Abstract
Objectives
The Burkholderia cepacia complex (Bcc) species are important opportunistic pathogens with intrinsic antibiotic resistance. They are also well known as contaminants of disinfectants, yet their biocide susceptibility has not been studied in detail. We investigated Bcc biocide susceptibility and correlated it to their taxonomy, antibiotic susceptibility and ability to form biofilms.
Methods
Genetically distinct Bcc strains belonging to 12 of the defined species were examined. Biocide susceptibility was assessed by (i) broth dilution MIC assays, (ii) agar growth-based MBC screens and (iii) suspension tests. Antibiotic MIC was determined by Etest® strips, and the ability to form biofilms was examined in a 96-well plate assay.
Results
Biocide susceptibility varied across the Bcc complex with high MIC recorded for chlorhexidine (>100 mg/L), cetylpyridinium chloride (>200 mg/L), triclosan (>500 mg/L), benzalkonium chloride (>400 mg/L) and povidone (>50 000 mg/L). Species-dependent differences were apparent only for cetylpyridinium chloride. There was no correlation between biocide susceptibility and (i) antibiotic susceptibility or (ii) the ability to form biofilms. Biocide MBC was considerably higher than the MIC (chlorhexidine, 6-fold greater; cetylpyridinium chloride, 20-fold greater). Cystic fibrosis outbreak strains (Burkholderia multivorans Glasgow strain and Burkholderia cenocepacia ET12) possessed elevated chlorhexidine resistance, and Bcc bacteria were also shown to remain viable in current commercial biocide formulations.
Conclusions
Bcc bacteria are resistant to a wide range of biocides and further representatives of this group should be included as reference strains in the development of new anti-infectives and commercial formulations.
Keywords: antibiotics, resistance and susceptibility, minimal inhibitory concentration
Introduction
The Burkholderia cepacia complex (Bcc) species are problematic Gram-negative opportunistic pathogens, and people with cystic fibrosis (CF) are particularly prone to chronic Bcc lung infection.1 The Bcc consists of multiple closely related species, all of which have been isolated from clinical infections.1,2 Until very recently, the following 10 species had been formally designated as members of the complex:3 B. cepacia, Burkholderia multivorans, Burkholderia cenocepacia, Burkholderia stabilis, Burkholderia vietnamiensis, Burkholderia dolosa, Burkholderia ambifaria, Burkholderia anthina, Burkholderia pyrrocinia and Burkholderia ubonensis. Two recent studies have extended the complex to 17 species by the addition of Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov., Burkholderia metallica sp. nov.,4 Burkholderia lata and Burkholderia contaminans.5 Of these 17 species, B. cenocepacia and B. multivorans are most associated with CF infection, which together account for ∼85% to 95% of all Bcc infections in this disease.1 The epidemiology of Bcc infection outside CF has not been extensively studied; however, the recent studies of Reik et al.6 have shown B. cenocepacia (25.6%), B. cepacia (18.9%) and B. multivorans (15.6%) to be the most common non-CF infections seen in the USA.
An important feature of Bcc bacteria is their very high innate antimicrobial resistance to both antibiotics and biocides.1,2 Bcc species are intrinsically resistant to many antibiotics such as aminoglycosides and polymyxin B and often require combination therapy to suppress infection in CF.7 The antibiotics polymyxin, gentamicin and vancomycin are used at high concentrations in B. cepacia Selective Agar, a highly effective medium for their isolation from CF sputum.8 Nzula et al.9 compared the antibiotic susceptibility of six Bcc species and concluded that it was highly variable except for innate polymyxin resistance and not linked to the taxonomic status of the isolates examined. Efflux, the secretion of chromosomal β-lactamases and the impermeability of the outer envelope of Bcc bacteria have been implicated in antibiotic resistance.1
In contrast, the molecular basis for biocide resistance in Bcc bacteria has been poorly studied despite these organisms being linked with many instances of contamination in disinfectants and other anti-infective solutions.10 Pharmaceutical formulations containing benzalkonium chloride,11 cetylpyridinium chloride12 and chlorhexidine13,14 have become contaminated with Bcc bacteria and led to large outbreaks of infection. Even during commercial manufacture, disinfectants such as povidone-iodine may become contaminated with Bcc bacteria.15 A review of commercial recalls in the USA from 1998 to 2006 demonstrated that Bcc bacteria were the most commonly isolated Gram-negative bacteria in both sterile and non-sterile products.10 Multiple instances of a single Bcc strain causing either contamination or clinical infection at geographically distinct sites have been identified using multilocus sequence typing (MLST);16–18 these data suggest that in our burgeoning world economy, the potential for global distribution of contaminating bacteria in anti-infective agents may exist.
The aim of this study was to provide an up-to-date survey of the biocide susceptibility profiles of a representative panel of Bcc species that accounts for taxonomy and genetic diversity. To ensure that a wide genetic diversity of Bcc bacteria were sampled, isolates were also selected on the basis of their MLST type.19 Biocide susceptibility was investigated using a standardized broth dilution method to determine MICs; this was also correlated to the MBC for resistant and susceptible strains. A possible link between biocide resistance and multidrug resistance has been proposed,20 and this hypothesis was also explored by correlating biocide and antibiotic resistance profiles of the Bcc isolates tested.
Material and methods
Bacterial collection
A panel of 101 genetically distinct Bcc strains were selected from the Cardiff University collection3 and the Belgium Coordinated Collection of Microorganisms (BCCM; http://bccm.belspo.be/about/lmg.php). The selected strains encompassed reference strains from the defined Bcc strain panels.21,22 Additional B. multivorans strains from the Glasgow outbreak23 were kindly provided by Dr Alan Brown and Prof. John R.W. Govan (Edinburgh University, UK). MLST was used to type and select strains spanning the genetic diversity of the Bcc.19 The strains spanned 11 of the most prevalent Bcc species (Table 1) and also two strains from the Bcc novel Group K (a distinct MLST cluster from the former Group K isolates re-named as the new species B. lata and B. contaminans); the source of the strains varied and included clinical (56), non-clinical (9) and environmental strains (36). Initially all strains were screened against chlorhexidine (ICN Biomedicals) and cetylpyridinium chloride (ICN Biomedicals) as representatives of agents that could be easily screened in a broth dilution assay. From this screen, a subcollection of 38 strains was selected that spanned the range of chlorhexidine and cetylpyridinium chloride susceptibility observed: 22 strains with low MICs and 16 with high MICs were selected to provide a spectrum of Bcc biocide resistance.
