain cell spreading and traction (Pelham and Wang, 1997). As shown by Lo et al. (2000), the points of higher stress in a cell adhering to a soft substrate, such as a hydrogel, are at the periphery and these forces generated by a cell are about 15%–25% of the substrate modulus.
Inert hydrogels, such as PEG, functionalized with adhesive motifs allow the investigation of adhesion on combined platforms for 3D culture and ligand concentrations. By using different elastic moduli for collagen-coated gels at varying concentration, Engler et al. (2004) investigated cell spreading and cytoskeletal organization in smooth muscle cells. Cells spreading on soft gels are unresponsive to collagen density, leading to the conclusion that cytoskeletal contractility plays a dominant role during cell spreading, while the regulation of adhesion by adhesive ligand concentrations is crucial for the reinforcement of the spreading process. Higher cell survival in PEG gels has been shown in the presence of RGD peptides, if presented with the proper conformation and spacer arm length (Salinas and Anseth, 2008).
To further introduce surface topographies onto these soft materials, micropattern transfer techniques based on photolithography have been recently employed; as such, both peptide functionalization and patterning strategies can be combined (Segura et al., 2005). The use of BCN transferred on PEG hydrogels has been successfully used to generate nanopatterns with flexible design and high precision of the pattern (Graeter et al., 2007). The gold nanoparticles embedded in the gel can be functionalized with adhesive peptides to couple ligand densities and spacing at the nanometer length with varying substrate stiffness. From these studies, it can be evinced that responses to physical cues of the ECM act synergistically with chemical factors, and only the combination of both physical and chemical aspects of the cell microenvironment strongly affects cellular responses [Figs. 5b, 5c, 5d, 5e, 5f, 5g].
CONCLUSION
The preparation of biointerfaces reached a quality and riches in molecule presentations to cells and in its surface physical properties. Identifying the relevant parameters, which regulate cell adhesion in vitro, is an essential aspect for mimicking functionality of in vivo scaffolds for cells. Here, we reviewed cell micro- and nanopatterning techniques based on chemically modified surfaces functionalized with adhesive molecules. The use of nanopatterned substrates, prepared by block copolymer nanolithography so far allowed great progress in the field of cell biology and tissue engineering. In particular, by using these types of synthetic substrates, it is possible to disentangle quantitatively the contribution of different material parameters to cell responses down to the level of controlling the clustering of individual cell transmembrane receptors, such as integrins.
In addition to substrates whose patterns are rigid, substrates have been designed to control the spatial and density distribution of ECM ligands. As such, they allow one to differentiate the global from local density and to determine whether the proximity of the receptors, rather than the number of bound receptors, is crucial for cell adhesion and focal adhesion formation. Substrates decorated with nanoscale gradients of ECM ligands could in the future represent useful platforms to determine the role of differential spatial clustering of adhesive receptors during cell adhesion and migration for screening the extracellular environment. Finally, dynamic substrates, whose cell adhesiveness can be changed by varying the compliance, have recently been the focus of attention because of their ability to guide cell differentiation and function. By combining both physical and chemical cues such in vitro complex synthetic materials will allow us to gain fundamental insights into adhesion-dependent cellular functions and to use these ECM mimics for therapeutic applications.
ACKNOWLEDGMENTS
We thank the Landesstiftung Baden-Württemberg in the frame of the program “Spitzenforschung Baden-Württemberg” and the Max-Planck-Society for the constant financial support.
