Abstract
It is commonly assumed that the ultrastructural organization of the rim region of outer segment (OS) discs in rods and lamellae in cones requires functional retinal degeneration slow/rod outer segment membrane protein 1 (Rds/Rom-1) complexes. Cysteine-150 (C150) in Rds has been implicated in intermolecular disulfide bonding essential for functional Rds complexes. Transgenic mice containing the Rds C150S mutation (C150S-Rds) failed to form higher-order Rds oligomers, although interactions between C150S-Rds and Rom-1 occurred in rods, but not in cones. C150S-Rds mice exhibited marked early-onset reductions in cone function and abnormal OS structure. In contrast, C150S-Rds expression in rods partly rescued the rds+/− phenotype. Although C150S-Rds was detected in the OSs in rods and cones, a substantial percentage of C150S-Rds and cone opsins were mislocalized to different cellular compartments in cones. The results of this study provide novel insights into the importance of C150 in Rds oligomerization and the differences in Rds requirements in rods versus cones. The apparent OS structural differences between rods and cones may cause cones to be more susceptible to the elimination of higher-order Rds/Rom-1 oligomers (e.g. as mediated by mutation of the Rds C150 residue).
INTRODUCTION
Highly evolved mammalian vision is made possible by the specialized organelles known as outer segments (OSs) found on both rod and cone photoreceptor cells. The microanatomy and structure of these modified cilia have been eloquently described (1–3). In rods, it was historically thought that the plasma membrane successively evaginates and invaginates with subsequent membrane fusion to form stacks of sealed organized discs encased on all sides by the plasma membrane. Although an alternative theory to evagination/invagination has recently been proposed (4), it remains quite clear that rod OSs contain stacks of discs completely distinct from the plasma membrane. On the contrary, in cones, membrane fusion does not take place; instead, stacks of lamellae are formed, which remain contiguous with the plasma membrane (2). As rods and cones mediate different types of vision, the structural and biochemical features of their OSs are necessarily distinct. Although the biochemistry of rod- and cone-mediated visual signaling is well understood, it remains a persistent question as to how cones and rods achieve their distinct differences in OS structure. This puzzle is made more complex by the observation that a key structural protein of the OS, called retinal degeneration slow (Rds), is expressed in both rods and cones.
Rds is a tetraspanin protein and is localized to the disc rim of the rod and cone OSs (5,6). Rds is necessary for the formation of OSs: retinal degeneration slow (rds−/−) mice lacking Rds completely fail to develop OS structures, whereas the heterozygous (rds+/−) mice exhibit disorganized OSs with improperly formed discs (5–9). In humans, over 80 mutations in the RDS gene have been found to associate with a variety of inherited retinal degenerative diseases, including autosomal-dominant retinitis pigmentosa, cone–rod dystrophy and multiple forms of macular dystrophy (10,11) (http://www.retina-international.org/sci-news/rdsmut.htm). Consistent with its role in the maintenance of rods and cones, Rds mutations manifest as rod and/or cone dystrophies with varying levels of severity, suggesting that the protein has distinct functions in rod and cone photoreceptors (12). Our recent work (13) using the cone-dominant mouse model lacking the transcription factor neural retina leucine zipper (nrl−/−) (14) supports this hypothesis. In the absence of Rds, cones in the nrl−/− retina retain the capacity for phototransduction and are able to elaborate some OS structure (albeit aberrant, dysmorphic structures that lack disc rims), in stark contrast to rods of the rds−/− retinas, which do not form OSs and have no detectable electroretinographic (ERG) responses (13). Clearly, rod and cone OS formation and maintenance both require Rds, but not in the same way.