Table 1.
Susceptibility of Bcc and other species to chlorhexidine and cetylpyridinium chloride
| Chlorhexidine MIC (mg/L) |
Cetylpyridinium chloride MIC (mg/L) |
|||
|---|---|---|---|---|
| Species (number of strains tested) | mean minimum | mean maximum | mean minimum | mean maximum |
| B. cepacia (10) | 57 | 65 | 170a | 175b |
| B. multivorans (30) | 36.6 | 46 | 77.7 | 101.3 |
| B. cenocepacia (31) | 47.4 | 54.8 | 175.5c | 184.5c |
| B. stabilis (3) | 40 | 46.7 | 146.7 | 166.7 |
| B. vietnamiensis (5) | 52 | 58 | 90 | 106 |
| B. dolosa (3) | 26.7 | 36.7 | 66.7 | 86.7 |
| B. ambifaria (6) | 33.3 | 41.7 | 98.3 | 116.7 |
| B. anthina (2) | 10 | 20 | 95 | 125 |
| B. pyrrocinia (2) | 15 | 25 | 50 | 70 |
| B. lata (4) | 15 | 25 | 183.3d | 200d |
| B. contaminans (3) | 13.3 | 23.3 | 200e | 200f |
| Novel Bcc Group K (2) | 50 | 60 | 200g | 200g |
| Mean Bcc (101) | 33.03 | 41.9 | 129.4 | 144.3 |
aAt the mean minimum cetylpyridinium chloride MIC, B. cepacia was significantly more resistant than B. multivorans, B. dolosa, B. ambifaria and B. pyrrocinia (P < 0.05).
bAt the mean maximum cetylpyridinium chloride MIC, B. cepacia was significantly more resistant than B. multivorans, B. dolosa and B. pyrrocinia (P < 0.05).
cAt the mean minimum and mean maximum cetylpyridinium chloride MICs, B. cenocepacia was significantly more resistant than B. multivorans, B. vietnamensis, B. dolosa, B. ambifaria, B. anthina and B. pyrrocinia (P < 0.05).
dAt the mean minimum and mean maximum cetylpyridinium chloride MICs, B. lata was significantly more resistant than B. multivorans (P < 0.05).
eAt the mean minimum cetylpyridinium chloride MIC, B. contaminans was significantly more resistant than B. multivorans and B. ambifaria (P < 0.05).
fAt the mean maximum cetylpyridinium chloride MIC, B. contaminans was significantly more resistant than B. multivorans (P < 0.05).
gAt the mean minimum and mean maximum cetylpyridinium chloride MICs, Bcc novel Group K was significantly more resistant than B. multivorans (P < 0.05).
The subcollection was then used (i) to examine the susceptibility to three additional biocides povidone (Sigma Aldrich), benzalkonium chloride (Sigma Aldrich) and triclosan (5-chloro-2-(2,4 dichlorophenoxy)phenol; Fluka, Biochemika) and (ii) to determine the MBC values of chlorhexidine and cetylpyridinium chloride. In addition, 10 non-Bcc species were also screened for chlorhexidine and cetylpyridinium chloride susceptibility as control species (which, except for Escherichia coli, can infect the CF lung): Staphylococcus aureus [both methicillin-resistant (MRSA, two isolates) and -sensitive (MSSA, four isolates)] and Pseudomonas aeruginosa (24 isolates), as well as one control isolate for Burkholderia gladioli, Stenotrophomonas maltophilia, Ralstonia mannitolytica, Pseudomonas stutzeri, Ralstonia picketti, Pseudomonas putida, Achromobacter xylosoxidans and E. coli. All strains were grown in Tryptone soya broth (TSB) or agar (TSA; Oxoid) and incubated for 24–72 h at 37°C.
Broth dilution MIC method
A broth dilution assay adapted to examine biocide susceptibility of Bcc bacteria was carried out as follows. Aqueous solutions (sterilized by filtration) of chlorhexidine (10 mg/mL) and cetylpyridinium chloride (5 mg/mL) were used to make up stock concentrations of 100 and 1000 mg/L biocides. These solutions were then added to TSB to make up biocide concentrations ranging from 0 to 100 mg/L for chlorhexidine and 0 to 200 mg/L for cetylpyridinium chloride. We found that growth of bacteria in a liquid medium when compared with that on slopes of solid agar produced consistent numbers of viable cells for the Bcc strains studied. Therefore, each Bcc strain was grown overnight (18 h) in 3 mL of TSB at 37°C (in a 14 mL tube, shaken horizontally at 200 rpm); the culture was diluted to an optical density (OD) of 1 (630 nm) corresponding to a viable count of ∼1 × 108 cfu/mL. Approximately 1 × 105 cfu of each test Bcc strain were then added to 1 mL of each biocide concentration, and 200 µL of this was added to three replicate wells within a 96-well plate. The microplates were incubated with shaking (200 rpm) at 37°C for 24 h and the OD was read using a Dynex Technologies MRX® microplate absorbance reader with Revelation application. An endpoint reading at 630 nm was taken with 10 s of shaking beforehand.
B. multivorans LMG 13010 was included in all assays as a control strain as preliminary experiments with this strain exhibited a reproducible effect. Addition of biocides to the growth media resulted in changes in OD. Therefore, the MIC value was designated as the concentration of biocides that resulted in an 80% knockdown of culture OD in comparison with the control wild-type growth with no added biocide. Viable counts performed on the control strain after running the broth dilution MIC assay demonstrated the presence of 9.7 × 105 cfu/mL at a chlorhexidine concentration of 20 mg/L, while no viable cells were detected above this concentration. Using the 80% knockdown in culture OD, the MIC for the control strain was found to be 30–40 mg/L, which corresponded well to the absence of viable cells in concentrations of chlorhexidine above 20 mg/L. The results were subjected to statistical analysis in order to determine levels of susceptibility.
Agar dilution MIC method
To screen the subset of 38 strains against the three additional biocides povidone, benzalkonium chloride and triclosan, an agar dilution method was used. The method allowed for higher concentrations of biocides to be examined when the broth dilution assay was prevented due to the biocides altering the OD reading. Stock solutions of the three biocides were made up as follows: 10 000 mg/L of triclosan in DMSO (this solvent did not inhibit the growth of Bcc bacteria at the concentrations used); 200 000 mg/L of povidone in water and 10 000 mg/L of benzalkonium chloride in water. After autoclaving, TSA was cooled to 50°C and the appropriate amount of each biocide was added to 40 mL of agar for concentration ranges of 0–500 mg/L for triclosan, 0–400 mg/L for benzalkonium chloride and 0–50 000 mg/L for povidone. The bacterial strains were cultured and diluted to 1 × 108 cfu/mL as described above and 200 µL of this suspension was then added to a 96-well plate in order to create a master plate containing eight replicates of each test strain. The strain suspensions from the master plate were then replicated onto the biocide-containing agar plates using a 96-well replicator and left to grow for 48 h at 37°C. The MIC was designated as the concentration of biocides of which no visible growth was apparent.