References
- Andrew, N, and Insall, R H (2007). “Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions.” Nat. Cell Biol. 9, 193–200. [DOI] [PubMed] [Google Scholar]
- Arnold, M, Cavalcanti-Adam, E A, Glass, R, Blummel, J, Eck, W, Kantlehner, M, Kessler, H, and Spatz, J P (2004). “Activation of integrin function by nanopatterned adhesive interfaces.” ChemPhysChem 10.1002/cphc.200301014 5, 383–388. [DOI] [PubMed] [Google Scholar]
- Arnold, M, Hirschfeld-Warneken, V C, Lohmuller, T, Heil, P, Blummel, J, Cavalcanti-Adam, E A, López-García, M, Walther, P, Kessler, H, Geiger, B, and Spatz, J P (2008). “Induction of cell polarization and migration by a gradient of nanoscale variations in adhesive ligand spacing.” Nano Lett. 8, 2063–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beningo, K A, and Wang, Y L (2002). “Flexible substrata for the detection of cellular traction forces.” Trends Cell Biol. 10.1016/S0962-8924(01)02205-X 12, 79–84. [DOI] [PubMed] [Google Scholar]
- Bergkvist, M, Carlsson, J, and Oscarsson, S (2003). “Surface-dependent conformations of human plasma fibronectin adsorbed to silica, mica, and hydrophobic surfaces, studied with use of atomic force microscopy.” J. Biomed. Mater. Res. 64, 349–356. [DOI] [PubMed] [Google Scholar]
- Biggs, M J, Richards, R G, Gadegaard, N, Wilkinson, C D, and Dalby, M J (2007). “The effects of nanoscale pits on primary human osteoblast adhesion formation and cellular spreading.” J. Mater. Sci.: Mater. Med. 18, 399–404. [DOI] [PubMed] [Google Scholar]
- Blümmel, J, Perschmann, N, Aydin, D, Drinjakovic, J, Surrey, T, López-García, M, Kessler, H, and Spatz, J P (2007). “Protein repellent properties of covalently attached PEG coatings on nanostructured SiO(2)-based interfaces.” Biomaterials 28, 4739–4747. [DOI] [PubMed] [Google Scholar]
- Cattaruzza, S, and Perris, R (2005). “Proteoglycan control of cell movement during wound healing and cancer spreading.” Matrix Biol. 24, 400–417. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam, E A, Micoulet, A, Blummel, J, Auernheimer, J, Kessler, H, and Spatz, J P (2006). “Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly.” Eur. J. Cell Biol. 10.1016/j.ejcb.2005.09.011 85, 219–224. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam, E A, Shapiro, I M, Composto, R J, Macarak, E J, and Adams, C S (2002). “RGD peptides immobilized on a mechanically deformable surface promote osteoblast differentiation.” J. Bone Miner. Res. 17, 2130–2140. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam, E A, Volberg, T, Micoulet, A, Kessler, H, Geiger, B, and Spatz, J P (2007). “Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands.” Biophys. J. 10.1529/biophysj.106.089730 92, 2964–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C S, Mrksich, M, Huang, S, Whitesides, G M, and Ingber, D E (1997). “Geometric control of cell life and death.” Science 10.1126/science.276.5317.1425 276, 1425–1428. [DOI] [PubMed] [Google Scholar]
- Comisar, W A, Hsiong, S X, Kong, H J, Mooney, D J, and Linderman, J J (2006). “Multi-scale modeling to predict ligand presentation within RGD nanopatterned hydrogels.” Biomaterials 27, 2322–2329. [DOI] [PubMed] [Google Scholar]
- Cukierman, E, Pankov, R, Stevens, D R, and Yamada, K M (2001). “Taking cell-matrix adhesions to the third dimension.” Science 10.1126/science.1064829 294, 1708–1712. [DOI] [PubMed] [Google Scholar]
- Curtis, A S, Gadegaard, N, Dalby, M J, Riehle, M O, Wilkinson, C D, and Aitchison, G (2004). “Cells react to nanoscale order and symmetry in their surroundings.” IEEE Trans. Nanobiosci. 10.1109/TNB.2004.824276 3, 61–65. [DOI] [PubMed] [Google Scholar]
- Dalby, M J, Giannaras, D, Riehle, M O, Gadegaard, N, Affrossman, S, and Curtis, A S (2004). “Rapid fibroblast adhesion to 27 nm high polymer demixed nano-topography.” Biomaterials 25, 77–83. [DOI] [PubMed] [Google Scholar]