Further understanding of the role of Rds in rods versus cones requires analysis of the function of Rds on the biochemical level. It is known that Rds forms non-covalent homo- and hetero-tetramers with its non-glycosylated homologue, rod outer segment membrane protein 1 (Rom-1) (15), another member of the tetraspanin family also localized to the disc rim region. These tetrameric complexes are assembled in the inner segment before trafficking to the rim region of the OS. Once in the OS, these tetramers can further interact with one another through intermolecular disulfide bonds to form octamers and higher-order oligomers (i.e. complexes larger than tetramers) (16). Rom-1 does not participate in these higher-order oligomers in rods or cones (17) and elimination of Rom-1 (rom-1−/−) does not lead to significant structural abnormalities in the photoreceptor OSs (18), leading us to hypothesize that higher-order Rds oligomers are the functional unit of Rds involved in the maintenance of rim structures. Early mutagenesis studies (19) suggest that the seven conserved Cys residues in the D2 loop of Rds play a critical role in maintaining the interactions between Rds and other proteins which are necessary for OS structural integrity. The role of Cys-mediated disulfide bonds in Rds/Rds interactions during complex formation is validated by the observation that a significant fraction of Rds migrates as a dimer on non-reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), but as a monomer in the presence of reducing agents (16). One of the D2 loop cysteines, in particular, C150 is thought to be functionally important for Rds–Rds disulfide bonding, based on in vitro studies with C150S-mutant Rds. In COS cells, C150S-Rds was unable to form disulfide bond-mediated dimers (19), but retained the ability to form heterotetramers with Rom-1. This suggests that C150 is not involved in tetrameric subunit assembly, but is required for intermolecular disulfide bonding and, thus, higher-order oligomer formation and, by extension, OS morphogenesis and maintenance.
In this study, we investigated the role of C150 using an in vivo model system in order to understand the role of intermolecular disulfide bonding in the function of Rds, particularly with regard to the formation and maintenance of rod versus cone OSs. As COS cells do not form OSs and, hence, are inadequate to model the structural complexity of rod and cone OSs, the findings presented here using transgenic mice are of particular importance. Our experiments provide critical new insights into how rods and cones differentially utilize Rds and shed some light on the question of how a single protein can be directly and critically involved in the formation of cellular structures as different as those of rod and cone OSs.
RESULTS
Expression of C150S-Rds in transgenic mice
To understand the differential role of C150-mediated Rds oligomerization in rod versus cone OS morphogenesis, we generated two transgenic models expressing C150S-Rds. Transgene expression was driven in rods by the mouse opsin promoter (named MOP-T) and in cones by the human red/green cone opsin promoter (named COP-T) (Fig. 1A). Both these promoters have been used in transgenic mice to drive cell-specific expression (20,21), with the COP-T promoter directing robust expression in both blue and red/green cones (21,22). Twenty MOP-T and 15 COP-T lines were generated and evaluated for site of integration and levels of transgene expression. General examination of these mice revealed no side effects of the transgene on animal weight or behavior. After eliminating the rd mutation (23), all founders were moved into the C57BL/6 background for several generations and then crossed with rds−/− mice on C57BL/6 background to obtain transgenic mice on all rds genetic backgrounds [wild-type (WT), rds+/− and rds−/−]. In this study, we present data from the highest expresser lines for the MOP and COP transgenes.
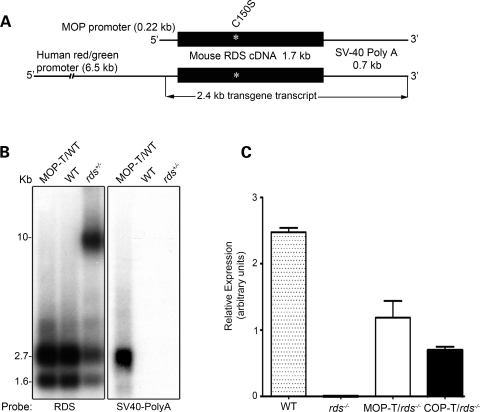
Figure 1.
Generation and expression of the C150S-Rds transgene. (A) Schematic diagram of C150S transgene constructs. The transgene is composed of the full-length mouse RDS cDNA with the C150S mutation followed by an SV40 poly-A tail and directed either to rods by the MOP or to cones by the human red/green opsin promoter (COP). (B) Northern blot analysis of whole retinal RNA extracts (10 µg/sample) taken from 1-month-old MOP-T transgenic, non-transgenic WT littermates and rds+/− mice. Two different probes were hybridized to the membrane: one from exon 1 of the RDS cDNA that recognizes both endogenous and transgene transcripts (left) and an SV40 poly-A probe that recognizes the transgenic message only (right). (C) qRT–PCR was used to measure levels of total RDS message from WT, rds−/−, MOP-T/rds−/− and COP-T/rds−/− retinas. Analysis was performed on retinas from 3- to 4-week-old mice using primer specific to the endogenous RDS gene.