MBC
Bacterial cultures were grown as described above and diluted to ∼106 cfu/mL. Chlorhexidine or cetylpyridinium chloride was added to diluted bacteria up to a maximum of 5000 mg/L; 200 µL of this suspension was placed into 96-well plates and incubated at 37°C with shaking for 24 h. After incubation, 20 µL of the bacterial suspension was placed into 100 µL of neutralizer solution (0.75% azolectin and 5% Tween)24 and left in contact for 5 min. The neutralizer solution was previously screened for efficiency of biocide termination and lack of toxicity towards the test bacteria (data not shown). After neutralization, the culture was replicated onto TSA and incubated for 24–48 h at 37°C; the MBC value was observed as the concentration at which growth ceased. Given the known starting viable count of bacteria, the dilution made in the neutralizer and the volume transferred by the replica plater (1.5 µL), in the absence of bacterial growth, the MBC assay was capable of detecting a 99.995% rate of bacterial kill.
Antibiotic MIC testing: Etest® strips
The susceptibility of Bcc strains to 10 antibiotics was determined using Antibiotic Etest® strips as described by the manufacturer (AB Biodisk). The strains (60 tested) were selected on the basis of their resistance to chlorhexidine and cetylpyridinium chloride. The 10 antibiotics examined were amikacin, azithromycin, ciprofloxacin, ceftazidime, chloramphenicol, imipenem, meropenem, piperacillin, tobramycin and trimethoprim/sulfamethoxazole; all these antibiotics may be used to treat microbial infection in individuals with CF. The selection of antibiotics used to treat Bcc infection in CF is dependent on multiple factors including strain antibiogram, patient tolerance and clinic practice. Double or triple antibiotic combination therapy is recommended with meropenem/high-dose tobramycin combined with ceftazidime, trimethoprim/sulfamethoxazole, chloramphenicol or amikacin.7 The bacterial strains tested were revived on agar plates, and fresh colony growth resuspended in Iso-Sensitest broth (Oxoid) to an OD of 0.5 at 630 nm. This culture was then swabbed onto Iso-Sensitest agar plates and left to dry for 15 min. Etest® strips were then placed on top of the lawn of bacteria and left at 37°C overnight. The MIC was read from the strips and the resistance/susceptibility determined using the manufacturer's guidelines. Spontaneous resistance of each Bcc strain was measured by counting the number of colonies that grew within the initial zone of clearing after 48 h of incubation; spontaneous resistance was defined by the appearance of one or more colonies after 48 h of growth.
Suspension Tests
Two commercial skin cleanser biocide formulations were assessed in a standardized suspension test assay:25 HibiSCRUB™ (containing an alcoholic solution of 4% chlorhexidine gluconate; Regent Medical Ltd) and Cuticura™ Waterless Hand Sanitizer (containing an unspecified amount of triclosan; Cuticura Labs Corp., NY, USA). Bacterial cultures were grown as described above and diluted to ∼108 cfu/mL, and 1 mL of this suspension was added to 9 mL of commercial biocide. After exposure for 5, 10, 20 and 60 min, 100 µL of this suspension was removed and placed into 900 µL of a neutralizer solution (0.75% azolectin and 5% Tween 80) for 5 min. A control experiment containing water instead of biocides was also carried out at the same time. To enumerate viable bacteria, serial dilutions and viable counts were conducted.
Biofilm assays
An adapted biofilm assay was used to determine the biofilm production for Bcc strains.26 Plates (96-well) were coated with 1 mg/well of porcine mucin (Sigma Aldrich, M1778; 200 µL of a 5000 mg/L solution was placed in each well) and left at 37°C overnight. Porcine mucin was used to aid the adherence of bacterial cells to the wells of the plate as mucin is a protein commonly found in the lungs. Plates were washed three times with PBS and 0.1% Tween 20 and then stored at 4°C until use. Bacterial cultures were grown in nutrient broth as described above and diluted to ∼106 cfu/mL; 200 µL of each strain was then placed into 96-well plates. Eight replicates of each strain were assessed with the control row containing only broth. The plates were incubated at 37°C statically for 72 h. After incubation, the plates were washed three times with PBS to remove planktonic growth. The remaining biofilm was fixed with methanol for 15 min. Once methanol was removed and plates were dried, biofilms were stained with 1% Crystal Violet for 5 min. The stain was removed by washing with water and plates were dried. Biofilm thickness was measured by adding 33% glacial acetic acid and taking an OD reading at 570 nm using an automated plate reader (see above).
Statistical analysis
Due to the multifactorial mechanisms of biocide action, the MIC values obtained in replicate experiments spanned a range; therefore the results were split into a mean minimum value and a mean maximum value. All the results were analysed using the statistical software Minitab V.14. Kruskal–Wallis and Mann–Whitney statistical tests were carried out on the datasets and significant differences were determined (P < 0.05 for Kruskal–Wallis and P < 0.05 for Mann–Whitney). An antibiotic profile score was also calculated as a measure of overall antibiotic resistance within a Bcc species as follows: (i) for a given antibiotic, the mean MIC for all the strains in the species tested was calculated and (ii) the mean for all 10 of the antibiotics tested was then calculated to produce an overall score value for the Bcc species. Mann–Whitney tests were applied to determine the significance of the species' mean antibiotic profile score (P < 0.05); however, the significance of the score was not determined for Bcc species represented by less than two strains. The antibiotic profile score did not correlate to clinically defined break points for resistance; it just served as a measure of the overall multiple drug resistance observed in each Bcc species.