- Dalby, M J, Yarwood, S J, Riehle, M O, Johnstone, H J, Affrossman, S, and Curtis, A S (2002). “Increasing fibroblast response to materials using nanotopography: morphological and genetic measurements of cell response to 13-nm-high polymer demixed islands.” Exp. Cell Res. 10.1006/excr.2002.5498 276, 1–9. [DOI] [PubMed] [Google Scholar]
- Dee, K C, Anderson, T T, and Bizios, R (1995). “Enhanced endothelialization of substrates modified with immobilized bioactive peptides.” Tissue Eng. 1, 135–145. [DOI] [PubMed] [Google Scholar]
- Dee, K C, Andersen, T T, and Bizios, R (1998). “Design and function of novel osteoblast-adhesive peptides for chemical modification of biomaterials.” J. Biomed. Mater. Res. 40, 371–377. [DOI] [PubMed] [Google Scholar]
- Dertinger, S K, Jiang, X, Li, Z, Murthy, V N, and Whitesides, G M (2002). “Gradients of substrate-bound laminin orient axonal specification of neurons.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.192457199 99, 12542–12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMilla, P A, Stone, J A, Quinn, J A, Albelda, S M, and Lauffenburger, D A (1993). “Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength.” J. Cell Biol. 10.1083/jcb.122.3.729 122, 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbert, D L, Pratt, A B, Lutolf, M P, Halstenberg, S, and Hubbell, J A (2001). “Protein delivery from materials formed by self-selective conjugate addition reactions.” J. Controlled Release 76, 11–25. [DOI] [PubMed] [Google Scholar]
- Engler, A, Bacakova, L, Newman, C, Hategan, A, Griffin, M, and Discher, D (2004). “Substrate compliance versus ligand density in cell on gel responses.” Biophys. J. 86, 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler, A J, Sen, S, Sweeney, H L, and Discher, D E (2006). “Matrix elasticity directs stem cell lineage specification.” Cell 10.1016/j.cell.2006.06.044 126, 677–689. [DOI] [PubMed] [Google Scholar]
- Garcia, A J, Vega, M D, and Boettiger, D (1999). “Modulation of cell proliferation and differentiation through substrate-dependent changes in fibronectin conformation.” Mol. Biol. Cell 10, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger, B (2001). “Cell biology. Encounters in space.” Science 294, 1661–1663. [DOI] [PubMed] [Google Scholar]
- Giancotti, F G (2000). “Complexity and specificity of integrin signalling.” Nat. Cell Biol. 2, E13–E14. [DOI] [PubMed] [Google Scholar]
- Giancotti, F G, and Ruoslahti, E (1999). “Integrin signaling.” Science 10.1126/science.285.5430.1028 285, 1028–1032. [DOI] [PubMed] [Google Scholar]
- Glass, R, Arnold, M, Blümmel, J, Küller, A, Möller, M, and Spatz, J P (2003). “Micronanostructured interfaces fabricated by the use of inorganic block copolymer micellar monolayers as negative resist for electron-beam lithography.” Adv. Funct. Mater. 10.1002/adfm.200304331 13, 569–575. [DOI] [Google Scholar]
- Glass, R, Möller, M., and Spatz, J P (2003). “Block copolymer micelle nanolithography.” Nanotechnology 10.1088/0957-4484/14/10/314 14, 1153–1160. [DOI] [Google Scholar]
- Gorzolnik, B, Mela, P, and Möller, M (2006). “Nano-structured micropatterns by combination of block copolymer self-assembly and UV photolithography.” Nanotechnology 10.1088/0957-4484/17/19/042 17, 5027–5032. [DOI] [Google Scholar]
- Graeter, S V, Huang, J, Perschmann, N, López-García, M, Kessler, H, Ding, J, and Spatz, J P (2007). “Mimicking cellular environments by nanostructured soft interfaces.” Nano Lett. 7, 1413–1418. [DOI] [PubMed] [Google Scholar]
- Grigoriou, V, Shapiro, I M, Cavalcanti-Adam, E A, Composto, R J, Ducheyne, P, and Adams, C S (2005). “Apoptosis and survival of osteoblast-like cells are regulated by surface attachment.” J. Biol. Chem. 280, 1733–1739. [DOI] [PubMed] [Google Scholar]
- Groll, J, Albrecht, K, Gasteier, P, Riethmueller, S, Ziener, U, and Moeller, M (2005). “Nanostructured ordering of fluorescent markers and single proteins on substrates.” ChemBioChem 10.1002/cbic.200500041 6, 1782–1787. [DOI] [PubMed] [Google Scholar]