To evaluate levels and integrity of the transgene transcript, northern blot and quantitative RT–PCR (qRT–PCR) analysis were performed on total retinal RNA isolated from transgenic retinas on different rds backgrounds (Fig. 1B and C). Northern blot hybridized with RDS cDNA probe detected two transcripts (1.6 and 2.7 kb) from the WT allele (Fig. 1B, left) and a large band (>9 kb) from the RDS allele in the rds+/− sample (Fig. 1B, left) (8,24,25). When the blot was hybridized to an SV40 poly-A probe, a 2.4 kb transgene transcript was only detected in MOP-T/WT (Fig. 1B, right). qRT–PCR analysis on MOP-T and COP-T retinas on rds−/− background revealed transgene transcript levels of 50 and 25% of the WT, respectively. This suggests that the amount of MOP-T message is equivalent to that of one WT allele, whereas the COP-T message is close to half of one WT allele (Fig. 1C).
C150S-Rds protein is stably expressed in rods and cones
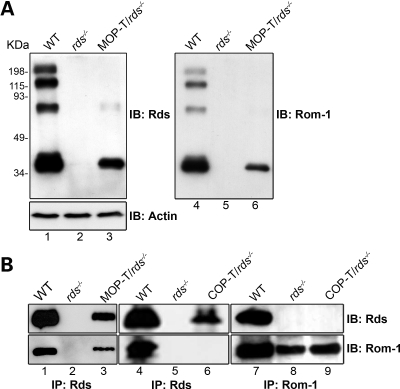
To determine the levels of C150S-Rds protein in the two selected lines, western blot analysis and immunoprecipitation (IP) were performed on retinal extracts from MOP-T and COP-T mice in the rds−/− background (Fig. 2). Figure 2A shows non-reducing western blots probed with anti-Rds-CT (left) and anti-Rom-1-CT (right) antibody. MOP-T C150S-Rds is expressed in the absence of endogenous Rds (lane 3, left) and stabilizes Rom-1 which is not usually detected in the rds−/− (lanes 5 and 6, right). In WT retinal extract, monomers, dimers and higher-order Rds complexes are readily detected, whereas the MOP-T extract in the rds−/− background shows monomers. Occasionally, a very faint dimer-sized band of unknown origin is observed in MOP-T/rds−/−. It is possible that alterations in the structure of the Rds-D2 loop arising from the C to S substitution have prompted a small fraction of C150S-Rds protein to form dimers mediated by one of the other D2 loop cysteines; however, very little dimer is ever observed, and higher-order complexes are never detected. These results confirm earlier in vitro observations that suggested that C150-mediated intermolecular disulfide bonding is necessary for normal Rds dimerization (19). Because the photoreceptor population of WT retinas is comprised of >95% rods and only ∼3–5% cones, we were not able to calculate COP-T protein levels by western blot analysis. As indicated below, we used IP in conjunction with the western blot analysis to examine C150S-Rds in COP-T retinas.
Figure 2.
C150S-Rds protein is stably formed in rods and cones. (A) Retinal extracts were isolated from 1-month-old MOP-T animals in the rds−/− background and controls and were analyzed by non-reducing western blot. The blot was sequentially probed with anti-Rds-CT (left), anti-actin (left bottom) and anti-Rom-1-CT antibodies (right). (B) Western blot analysis of anti-Rds-CT (left and middle) or anti-Rom-1-CT (right) immunoprecipitants from retinal extracts of WT, rds−/−, MOP-T/rds−/− and COP-T/rds−/− mice. Blots were probed with anti-Rds-CT (top) and anti-Rom-1-CT antibodies (bottom). Both MOP-T and COP-T Rds are expressed, although Rom-1 binds to MOP-T, but not to COP-T, Rds.