Results
Chlorhexidine and cetylpyridinium chloride susceptibility
MICs of chlorhexidine and cetylpyridinium chloride were initially determined for 101 strains of the Bcc; after replicate analysis, the mean minimum and mean maximum MIC values for each species were calculated (Table 1). For chlorhexidine, a variable range of growth inhibition was observed around the Bcc species mean (33–42 mg/L), from 10 mg/L (B. lata) to 65 mg/L (B. cepacia; Table 1). Although there were no significant differences observed between species, strains of B. cepacia, B. vietnamiensis and B. cenocepacia possessed the highest chlorhexidine MIC values observed (Table 1). Approximately 3-fold larger amounts of cetylpyridinium chloride were required to inhibit the growth of Bcc compared with chlorhexidine (mean MIC range, 129–144 mg/L; Table 1). In addition, species differences were apparent, with B. cepacia, B. cenocepacia, B. lata, B. contaminans and novel Bcc Group K strains being significantly less susceptible to cetylpyridinium chloride than other members of the Bcc (Table 1; P < 0.05%).
The collection of isolates examined contained nine examples of Bcc strains that were involved in outbreaks involving multiple CF patients; interestingly, two of these epidemic strains demonstrated very high levels of chlorhexidine resistance. The B. cenocepacia ET12 epidemic strain J2315 and two additional strains that were single-locus variants (part of the same epidemic cable pilus and B. cepacia Epidemic Strain Marker-positive lineage) of J2315 had high MIC values for chlorhexidine that were >100 mg/L (one of these, LMG 18827, is shown in Table 2). Of the remaining 28 B. cenocepacia strains tested, only one other recA Group III-A outbreak strain (MU1; Table 2) and one non-epidemic III-B group strain (LMG 18832; Table 2) possessed a chlorhexidine MIC in excess of 100 mg/L. Other epidemic strains within B. cenocepacia III-B such as the Philadelphia–District Columbia27 and Mid-Western strains28 were prevented from growing by <60 mg/L chlorhexidine.
Table 2.
Biocide susceptibility and MBCs for selected Bcc strains
| Chlorhexidine |
Cetylpyridinium chloride |
|||||
|---|---|---|---|---|---|---|
| Species and strain name | MIC (mg/L) | MBC (mg/L) | MIC (mg/L) | MBC (mg/L) | Triclosan MICa (mg/L) | Benzalkonium chloride MICa (mg/L) |
| B. cepacia (7) | ||||||
| LMG 17997 | 100 | 100 | 70–100 | 2000 | 400 | 200 |
| ATCC 49709 | 100 | 100 | >200 | 500 | 300 | 50 |
| IST431 | 90–100 | — | >200 | 3000 | 400–450 | 200 |
| LMG 18821 | 90–100 | 300 | >200 | 2000 | >500 | 100 |
| AVC1717 | 70–80 | 300 | >200 | >5000 | 500 | 200 |
| ATCC 17759 | 20–30 | 100 | >200 | 3000 | >500 | 100 |
| Bcc0176 | 10–20 | 50 | 30–40 | 50 | 500 | 100 |
| B. multivorans (5) | ||||||
| LMG 16660 | 90–100 | 300 | >200 | >5000 | >500 | 200–250 |
| ATCC 17616 | 10–20 | 300 | 50–150 | 1000 | 50 | 150 |
| C1582 | 100 | 300 | >200 | 1000 | 450–500 | 150 |
| C1607 | 100 | 500 | 200 | >5000 | 500 | 250 |
| LMG 18825 | 20–30 | 100 | 50–100 | 50 | 50 | 100 |
| B. cenocepacia (12) | ||||||
| J2315b | >100 | 100 | >200 | >5000 | >500 | 350 |
| X100 | 90–100 | 500 | 70–100 | >5000 | >500 | 350 |
| LMG 18827b | >100 | — | >200 | — | 500 | 150 |
| MU1 | >100 | 1000 | >200 | >5000 | 10 | 350 |
| WH1 | 10–20 | 100 | 150–180 | 4000 | 450–500 | 200 |
| LMG 16654 | 80–90 | 300 | >200 | >5000 | >500 | 50 |
| DN | 100 | 500 | >200 | 2000 | 450–500 | 50 |
| K56-2b | 90–100 | 300 | >200 | >5000 | >500 | 150 |
| PC002 | 100 | 300 | >200 | >5000 | >500 | >400 |
| ATCC 25609 | 50–60 | 300 | >200 | >5000 | 400 | 350 |
| LMG 18832 | >100 | 1000 | >200 | >5000 | >500 | 350 |
| PC523 | 10–20 | 100 | 150–190 | 3000 | 500 | 200 |
| B. stabilis (2) | ||||||
| ATCC 35254 | 10–20 | 100 | 200 | >5000 | 500 | 200 |
| LMG 14294 | 100 | 100 | 180–200 | >5000 | 400 | 200 |
| B. vietnamiensis (4) | ||||||
| LMG 16232 | 100 | 100 | 180–200 | 4000 | 450–500 | 100 |
| G4 | 10–20 | 300 | 30–50 | 500 | 50 | 50 |
| J1738 | 20–30 | 100 | 10–30 | 50 | 50 | 50 |
| LMG 10929 | 100 | 100 | 30–50 | 300 | 100 | 50 |
| B. dolosa (2) | ||||||
| LMG 18943 | 20–30 | 300 | 50–70 | 2000 | 450–500 | 200 |
| AU0090 | 40–50 | 100 | 100–120 | 5000 | 450–500 | 100 |
| B. ambifaria (3) | ||||||
| MC40-6 | 20–30 | 50 | 100–120 | 2000 | 50 | 50 |
| ATCC 53267 | 100 | — | 180–200 | 5000 | >500 | 50 |
| LMG 19467 | 20–30 | 50 | 70–100 | 1000 | >500 | 150 |
| B. anthina (2) | ||||||
| LMG 16670 | 10–20 | 50 | 70–100 | 500 | 150–200 | 50 |
| LMG 20980 | 10–20 | 50 | 120–150 | 2000 | 150 | 50–100 |
| B. pyrrocinia (1) | ||||||
| LMG 14191 | 10–20 | 50 | 50–70 | 2000 | 50 | 50 |
| Mean maximum value (mg/L) | 74 | 229 | 155 | 2998 | 374 | 164 |
aThe MIC of triclosan and benzalkonium chloride was determined on agar media.
bIsolates representative of the B. cenocepacia ET12 epidemic CF strain.