- Healy, K E, Rezania, A, and Stile, R A (1999). “Designing biomaterials to direct biological responses.” Ann. N.Y. Acad. Sci. 875, 24–35. [DOI] [PubMed] [Google Scholar]
- Herbert, C B, McLernon, T L, Hypolite, C L, Adams, D N, Pikus, L, Huang, C C, Fields, G B, Letourneau, P C, Distefano, M D, and Hu, W S (1997). “Micropatterning gradients and controlling surface densities of photoactivatable biomolecules on self-assembled monolayers of oligo(ethylene glycol) alkanethiolates.” Chem. Biol. 4, 731–737. [DOI] [PubMed] [Google Scholar]
- Hirschfeld-Warneken, V C, Arnold, M, Cavalcanti-Adam, A, López-García, M, Kessler, H, and Spatz, J P (2008). “Cell adhesion and polarisation on molecularly defined spacing gradient surfaces of cyclic RGDfK peptide patches.” Eur. J. Cell Biol. 87, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes, R O (1992). “Integrins: versatility, modulation, and signaling in cell adhesion.” Cell 10.1016/0092-8674(92)90115-S 69, 11–25. [DOI] [PubMed] [Google Scholar]
- Jung, D R, Kapur, R, Adams, T, Giuliano, K A, Mrksich, M, Craighead, H G, and Taylor, D L (2001). “Topographical and physicochemical modification of material surface to enable patterning of living cells.” Crit. Rev. Biotechnol. 10.1080/20013891081700 21, 111–154. [DOI] [PubMed] [Google Scholar]
- Kaehler, J, Zilla, P, Fasol, R, Deutsch, M, and Kadletz, M (1989). “Precoating substrate and surface configuration determine adherence and spreading of seeded endothelial cells on polytetrafluoroethylene grafts.” J. Vasc. Surg. 9, 535–541. [PubMed] [Google Scholar]
- Kang, C E, Gemeinhart, E J, and Gemeinhart, R A (2004). “Cellular alignment by grafted adhesion peptide surface density gradients.” J. Biomed. Mater. Res. 71, 403–411. [DOI] [PubMed] [Google Scholar]
- Katz, B Z, Zamir, E, Bershadsky, A, Kam, Z, Yamada, K M, and Geiger, B (2000). “Physical state of the extracellular matrix regulates the structure and molecular composition of cell-matrix adhesions.” Mol. Biol. Cell 11, 1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, R R, Langridge, P, Traynor, D, and Hoeller, O (2008). “Changing directions in the study of chemotaxis.” Nat. Rev. Mol. Cell Biol. 9, 455–463. [DOI] [PubMed] [Google Scholar]
- Keselowsky, B G, Collard, D M, and Garcia, A J (2004). “Surface chemistry modulates focal adhesion composition and signaling through changes in integrin binding.” Biomaterials 25, 5947–5954. [DOI] [PubMed] [Google Scholar]
- Keselowsky, B G, Collard, D M, and Garcia, A J (2005). “Integrin binding specificity regulates biomaterial surface chemistry effects on cell differentiation.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0407356102 102, 5953–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman, H K, Klebe, R J, and Martin, G R (1981). “Role of collagenous matrices in the adhesion and growth of cells.” J. Cell Biol. 10.1083/jcb.88.3.473 88, 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, L Y, Irvine, D J, Mayes, A M, Lauffenburger, D A, and Griffith, L G (2002). “Coregulation of cell adhesion by nanoscale RGD organization and mechanical stimulus.” J. Cell. Sci. 115, 1423–1433. [DOI] [PubMed] [Google Scholar]
- Lauffenburger, D A, and Horwitz, A F (1996). “Cell migration: a physically integrated molecular process.” Cell 10.1016/S0092-8674(00)81280-5 84, 359–369. [DOI] [PubMed] [Google Scholar]
- Lee, K B, Park, S J, Mirkin, C A, Smith, J C, and Mrksich, M (2002). “Protein nanoarrays generated by dip-pen nanolithography.” Science 10.1126/science.1067172 295, 1702–1705. [DOI] [PubMed] [Google Scholar]
- Lehnert, D, Wehrle-Haller, B, David, C, Weiland, U, Ballestrem, C, Imhof, B A, and Bastmeyer, M (2004). “Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion.” J. Cell. Sci. 10.1242/jcs.00836 117, 41–52. [DOI] [PubMed] [Google Scholar]