C150S-Rds associates with Rom-1 in rods, but not in cones
Rds and Rom-1 are known to form hetero- and homomeric complexes of different sizes (16,17). To evaluate the role of Rds C150 in these associations in rods versus cones, co-immunoprecipitation (co-IP) with Rds-CT and Rom-1-CT antibodies was performed on retinal extracts from MOP-T and COP-T mice on the rds−/− background and from WT and rds−/− extracts as controls (Fig. 2B). The blots were probed with anti-Rds-CT (Fig. 2B, top) and with anti-Rom-1-CT (Fig. 2B, bottom). As in the WT, IP with Rds-CT antibody from MOP-T/rds−/− extract brought down Rom-1 (lane 3). On the contrary, IP from COP-T/rds−/− failed to bring down Rom-1 (lane 6). Reciprocal IP with Rom-1 antibody confirmed that Rom-1 is present in COP-T/rds−/−, but does not associate with C150S-Rds in cones (lane 9). These results clearly demonstrate that C150S-Rds can bind to Rom-1 in rods, but not in cones, and serve as further confirmation that Rds has different roles in the two photoreceptor cell types. In addition, these results suggest that different conditions are necessary for Rds complex formation in rods versus cones.
C150S-Rds is not capable of forming higher-order complexes in rods
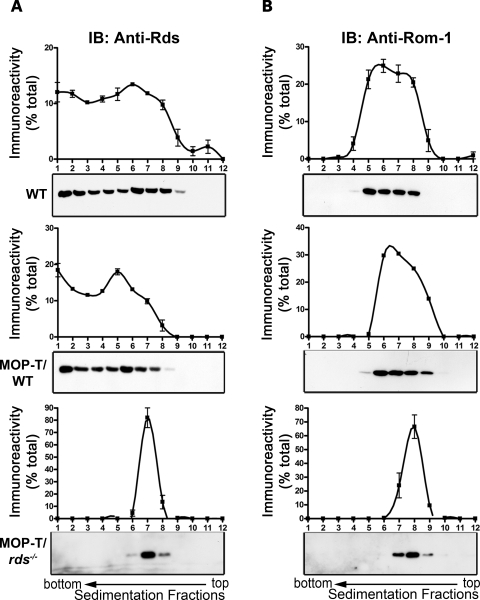
Rds molecules are normally assembled into tetramers before trafficking to the OS and into oligomers in the OS, likely at the site of disc rim morphogenesis (17). Mutations can impair these processes and result in photoreceptor degeneration (9). To better understand what role C150 plays in tetrameric/oligomeric complex assembly, we performed non-reducing sucrose gradient velocity sedimentation on retinal extracts from MOP-T/WT, MOP-T/rds−/− and WT non-transgenic controls. Samples were layered onto a 5–20% non-reducing sucrose gradient, and 12 fractions were collected from each gradient and then subjected to reducing SDS–PAGE/western blot analysis (Fig. 3). We have recently reported that, under these conditions, higher-order homo–oligomeric Rds complexes are detected mainly in fractions 1–4, intermediate complexes in fractions 5–6 and core tetrameric complexes in fractions 6–9 (17). In contrast, Rom-1 does not participate in higher-order oligomeric complex formation and is only detected as intermediate oligomers or tetrameric forms (16,17). Sedimentation profiles for Rds and Rom-1 from MOP-T/WT are not significantly different from the non-transgenic WT control (Fig. 3A and B, top two), suggesting that C150S does not interfere with the ability of WT Rds to form complexes. In contrast, we observed a significantly different pattern of Rds/Rom-1 complexes in MOP-T/rds−/−; in the absence of endogenous Rds, no intermediate or higher-order Rds oligomers were formed, and all Rds and Rom-1 were found as core tetrameric complexes in fractions 6–9 (Fig. 3A, bottom). In the absence of WT Rds, Rom-1 is distributed in fewer fractions than normal (7–9 versus 4–8 in the WT, Fig. 3B, bottom). Consistent with the results of previous in vitro work (19), these data clearly indicate that C150S-Rds can assemble into non-covalent core tetrameric complexes with itself and Rom-1, but cannot participate in higher-order complex formation. Furthermore, our sedimentation results from MOP-T confirm that in the absence of native Rds, Rom-1 alone is not sufficient for the production of intermediate or higher-order oligomers.