The B. multivorans isolate LMG 16660, a strain that caused a major outbreak among CF patients in Glasgow,23 possessed a very high chlorhexidine MIC value (90–100 mg/L; Table 2) compared with the other B. multivorans strains. To determine whether this was a feature of the outbreak isolates, 19 additional Glasgow strains (from 17 CF patients involved in the outbreak) were examined and their chlorhexidine MICs were found to range from 44 to 100 mg/L. These minimum and maximum chlorhexidine MICs were significantly higher than the respective mean levels observed for all the other B. multivorans strains examined (22–33 mg/L; P < 0.05). However, in contrast to the elevated chlorhexidine MIC, the Glasgow outbreak strains were significantly more susceptible to cetylpyridinium chloride (76.5–89.5 mg/L; P < 0.05) than the other B. multivorans strains (105–127 mg/L; P < 0.05).
The Bcc strains were also compared with a control group of non-Bcc species commonly encountered in CF infection. With chlorhexidine, there were no significant differences between the MIC values for Bcc compared with non-Bcc species, although 6 of 10 species tested were prevented from growth by <20 mg/L chlorhexidine. R. pickettii, R. mannitolytica, S. aureus and P. aeruginosa required more than 20 mg/L chlorhexidine to inhibit growth. Cetylpyridinium chloride resistance was significantly elevated in several Bcc species compared with other CF colonizers, with B. cepacia, B. cenocepacia and Bcc novel Group K being significantly less susceptible (P < 0.05). Overall, P. aeruginosa was the only CF pathogen that demonstrated chlorhexidine MICs (33–42 mg/L) and cetylpyridinium chloride MICs (200 mg/L) equivalent to those found in Bcc bacteria.
MBC
After screening 101 strains from the Bcc for growth inhibition, 38 strains spanning the observed MIC ranges for chlorhexidine and cetylpyridinium chloride were selected for MBC analysis (Table 2). The mean Bcc chlorhexidine MBC was 229 mg/L (Table 2), ∼6-fold greater than the mean MIC for this biocide. The endpoint difference between the MIC and MBC assays demonstrated that the MIC concentration should not be taken as a measure of biocide efficacy, since for all but two of the strains examined it was below the bactericidal concentration. The majority of strains tested (35 out of 38) required no more than 300 mg/L for a bactericidal outcome. One B. multivorans and two B. cenocepacia strains had MBCs of 500 mg/L; two B. cenocepacia strains needed 1000 mg/L of chlorhexidine to achieve killing, 10-fold above their respective MIC concentration (Table 2). In contrast, 27 of the 38 Bcc strains required over 20-fold more cetylpyridinium chloride than the MIC for a bactericidal outcome, with a mean cetylpyridinium chloride MBC of 2998 mg/L (Table 2). In addition, 13 Bcc strains (eight of which were B. cenocepacia) were still viable after a 24 h exposure to 5000 mg/L of cetylpyridinium chloride.
Triclosan, povidone and benzalkonium chloride susceptibility
The same subgroup of Bcc strains selected for MBC analysis was tested for their growth susceptibility to triclosan, povidone and benzalkonium chloride (Table 2). All Bcc strains tested possessed povidone MICs in excess of 50 000 mg/L, except for one B. pyrrocinia strain (MIC 2000 mg/L). The mean maximum MIC observed for triclosan was 374 mg/L with 11 Bcc strains having MICs >500 mg/L. The mean maximum MIC of benzalkonium chloride was 164 mg/L, similar to that of the other quaternary ammonium compound tested, cetylpyridinium chloride (155 mg/L for the subgroup of strains tested). Both B. cenocepacia J2315 and K56-2 from the ET12 epidemic lineage (which possessed high chlorhexidine and cetylpyridinium chloride MICs; Table 1) had high MICs of triclosan (>500 mg/L), povidone [>50 000 mg/L (5%)] and benzalkonium chloride (350 mg/L).
Antibiotic susceptibilities
Antibiotic susceptibility to 10 antibiotics commonly used in CF was determined for 60 Bcc strains (Table 3). Clinically defined resistance or susceptibility to individual antibiotics varied widely for the Bcc strains tested. However, when the total antibiotic MIC score was examined, B. cepacia, B. multivorans, B. cenocepacia and B. dolosa possessed the greatest levels of multiple drug resistance (Table 3). B. dolosa, in particular, possessed very high levels of antibiotic resistance (Table 3); however, this did not correlate to elevated biocide resistance. For example, the B. dolosa strain LMG 18943, a strain associated with an outbreak in a hospital in Boston,29 had a chlorhexidine MIC value of 20–30 mg/L and a cetylpyridinium chloride MIC value of 50–70 mg/L, but was resistant to all 10 antibiotics tested.
Table 3.
Mean MIC (mg/L) values for Bcc species to 10 antibiotics
| Organism (number of strains tested) | AMK | AZM | CAZ | CHL | CIP | IPM | MEM | PIP | TOB | SXT | Antibiotic profile score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| B. cepacia (5) | 109.6 | 125.3 | 5.98 | 13.3 | 5.2 | 17.6 | 0.9 | 13.1 | 88.23 | 0.5 | 37.9a |
| B. multivorans (24) | 81.6 | 110.4 | 52.9 | 101.6 | 1.976 | 25.8 | 2.2 | 21.4 | 30 | 0.4 | 42.8b |
| B. cenocepacia (14) | 111.5 | 81.2 | 2.6 | 12.2 | 1 | 35.6 | 3.1 | 23.4 | 37.5 | 9.1 | 31.7c |
| B. stabilis (2) | 8 | 32.7 | 1.6 | 22 | 16.3 | 3 | 1.5 | 3 | 4.5 | 2 | 9.5d |
| B. vietnamiensis (4) | 6.3 | 13.5 | 2.5 | 8 | 0.2 | 0.4 | 0.5 | 1.25 | 4.6 | 0.3 | 3.8 |
| B. dolosa (4) | 224 | 256 | 66.7 | 192.7 | 10.3 | 26.2 | 17.5 | 82.3 | 118 | 12 | 100.6e |
| B. ambifaria (4) | 9.8 | 23.6 | 1.2 | 3.25 | 0.1 | 1.1 | 0.2 | 1.1 | 6.6 | 0.1 | 4.7 |
| B. anthina (1) | 16 | 32 | 1.5 | 4 | 0.19 | 2 | 0.38 | 1 | 8 | 0.19 | 6.5d |
| B. pyrrocinia (1) | 0.094 | 24 | 0.75 | 1.5 | 0.003 | 0.19 | 0.002 | 0.25 | 2 | 0.002 | 2.9d |
| B. contaminans (1) | 1 | 16 | 1.5 | 8 | 0.25 | 24 | 0.75 | 1 | 0.38 | 0.125 | 5.3d |
AMK, amikacin; AZM, azithromycin; CIP, ciprofloxacin; CAZ, ceftazidime; CHL, chloramphenicol; IPM, imipenem; MEM, meropenem; PIP, piperacillin; TOB, tobramycin; SXT, trimethoprim/sulfamethoxazole.
aB. cepacia was significantly more resistant than B. vietnamensis and B. ambifaria (P < 0.05).
bB. multivorans was significantly more resistant than B. ambifaria (P < 0.05).
cB. cenocepacia was significantly more resistant than B. vietnamensis and B. ambifaria (P < 0.05).
dB. stabilis, B. anthina, B. pyrrocinia and B. contaminans were not statistically significant owing to small sample size.
eB. dolosa was significantly more resistant than B. stabilis, B. vietnamensis and B. ambifaria (P < 0.05).