- Liu, Y, Li, J P, Hunziker, E B, and de Groot, K (2006). “Incorporation of growth factors into medical devices via biomimetic coatings.” Philos. Trans. R. Soc. London, Ser. A 364, 233–248. [DOI] [PubMed] [Google Scholar]
- Lo, C M, Wang, H B, Dembo, M, and Wang, Y L (2000). “Cell movement is guided by the rigidity of the substrate.” Biophys. J. 79, 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari, G, Brown, G, Lauffenburger, D A, Wells, A, and Griffith, L G (2000). “Cell adhesion and motility depend on nanoscale RGD clustering.” J. Cell. Sci. 113(Pt 10), 1677–1686. [DOI] [PubMed] [Google Scholar]
- Massia, S P, and Hubbell, J A (1991). “An RGD spacing of 440 nm is sufficient for integrin alpha V beta 3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation.” J. Cell Biol. 10.1083/jcb.114.5.1089 114, 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massia, S P, Rao, S S, and Hubbell, J A (1993). “Covalently immobilized laminin peptide Tyr-Ile-Gly-Ser-Arg (YIGSR) supports cell spreading and co-localization of the 67-kilodalton laminin receptor with alpha-actinin and vinculin.” J. Biol. Chem. 268, 8053–8059. [PubMed] [Google Scholar]
- McFarland, C D, Mayer, S, Scotchford, C, Dalton, B A, Steele, J G, and Downes, S (1999). “Attachment of cultured human bone cells to novel polymers.” J. Biomed. Mater. Res. 44, 1–11. [DOI] [PubMed] [Google Scholar]
- Mela, P, Gorzolnik, B, Bueckins, M, Mourran, A, Mayer, J, and Möller, M (2007). “Low-ion-dose FIB modification of monomicellar layers for the creation of highly ordered metal nanodot arrays.” Small 10.1002/smll.200600338 3, 1368–1373. [DOI] [PubMed] [Google Scholar]
- Memmo, L M, and McKeown-Longo, P (1998). “The alphavbeta5 integrin functions as an endocytic receptor for vitronectin.” J. Cell. Sci. 111(Pt 4), 425–433. [DOI] [PubMed] [Google Scholar]
- Meyer, A, Auernheimer, J, Modlinger, A, and Kessler, H (2006). “Targeting RGD recognizing integrins: drug development, biomaterial research, tumor imaging and targeting.” Curr. Pharm. Des. 12, 2723–2747. [DOI] [PubMed] [Google Scholar]
- Pelham, R J, Jr., and Wang, Y (1997). “Cell locomotion and focal adhesions are regulated by substrate flexibility.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.94.25.13661 94, 13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff, M, Tangemann, K, Muller, B, Gurrath, M, Muller, G, Kessler, H, Timpl, R, and Engel, J (1994). “Selective recognition of cyclic RGD peptides of NMR defined conformation by alpha IIb beta 3, alpha V beta 3, and alpha 5 beta 1 integrins.” J. Biol. Chem. 269, 20233–20238. [PubMed] [Google Scholar]
- Pierschbacher, M D, Polarek, J W, Craig, W S, Tschopp, J F, Sipes, N J, and Harper, J R (1994). “Manipulation of cellular interactions with biomaterials toward a therapeutic outcome: a perspective.” J. Cell. Biochem. 56, 150–154. [DOI] [PubMed] [Google Scholar]
- Pierschbacher, M D, and Ruoslahti, E (1987). “Influence of stereochemistry of the sequence Arg-Gly-Asp-Xaa on binding specificity in cell adhesion.” J. Biol. Chem. 262, 17294–17298. [PubMed] [Google Scholar]
- Pizzo, A M, Kokini, K, Vaughn, L C, Waisner, B Z, and Voytik-Harbin, S L (2005). “Extracellular matrix (ECM) microstructural composition regulates local cell-ECM biomechanics and fundamental fibroblast behavior: a multidimensional perspective.” J. Appl. Physiol. 98, 1909–1921. [DOI] [PubMed] [Google Scholar]
- Pytela, R, Pierschbacher, M D, and Ruoslahti, E (1985). “A 125∕115-kDa cell surface receptor specific for vitronectin interacts with the arginine-glycine-aspartic acid adhesion sequence derived from fibronectin.” Proc. Natl. Acad. Sci. U.S.A. 82, 5766–5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania, A, and Healy, K E (1999). “Biomimetic peptide surfaces that regulate adhesion, spreading, cytoskeletal organization, and mineralization of the matrix deposited by osteoblast-like cells.” Biotechnol. Prog. 15, 19–32. [DOI] [PubMed] [Google Scholar]