Figure 3.
The effect of C150S-Rds on the pattern of complex assembly in rods. Non-reducing velocity sedimentation was performed on WT, MOP-T/WT and MOP-T/rds−/− retinal extracts in the presence of NEM and Triton X-100. Fractionated gradients (1–12) were collected and evaluated by western blot using anti-Rds-CT (A) and anti-Rom-1-CT (B) antibodies. Image analysis was performed on blots from three independent experiments for each genotype and corresponding densitometry plots (mean±S.E.M.) are presented to show the distribution patterns of Rds and Rom-1 complexes. The distribution of these complexes is similar in MOP-T/WT and WT retinal extracts. However, in the absence of native Rds, C150S Rds was unable to make higher-order or intermediate complexes but retained the ability to make tetramers (bottom).
C150S-Rds exerts a severe dominant negative effect on cone, but not rod, function
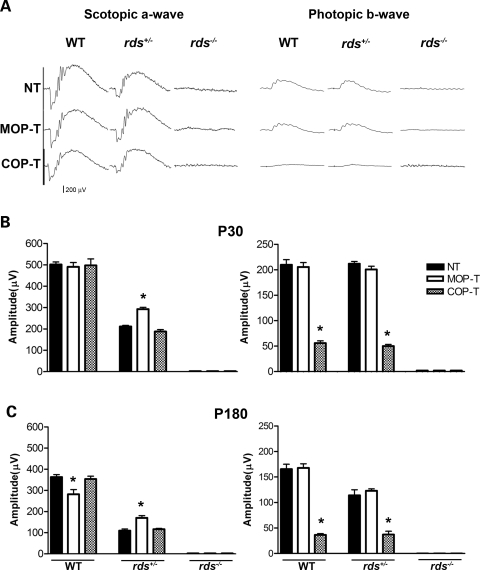
We next determined the role of Rds intermolecular disulfide bonding in rod- and cone- mediated vision. To address this, we assessed retinal function using scotopic and photopic full-field ERG analysis in the MOP-T and COP-T transgenic mice on the WT, rds+/− and rds−/− genetic backgrounds (Fig. 4). At 1 month of age, neither transgene affected rod function in the WT background, although the MOP-T mice exhibited a 22% reduction in maximum scotopic a-wave amplitude at 6 months of age (Fig. 4B and C, left). In the rds+/− background, the MOP-T transgene provided some rod functional rescue at 1 and 6 months of age (Fig. 4B and C, left). In contrast to the mild functional defect in MOP-T mice, cone function in COP-T mice was devastated in both WT and rds+/− backgrounds at all time points (∼75% reduction in photopic b-wave amplitude, Fig. 4B and C, right). Neither transgene was capable of rescuing retinal function in the absence of endogenous Rds (rds−/−). Consistent with the cellular specificity of the promoters, the COP transgene had no effect on rod function, and likewise the MOP transgene had no effect on cone function. The severe dominant negative effect on cones in COP-T mice combined with the partial rescue of rods in MOP-T mice confirms that differences in the ability of Rds to form complexes have severe functional consequences.
Figure 4.
Functional defects associated with the C150S mutation. (A) Scotopic (left) and photopic (right) ERG wave forms from 1-month-old MOP-T, COP-T and non-transgenic animals in the WT, rds+/− and rds−/− backgrounds. (B) Quantitation of scotopic (left) and photopic (right) ERG amplitudes of 1-month-old C150S mice and controls in the WT, rds+/− and rds−/− backgrounds. At P30 a significant cone defect is associated with the expression of C150S-Rds in WT and rds+/− backgrounds and no rod or cone function was seen when C150S is expressed in the rds−/− background. (C) ERG analysis of 6-month-old C150S mice in all genetic backgrounds. Expression of C150S (MOP-T) exerted a late-onset functional defect in rods. However, expression of C150S-Rds in cones (COP-T) continued to dramatically affect cone function. Shown are means±S.E.M. from 5–7 mice. Significant differences as measured by one-way ANOVA (P < 0.001) between transgenic and non-transgenic littermates are marked with asterisks (*).