Overall, the Bcc strains examined were most susceptible to meropenem, imipenem, piperacillin and trimethoprim/sulfamethoxazole; B. cenocepacia strains such as J2315 were, however, innately resistant to this antibiotic combination. Spontaneous resistant colonies appearing within the zones of clearing on the Etest® strips occurred most frequently with the β-lactam antibiotics (meropenem, imipenem and piperacillin). In addition, B. multivorans and B. cenocepacia strains (41% and 33%, respectively, of the strains tested) produced the most spontaneous resistance to the β-lactams. All other Bcc species tested demonstrated low rates of this phenomenon with B. vietnamiensis (at 11% of strains tested) being the next highest observed.
Susceptibility to commercial biocide formulations
To determine whether commercial biocide formulation was bactericidal for Bcc strains that possessed high MIC and MBC to specific disinfectants, suspension tests were performed. Two biocides were evaluated: Hibiscrub, a chlorhexidine-based disinfectant used in clinical settings for pre-operative hand disinfection and skin antiseptisis, and Cuticura, a hand gel sold for both home and clinical use. Four Bcc strains with high resistance to chlorhexidine (90–100 mg/L) were tested: B. cepacia LMG 18821, B. multivorans LMG 16660, B. cenocepacia J2315 and Bcc novel Group K strain 24. B. multivorans ATCC 17616 was used as a susceptible control isolate. Five minutes of exposure to the commercial biocide formulation was sufficient to achieve killing of three Bcc strains (strain LMG 16660, Group K strain 24 and ATCC 17616). However, B. cepacia LMG 18821 remained viable after 1 h of exposure to Cuticura (only a 1 log reduction in viable count was observed); the strain was killed within 5 min in Hibiscrub. B. cenocepacia J2315 remained viable in Hibiscrub after 1 h of exposure; however, it was more susceptible to Cuticura with complete killing observed after 20 min.
Biofilm production
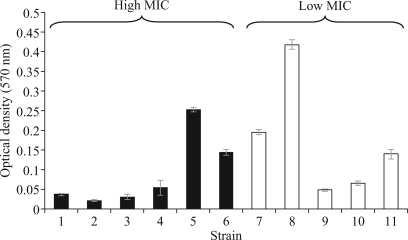
Bacteria within biofilms are less susceptible to killing by antimicrobial agents than planktonic cells.30 To determine whether there was a correlation between biofilm formation and biocide resistance in a planktonic state, 11 Bcc strains were screened for biofilm production. Six strains were selected with high (>80 mg/L) and five with low (<30 mg/L) resistance to chlorhexidine (Figure 1). No correlation between chlorhexidine MIC and the ability to form a biofilm was observed. For example, B. multivorans strain ATCC 17616, which possessed a low chlorhexidine MIC (10–20 mg/L) and low MICs to the other biocide screens (Table 1), produced the most biofilms. In contrast, B. cenocepacia J2315, which was representative of a highly transmissible lineage associated with major clinical infections and possessed high biocide (Table 2) and antibiotic resistance (see above), was a poor biofilm former.
Figure 1.
Biofilm production by selected Bcc strains. Six strains with high (black bars) and five with low (open bars) chlorhexidine MICs were tested for biofilm formation. The biofilm biomass readout is shown on the vertical axis. Strains are as follows: 1, B. cepacia LMG 18821; 2, B. multivorans LMG 16660; 3, B. cenocepacia LMG 18832; 4, B. cenocepacia J2315; 5, B. cenocepacia C4455; 6, B. cepacia novel Group K 24; 7, B. multivorans LMG 13010; 8, B. multivorans ATCC 17616; 9, B. dolosa LMG 18943; 10, B. ambifaria AMMD; 11, Burkholderia vietnamensis G4.
Discussion
Bcc bacteria are known to possess high levels of antimicrobial resistance; however, a survey of their biocide resistance that accounts for the recent changes in their taxonomy has not been performed. Our analysis of the biocides chlorhexidine, cetylpyridinium chloride, triclosan and benzalkonium chloride has demonstrated that susceptibility to these agents varies considerably across a panel of genetically distinct Bcc strains. Species-dependent differences were apparent only for cetylpyridinium chloride. No specific correlations between biocide susceptibility, antibiotic susceptibility and the ability to form biofilms were seen; however, certain CF outbreak strains possessed very high chlorhexidine resistance. The ability of biocide-resistant Bcc strains to survive exposure to two widely used commercial skin washing biocides was also demonstrated, suggesting that there is still room to improve disinfectant formulations to ensure they kill this group of bacteria.