- Rezania, A, Thomas, C H, Branger, A B, Waters, C M, and Healy, K E (1997). “The detachment strength and morphology of bone cells contacting materials modified with a peptide sequence found within bone sialoprotein.” J. Biomed. Mater. Res. 37, 9–19. [DOI] [PubMed] [Google Scholar]
- Rhoads, D S, and Guan, J L (2007). “Analysis of directional cell migration on defined FN gradients: role of intracellular signaling molecules.” Exp. Cell Res. 313, 3859–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoslahti, E (1996). “RGD and other recognition sequences for integrins.” Annu. Rev. Cell Dev. Biol. 10.1146/annurev.cellbio.12.1.697 12, 697–715. [DOI] [PubMed] [Google Scholar]
- Salinas, C N, and Anseth, K S (2008). “The influence of the RGD peptide motif and its contextual presentation in PEG gels on human mesenchymal stem cell viability.” J. Tissue Eng. Regen. Med. 2, 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura, T B, Anderson, C, Chung, P H, Webber, R E, Shull, K R, and Shea, L D (2005). “Crosslinked hyaluronic acid hydrogels: a strategy to functionalize and pattern.” Biomaterials 26, 359–371. [DOI] [PubMed] [Google Scholar]
- Selhuber-Unkel, C, López-García, M, Kessler, H, and Spatz, J P (2008). “Cooperativity in adhesion cluster formation during initial cell adhesion.” Biophys. J. (in press). [DOI] [PMC free article] [PubMed]
- Smith, J T, Elkin, J T, and Reichert, W M (2006). “Directed cell migration on fibronectin gradients: effect of gradient slope.” Exp. Cell Res. 312, 2424–2432. [DOI] [PubMed] [Google Scholar]
- Smith, J T, Tomfohr, J K, Wells, M C, Beebe, T P, Jr., Kepler, T B, and Reichert, W M (2004). “Measurement of cell migration on surface-bound fibronectin gradients.” Langmuir 10.1021/la0489763 20, 8279–8286. [DOI] [PubMed] [Google Scholar]
- Thibault, M M, Hoemann, C D, and Buschmann, M D (2007). “Fibronectin, vitronectin, and collagen I induce chemotaxis and haptotaxis of human and rabbit mesenchymal stem cells in a standardized transmembrane assay.” Stem. Cells Dev. 16, 489–502. [DOI] [PubMed] [Google Scholar]
- Wang, Q, Jakubowski, J A, Sweedler, J V, and Bohn, P W (2004). “Quantitative submonolayer spatial mapping of Arg-Gly-Asp-containing peptide organomercaptan gradients on gold with matrix-assisted laser desorption∕ionization mass spectrometry.” Anal. Chem. 76, 1–8. [DOI] [PubMed] [Google Scholar]
- Wolfram, T, Belz, F, Schön, T, and Spatz, J P (2007). “Site-specific presentation of single recombinant proteins in defined nanoarrays.” BioInterphases 10.1116/1.2713991 2, 44–48. [DOI] [PubMed] [Google Scholar]
- Xiong, J P, Stehle, T, Diefenbach, B, Zhang, R, Dunker, R, Scott, D L, Joachimiak, A, Goodman, S L, and Arnaout, M A (2001). “Crystal structure of the extracellular segment of integrin alpha Vbeta3.” Science 294, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, J P, Stehle, T, Zhang, R, Joachimiak, A, Frech, M, Goodman, S L, and Arnaout, M A (2002). “Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand.” Science 296, 151–155. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A, Mishima, S, Maruyama, N, and Sumita, M (2000). “Quantitative evaluation of cell attachment to glass, polystyrene, and fibronectin- or collagen-coated polystyrene by measurement of cell adhesive shear force and cell detachment energy.” J. Biomed. Mater. Res. 50, 114–124. [DOI] [PubMed] [Google Scholar]
- Yamamoto, S, Tanaka, M, Sunami, H, Ito, E, Yamashita, S, Morita, Y, and Shimomura, M (2007). “Effect of honeycomb-patterned surface topography on the adhesion and signal transduction of porcine aortic endothelial cells.” Langmuir 23, 8114–8120. [DOI] [PubMed] [Google Scholar]
- Zhelev, D V, Alteraifi, A M, and Chodniewicz, D (2004). “Controlled pseudopod extension of human neutrophils stimulated with different chemoattractants.” Biophys. J. 10.1529/biophysj.103.036699 87, 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]