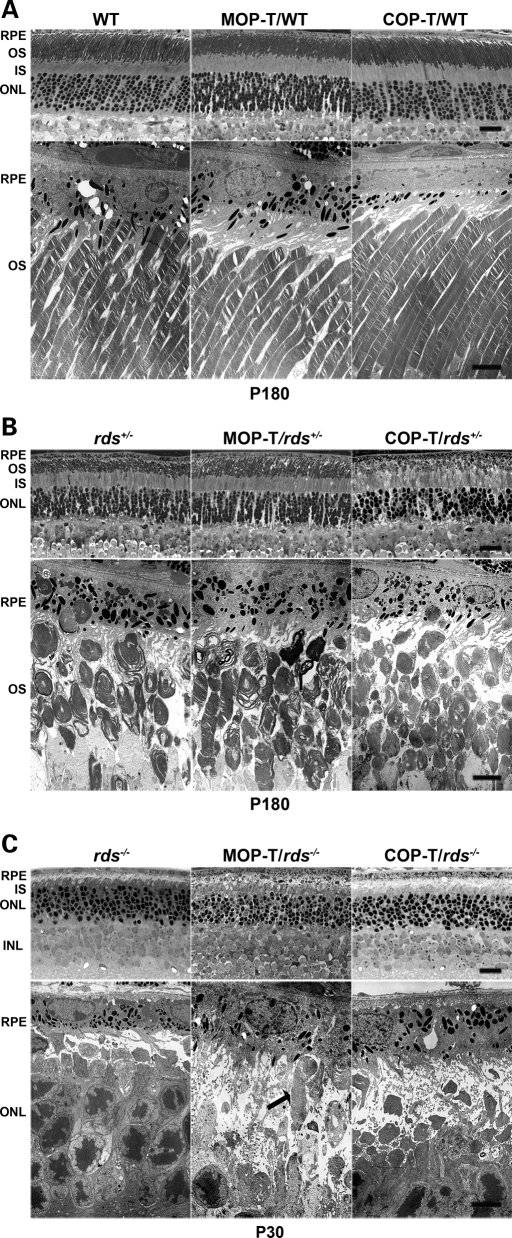
C150S-Rds partially rescues rod OS morphogenesis in rds+/− mice
We next examined what effect C150S-Rds has on retinal structure using light and transmission electron microscopy. In the WT background, neither outer nuclear layer (ONL) thickness (Fig. 5A, top) nor OS length or ultrastructure (Fig. 5A, bottom) was affected by the transgenes when compared with non-transgenic littermates, suggesting that rod viability is unaltered. Expression of C150S-Rds in rods in the rds+/− background (MOP-T/rds+/−) resulted in partial improvements in OS disc formation and alignment, increased OS length and decreased frequency of aberrant, whorl-like OS structures in comparison to non-transgenic controls (Fig. 5B, middle). As expected, expression of C150S-Rds in cones on the rds+/− background (COP-T/rds+/−) did not provide any improvement in rod structure (Fig. 5B, right). As shown in Fig. 5C, C150S-Rds was not capable of supporting significant OS formation in the absence of endogenous Rds. We occasionally noticed the presence of single abnormal rod OS within MOP-T/rds−/− retinas (Fig. 5C, arrow), but never in COP-T/rds−/− or non-transgenic rds−/− retinas. This suggests that some small amount of C150S-Rds may make it to the OS of rods, but not cones and that only in rare instances is this protein sufficient to support the formation of OSs.
Figure 5.
C150S improves rod photoreceptor OS structure in rds+/− retina. Shown are representative light (top) and electron (bottom) microscopy from retinal sections of MOP-T, COP-T and non-transgenic controls in the WT (A), rds+/− (B) and rds−/− (C) backgrounds. Eyes presented in A and B were taken from mice at P180 and in C from mice at P30. Expression of C150S-Rds had no effect on rod OS structure in the WT background and led to partial improvement in OS structure in the rds+/− background. As expected, no change in rod OS structure was noticed in COP-T/rds+/− or COP-T/WT retinas compared with non-transgenic controls. (C) Except in rare instances (arrow) C150S-Rds was not sufficient to support OS formation in the absence of endogenous Rds. RPE, retinal pigment epithelium; OS, outer segment; IS, inner segment; ONL, outer nuclear layer; INL, inner nuclear layer. Scale bar, 4 µm (EM) and 20 µm (light).