While the correlation of antibiotic susceptibility to six genomovars has been reported,9 the influence of Bcc taxonomy on biocide susceptibility has not been examined. The MIC for Bcc strains was investigated in great depth for chlorhexidine and cetylpyridinium chloride, with B. cepacia, B. cenocepacia, B. lata, B. contaminans and Bcc novel Group K being significantly less sensitive to cetylpyridinium chloride than other species. Previous work correlating clinical antibiotic resistance to Bcc taxonomy did not find any significant trends,9 however, we found that if an overall antibiotic resistance score was calculated based on the MIC value, then B. cepacia, B. multivorans, B. cenocepacia and B. dolosa were significantly more resistant. Interestingly, B. cenocepacia, which overall possessed both high biocide (Tables 1 and 2) and antibiotic resistance (Table 3), was also the most prevalent Bcc species encountered in a recent survey of both CF and other clinical infections.6
Correlations between antibiotic resistance and biocide resistance were not obvious among the Bcc bacteria. Certain epidemic Bcc strains such as those of the ET12 group (e.g. J2315, Table 2) possessed the highest levels of resistance to both classes of antimicrobial agents. The Glasgow outbreak B. multivorans strain23 possessed chlorhexidine MICs that were nearly double that of other B. multivorans strains (Table 2), yet had similar levels of antibiotic resistance within this species group. Other strains, such as the epidemic B. dolosa LMG 18943,29 possessed low chlorhexidine and cetylpyridinium chloride MICs (Table 2) but were resistant to all 10 antibiotics tested. There was also a lack of correlation between planktonic biocide resistance and the ability to form biofilms (Figure 1) for Bcc bacteria. This suggests that despite the advantages biofilm growth confers in terms of survival under antimicrobial stress,30 not all Bcc bacteria have adopted this phenotype, but instead have other enhanced resistance mechanisms that function during planktonic growth. However, strains such as B. cenocepacia C4455 and Bcc novel Group K 24 (Figure 1), which have both high intrinsic biocide and a good ability to form biofilms, have the potential to be particularly problematic in contamination or outbreak scenarios. The lipopolysaccharide (LPS) structure of Bcc bacteria is very unusual and also varies between species.31 Given the important role this part of the Bcc cell envelope plays in its resistance to cationic antimicrobial agents, it would be interesting in future studies to compare the LPS structures of strains that are resistant or susceptible to membrane-active biocides such as chlorhexidine.
Maintaining product sterility is a problem faced by many manufacturing industries and a recent survey of the pharmaceutical industry has shown that Bcc bacteria are one of the leading causes of pharmaceutical product recalls.10 Our analysis of MBC data showed that Bcc bacteria may need more than 25 times more biocides over the MIC value to achieve killing. The concentrations of biocides used in commercial products are dependent on the biocide and type of product application. Chlorhexidine, cetylpyridinium chloride and triclosan are used in a variety of commercial products in concentrations ranging from 0.1% to 4% (1000–40 000 mg/L).32 At the lowest level, this is insufficient to kill Bcc bacteria. For example, two B. cenocepacia strains had chlorhexidine MBCs of 1000 mg/L and 31 Bcc strains had cetylpyridinium chloride MBCs in excess of 1000 mg/L (Table 2). Dilute solutions of chlorhexidine appear particularly prone to contamination with Bcc bacteria, and outbreaks associated with mouthwash (0.2%)33 or blood-product disinfectant solutions (0.5% and lower concentrations)13 have been reported. The MIC of five disinfectants used for organ irrigation in CF lung-transplant procedures was evaluated by Perry et al.34 Taurolin and Noxyflex were found to be more inhibitory towards the 19 Bcc isolates tested than chlorhexidine, triclosan and povidone, and were specifically recommended for use where these bacteria may be present.34
The efficacy of skin disinfection products and procedures may be tested according to standard European guidelines using bacterial species such as E. coli, P. aeruginosa, S. aureus and Enterococcus hirae.25 Preservative challenge test guidelines include the B. cepacia Type strain ATCC 25416, among the recommended test strains.35 However, although this strain possessed the maximal cetylpyridinium chloride MIC (>200 mg/L), its chlorhexidine MIC was 20–30 mg/L and was not among the most resistant encountered in our survey. Three well-defined strains stand out in our analysis as being resistant to multiple biocides (Table 2): (i) B. cenocepacia LMG 18832 (ATCC 17765), a urinary tract infection isolate;22 (ii) B. cenocepacia J2315 (LMG 16656), an epidemic CF isolate,22 and (iii) B. multivorans LMG 16660 (C1576), a CF isolate from the Glasgow outbreak.23 All the latter strains were included in a historical reference panel of Bcc strains22 and are available from recognized strain collections. Given the major role Bcc organisms may play in the contamination of such products,10 future testing should also include these bacteria.
In summary, the characterization of Bcc as a group of problematic opportunistic pathogens has advanced considerably in the last decade.3 The ability to genetically define strains17,19 and systematically identify new species within the group4,5 has improved considerably. We have shown that as a closely related group of Gram-negative pathogens, Bcc bacteria possess very high resistance to biocides and commercial formulations of these agents. This understanding, together with the potential use of the biocide-resistant reference strains we have defined, can be used to develop new anti-infectives capable of killing Bcc bacteria and preventing their occurrence in disease2 and contamination of commercial products.10
Funding
This work was funded via the UK Cystic Fibrosis Microbiology Consortium with grants from the CF Trust (Grant No. MbC003) and Big Lottery. MLST analysis was funded by grants from the Wellcome Trust (Grant No. 72853) and CF Trust (PJ535).
Transparency declarations
None to declare.
Acknowledgements
We thank Alan Brown and John R.W. Govan (Edinburgh University) for providing additional Bcc strains and Gisli Einarsson (Queens University, Belfast) for information on the biofilm assay.
