Abstract
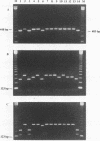
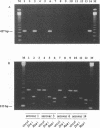
Ureaplasma urealyticum is a commensal organism of the lower genital tract of females, but in a subpopulation of individuals, it can invade the upper genital tract. It is a significant cause of chorioamnionitis and neonatal morbidity and mortality. There are 14 recognized serovars of U. urealyticum; these can be divided into two distinct clusters or biovars. Biovar 1 is composed of serovars 1, 3, 6, and 14, Biovar 2 is composed of serovars 2, 4, 5, 7, 8, 9, 10, 11, 12, and 13. We previously identified a surface antigen, the multiple-banded (MB) antigen, which contains both serovar-specific and cross-reactive epitopes. Genotypic characterization of the C-terminal region of the MB antigen of serovar 3 indicates that serovar specificity and MB antigen size variation reside in that domain. In the present study, we used PCR analysis with primers derived from the serovar 3 MB antigen gene DNA sequence to determine if the MB antigen gene was present in the remaining 13 reference serovars as well as in invasive clinical isolates. The results indicated that not only was the MB antigen gene present in all serovars but that the genes' 5' regions were markers of biovar specificity and diversity. Further analysis of this region should reveal the phylogenetic relationship among serovars of U. urealyticum and, possibly, their invasive potential.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blanchard A., Gautier M., Mayau V. Detection and identification of mycoplasmas by amplification of rDNA. FEMS Microbiol Lett. 1991 Jun 1;65(1):37–42. doi: 10.1016/0378-1097(91)90467-o. [DOI] [PubMed] [Google Scholar]
- Blanchard A., Hentschel J., Duffy L., Baldus K., Cassell G. H. Detection of Ureaplasma urealyticum by polymerase chain reaction in the urogenital tract of adults, in amniotic fluid, and in the respiratory tract of newborns. Clin Infect Dis. 1993 Aug;17 (Suppl 1):S148–S153. doi: 10.1093/clinids/17.supplement_1.s148. [DOI] [PubMed] [Google Scholar]
- Blanchard A. Ureaplasma urealyticum urease genes; use of a UGA tryptophan codon. Mol Microbiol. 1990 Apr;4(4):669–676. doi: 10.1111/j.1365-2958.1990.tb00636.x. [DOI] [PubMed] [Google Scholar]
- Cassell G. H., Waites K. B., Crouse D. T. Perinatal mycoplasmal infections. Clin Perinatol. 1991 Jun;18(2):241–262. [PubMed] [Google Scholar]
- Cassell G. H., Waites K. B., Watson H. L., Crouse D. T., Harasawa R. Ureaplasma urealyticum intrauterine infection: role in prematurity and disease in newborns. Clin Microbiol Rev. 1993 Jan;6(1):69–87. doi: 10.1128/cmr.6.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Naessens A., Lauwers S. Identification and characterization of serotype 4-specific antigens of Ureaplasma urealyticum by use of monoclonal antibodies. Infect Immun. 1993 May;61(5):2253–2256. doi: 10.1128/iai.61.5.2253-2256.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ey P. L., Andrews R. H., Mayrhofer G. Differentiation of major genotypes of Giardia intestinalis by polymerase chain reaction analysis of a gene encoding a trophozoite surface antigen. Parasitology. 1993 May;106(Pt 4):347–356. doi: 10.1017/s0031182000067081. [DOI] [PubMed] [Google Scholar]
- Fischetti V. A., Hodges W. M., Hruby D. E. Protection against streptococcal pharyngeal colonization with a vaccinia: M protein recombinant. Science. 1989 Jun 23;244(4911):1487–1490. doi: 10.1126/science.2660266. [DOI] [PubMed] [Google Scholar]
- Hollingshead S. K., Fischetti V. A., Scott J. R. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol Gen Genet. 1987 May;207(2-3):196–203. doi: 10.1007/BF00331578. [DOI] [PubMed] [Google Scholar]
- Kemp D. J., Coppel R. L., Anders R. F. Repetitive proteins and genes of malaria. Annu Rev Microbiol. 1987;41:181–208. doi: 10.1146/annurev.mi.41.100187.001145. [DOI] [PubMed] [Google Scholar]
- Michel J. L., Madoff L. C., Olson K., Kling D. E., Kasper D. L., Ausubel F. M. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10060–10064. doi: 10.1073/pnas.89.21.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouches C., Taylor-Robinson D., Stipkovits L., Bove J. M. Comparison of human and animal Ureaplasmas by one- and two-dimensional protein analysis on polyacrylamide slab gel. Ann Microbiol (Paris) 1981 Sep-Oct;132B(2):171–196. [PubMed] [Google Scholar]
- Razin S., Tully J. G., Rose D. L., Barile M. F. DNA cleavage patterns as indicators of genotypic heterogeneity among strains of Acholeplasma and Mycoplasma species. J Gen Microbiol. 1983 Jun;129(6):1935–1944. doi: 10.1099/00221287-129-6-1935. [DOI] [PubMed] [Google Scholar]
- Razin S., Yogev D. Genetic relatedness among Ureaplasma urealyticum serotypes (serovars). Pediatr Infect Dis. 1986 Nov-Dec;5(6 Suppl):S300–S304. doi: 10.1097/00006454-198611010-00022. [DOI] [PubMed] [Google Scholar]
- Robertson J. A., Chen M. H. Effects of manganese on the growth and morphology of Ureaplasma urealyticum. J Clin Microbiol. 1984 Jun;19(6):857–864. doi: 10.1128/jcm.19.6.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. A., Pyle L. E., Stemke G. W., Finch L. R. Human ureaplasmas show diverse genome sizes by pulsed-field electrophoresis. Nucleic Acids Res. 1990 Mar 25;18(6):1451–1455. doi: 10.1093/nar/18.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. A., Vekris A., Bebear C., Stemke G. W. Polymerase chain reaction using 16S rRNA gene sequences distinguishes the two biovars of Ureaplasma urealyticum. J Clin Microbiol. 1993 Apr;31(4):824–830. doi: 10.1128/jcm.31.4.824-830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemke G. W., Robertson J. A. Modified colony indirect epifluorescence test for serotyping Ureaplasma urealyticum and an adaptation to detect common antigenic specificity. J Clin Microbiol. 1981 Nov;14(5):582–584. doi: 10.1128/jcm.14.5.582-584.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson H. L., Blalock D. K., Cassell G. H. Variable antigens of Ureaplasma urealyticum containing both serovar-specific and serovar-cross-reactive epitopes. Infect Immun. 1990 Nov;58(11):3679–3688. doi: 10.1128/iai.58.11.3679-3688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods J. P., Spinola S. M., Strobel S. M., Cannon J. G. Conserved lipoprotein H.8 of pathogenic Neisseria consists entirely of pentapeptide repeats. Mol Microbiol. 1989 Jan;3(1):43–48. doi: 10.1111/j.1365-2958.1989.tb00102.x. [DOI] [PubMed] [Google Scholar]
- Yogev D., Rosengarten R., Watson-McKown R., Wise K. S. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5' regulatory sequences. EMBO J. 1991 Dec;10(13):4069–4079. doi: 10.1002/j.1460-2075.1991.tb04983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yother J., Briles D. E. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J Bacteriol. 1992 Jan;174(2):601–609. doi: 10.1128/jb.174.2.601-609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Watson H. L., Waites K. B., Cassell G. H. Serotype diversity and antigen variation among invasive isolates of Ureaplasma urealyticum from neonates. Infect Immun. 1992 Aug;60(8):3472–3474. doi: 10.1128/iai.60.8.3472-3474.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]