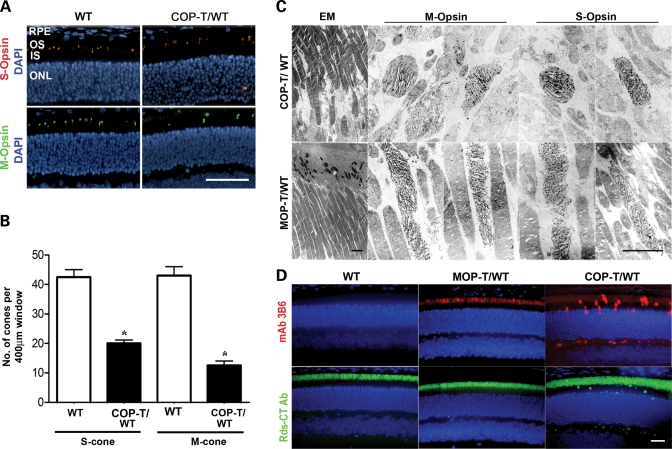
COP-T causes dominant negative cone degeneration and protein mislocalization
Since cones are so scarce in the COP-T retinas, we used immunohistochemistry and EM immunogold labeling with S- or M- cone opsin antibodies to study the effects of C150S-Rds on cone structure and to evaluate the number of cone cells. Although both S- and M- cones were detected in COP-T/WT retinas (Fig. 6A, right), there was a significant decrease in cell number for both S (54% reduction) and M (71%) cones as early as P30 (Fig. 6B). In addition to the decrease in cone number in COP-T/WT retinas, we also noticed considerable structural defects in the OSs of the remaining cones. EM immunogold labeling of both S and M cones (with the opsin antibodies mentioned above) showed that the OSs of both cone cell types have elongated, slightly swirly lamellae and are significantly shorter and more rounded in COP-T/WT compared with the normal cones OSs seen in the MOP-T/WT (Fig. 6C).
Figure 6.
(A) Paraffin-embedded retinal sections from WT and COP-T/WT mice were labeled with anti-S-opsin (red) or anti-M-opsin (green) and counterstained with DAPI (blue). RPE, retinal pigment epithelium; OS, outer segment; IS, inner segment; ONL, outer nuclear layer. Scale bar, 50 µm. (B) The number of cones in retinas of COP-T/WT mice at P30 was significantly decreased compared with non-transgenic controls (mean±S.E.M. n = 3, *=P < 0.001 by Student’s t-test). (C) Immunogold labeling of M- and S-cones (with the corresponding opsin antibodies) in COP-T/WT (top) and MOP-T/WT (bottom) retinas at P30. Expression of C150S Rds in cones (COP-T/WT) results in shorter, abnormal cone lamellae compared with the normal cones seen in the MOP-T/WT. Scale bars: EM, 4 µm; IM, 2 µm. (D) Paraffin-embedded retinal sections were labeled with mAb 3B6 to visualize C150S Rds or with anti-Rds-CT polyclonal antibody to visualize both endogenous and transgenic Rds. C150S-Rds is properly localized to the OS of MOP-T/WT retinas, whereas mislocalized to the inner segments, outer nuclear layer and the outer plexiform layer of COP-T/WT retinas. Scale bar, 20 µm.
To enable us to determine the localization of the C150S-Rds, it was necessary to distinguish between native Rds and transgenic Rds protein. We therefore introduced a P341Q modification at the C-terminus of both transgenes to enable the use of the monoclonal antibody 3B6 (mAb 3B6). Glutamine at position 341 is the only residue of the 3B6 antigenic site shared by human, bovine and rat Rds, but is substituted by a proline in mouse. In a previously published study, we demonstrated that the P341Q modification does not have any negative effect on the structure, function or localization of Rds, but does render it detectable by mAb 3B6 (26). We have also introduced this modification into our other Rds transgenic lines (C214S and R172W) and further documented the specificity of the mAB 3B6 to Rds with the P341Q modification (25,27).