References
- 1.Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol. 2005;3:144–56. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 2.Govan JR, Brown AR, Jones AM. Evolving epidemiology of Pseudomonas aeruginosa and the Burkholderia cepacia complex in cystic fibrosis lung infection. Future Microbiol. 2007;2:153–64. doi: 10.2217/17460913.2.2.153. [DOI] [PubMed] [Google Scholar]
- 3.Mahenthiralingam E, Baldwin A, Dowson CG. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol. 2008;104:1539–51. doi: 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- 4.Vanlaere E, Lipuma JJ, Baldwin A, et al. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int J Syst Evol Microbiol. 2008;58:1580–90. doi: 10.1099/ijs.0.65634-0. [DOI] [PubMed] [Google Scholar]
- 5.Vanlaere E, Baldwin A, Gevers D, et al. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species: Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol. 2009;59:102–111. doi: 10.1099/ijs.0.001123-0. [DOI] [PubMed] [Google Scholar]
- 6.Reik R, Spilker T, Lipuma JJ. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J Clin Microbiol. 2005;43:2926–8. doi: 10.1128/JCM.43.6.2926-2928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chernish RN, Aaron SD. Approach to resistant gram-negative bacterial pulmonary infections in patients with cystic fibrosis. Curr Opin Pulm Med. 2003;9:509–15. doi: 10.1097/00063198-200311000-00011. [DOI] [PubMed] [Google Scholar]
- 8.Henry DA, Campbell ME, LiPuma JJ, et al. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J Clin Microbiol. 1997;35:614–9. doi: 10.1128/jcm.35.3.614-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nzula S, Vandamme P, Govan JR. Influence of taxonomic status on the in vitro antimicrobial susceptibility of the Burkholderia cepacia complex. J Antimicrob Chemother. 2002;50:265–9. doi: 10.1093/jac/dkf137. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez L. Microbial diversity in pharmaceutical product recalls and environments. PDA J Pharm Sci Technol. 2007;61:383–99. [PubMed] [Google Scholar]
- 11.Frank MJ, Schaffner W. Contaminated aqueous benzalkonium chloride. An unnecessary hospital infection hazard. JAMA. 1976;236:2418–9. [PubMed] [Google Scholar]
- 12.Kutty PK, Moody B, Gullion JS, et al. Multistate outbreak of Burkholderia cenocepacia colonization and infection associated with the use of intrinsically contaminated alcohol-free mouthwash. Chest. 2007;132:1825–31. doi: 10.1378/chest.07-1545. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Erce JA, Grasa JM, Solano VM, et al. Bacterial contamination of blood components due to Burkholderia cepacia contamination from chlorhexidine bottles. Vox Sang. 2002;83:70–1. doi: 10.1046/j.1423-0410.2002.00195.x. [DOI] [PubMed] [Google Scholar]
- 14.Romero-Gomez MP, Quiles-Melero MI, Pena Garcia P, et al. Outbreak of Burkholderia cepacia bacteremia caused by contaminated chlorhexidine in a hemodialysis unit. Infect Control Hosp Epidemiol. 2008;29:377–8. doi: 10.1086/529032. [DOI] [PubMed] [Google Scholar]
- 15.Anderson RL, Vess RW, Carr JH, et al. Investigations of intrinsic Pseudomonas cepacia contamination in commercially manufactured povidone-iodine. Infect Control Hosp Epidemiol. 1991;12:297–302. doi: 10.1086/646342. [DOI] [PubMed] [Google Scholar]
- 16.Baldwin A, Mahenthiralingam E, Drevinek P, et al. Elucidating global epidemiology of Burkholderia multivorans in cases of cystic fibrosis by multilocus sequence typing. J Clin Microbiol. 2008;46:290–5. doi: 10.1128/JCM.01818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldwin A, Mahenthiralingam E, Drevinek P, et al. Environmental Burkholderia cepacia complex isolates in human infection. Emerg Infect Dis. 2007;13:458–61. doi: 10.3201/eid1303.060403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahenthiralingam E, Baldwin A, Drevinek P, et al. Multilocus sequence typing breathes life into a microbial metagenome. PLoS One. 2006;1:e17. doi: 10.1371/journal.pone.0000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldwin A, Mahenthiralingam E, Thickett KM, et al. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J Clin Microbiol. 2005;43:4665–73. doi: 10.1128/JCM.43.9.4665-4673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert P, McBain AJ. Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin Microbiol Rev. 2003;16:189–208. doi: 10.1128/CMR.16.2.189-208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coenye T, Vandamme P, LiPuma JJ, et al. Updated version of the Burkholderia cepacia complex experimental strain panel. J Clin Microbiol. 2003;41:2797–8. doi: 10.1128/JCM.41.6.2797-2798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahenthiralingam E, Coenye T, Chung JW, et al. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol. 2000;38:910–3. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whiteford ML, Wilkinson JD, McColl JH, et al. Outcome of Burkholderia (Pseudomonas) cepacia colonisation in children with cystic fibrosis following a hospital outbreak. Thorax. 1995;50:1194–8. doi: 10.1136/thx.50.11.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lear JC, Maillard JY, Dettmar PW, et al. Chloroxylenol- and triclosan-tolerant bacteria from industrial sources. J Ind Microbiol Biotechnol. 2002;29:238–42. doi: 10.1038/sj.jim.7000320. [DOI] [PubMed] [Google Scholar]
- 25.Marchetti MG, Kampf G, Finzi G, et al. Evaluation of the bactericidal effect of five products for surgical hand disinfection according to prEN 12054 and prEN 12791. J Hosp Infect. 2003;54:63–7. doi: 10.1016/s0195-6701(03)00039-2. [DOI] [PubMed] [Google Scholar]
- 26.Stepanovic S, Vukovic D, Dakic I, et al. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40:175–9. doi: 10.1016/s0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 27.Coenye T, Spilker T, Van Schoor A, et al. Recovery of Burkholderia cenocepacia strain PHDC from cystic fibrosis patients in Europe. Thorax. 2004;59:952–4. doi: 10.1136/thx.2003.019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coenye T, LiPuma JJ. Multilocus restriction typing: a novel tool for studying global epidemiology of Burkholderia cepacia complex infection in cystic fibrosis. J Infect Dis. 2002;185:1454–62. doi: 10.1086/340279. [DOI] [PubMed] [Google Scholar]
- 29.Kalish LA, Waltz DA, Dovey M, et al. Impact of Burkholderia dolosa on lung function and survival in cystic fibrosis. Am J Respir Crit Care Med. 2006;173:421–5. doi: 10.1164/rccm.200503-344OC. [DOI] [PubMed] [Google Scholar]
- 30.Parsek MR, Fuqua C. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J Bacteriol. 2004;186:4427–40. doi: 10.1128/JB.186.14.4427-4440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Soyza A, Silipo A, Lanzetta R, et al. Chemical and biological features of Burkholderia cepacia complex lipopolysaccharides. Innate Immun. 2008;14:127–44. doi: 10.1177/1753425908093984. [DOI] [PubMed] [Google Scholar]
- 32.Lambert RJ. Evaluation of antimicrobial efficacy. In: Fraise AP, Lambert PA, Maillard JY, editors. Russell, Hugo and Ayliffe's Principles and Practice of Disinfection, Preservation and Sterilization. Oxford: Blackwell Science Ltd; 2004. p. 345. [Google Scholar]
- 33.Sobel JD, Hashman N, Reinherz G, et al. Nosocomial Pseudomonas cepacia infection associated with chlorhexidine contamination. Am J Med. 1982;73:183–6. doi: 10.1016/0002-9343(82)90176-0. [DOI] [PubMed] [Google Scholar]
- 34.Perry JD, Riley G, Johnston S, et al. Activity of disinfectants against Gram-negative bacilli isolated from patients undergoing lung transplantation for cystic fibrosis. J Heart Lung Transplant. 2002;21:1230–1. doi: 10.1016/s1053-2498(02)00434-5. [DOI] [PubMed] [Google Scholar]
- 35.English DJ. Factors in selecting and testing preservatives in product formulations. In: Orth DS, Kabara JJ, Denyer SP, et al., editors. Cosmetic and Drug Microbiology. New York, London: Informa Healthcare; 2006. pp. 57–101. [Google Scholar]