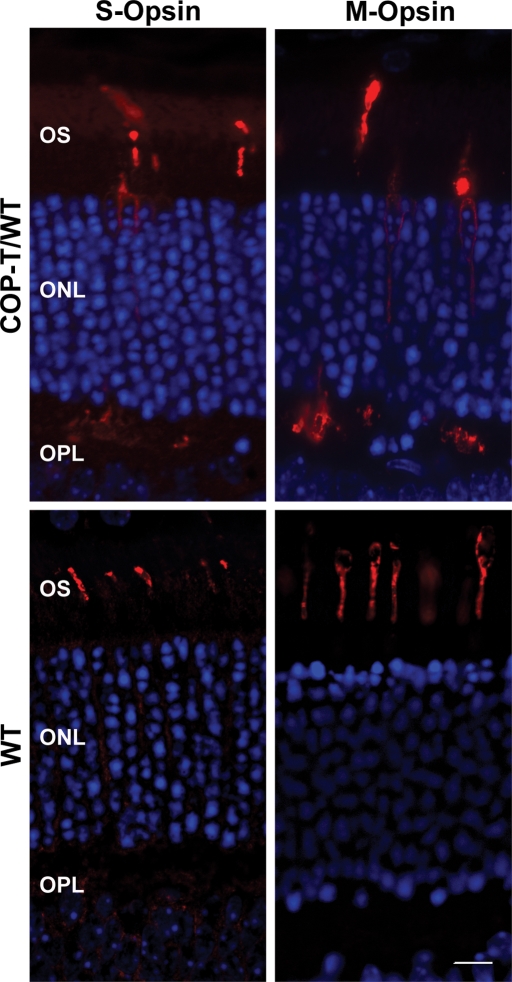
Although both transgenes were expressed in the OS, we observed that C150S-Rds was also mislocalized to the inner segment, outer nuclear layer and outer plexiform layer in COP-T/WT retinas (Fig. 6D, top). When we used an anti-Rds polyclonal antibody that recognizes endogenous Rds and, to a lesser extent, transgenic Rds (Fig. 6D, bottom), we observed robust immunolabeling throughout the OS layer of MOP-T, COP-T transgenic and non-transgenic retinas, with only a small amount of mislocalization in the COP-T retina. This suggests that a substantial portion of the transgenic protein is mislocalized when expressed in cones but that the overwhelming majority (if not all) of the native Rds is properly trafficked to the OSs. As the transgenic C150S protein was partially mislocalized when expressed in cones, we decided to see whether the cone opsins were also mislocalized. Sections from COP-T/WT and WT non-transgenic controls were stained with short wavelength (S-) opsin or medium wavelength (M-) opsin. In Figure 7, we show that both S-opsin and M-opsin are expressed not only in the OS, but are mislocalized throughout the photoreceptor and are particularly noticeable in the synapses in the outer plexiform layer.
Figure 7.
Paraffin-embedded sections from COP-T mice and non-transgenic littermates were collected at P30 and stained with anti-S-opsin or anti-M-opsin to label the cone opsin proteins (left and right). In the presence of the C150S-Rds protein in the COP-T retina (top), both S- and M-opsin are distributed throughout the photoreceptor in contrast to the WT (bottom) in which the protein is restricted to the OS. Scale bar 10 µm.
DISCUSSION
Unlike other tetraspanin proteins, Rds and Rom-1 have only one free Cys that can participate in intermolecular disulfide bond formation (19,28). In the present study, we used mice that express a transgene lacking this free Cys residue (C150S) to study the role of intermolecular disulfide bonding separately in rods (MOP-T) and cones (COP-T). Our initial characterization shows that the C150S-Rds mutant protein is properly folded, stable and capable of forming tetramers, but cannot mediate higher-order oligomer (i.e. larger than tetramer) formation. The lack of C150 leads to a severe dominant negative functional and structural defect in cones, including accelerated cone degeneration and cone opsin mislocalization. In contrast, rods are much more resistant to the lack of C150 in Rds. These phenotypic changes are supported by distinct differences in the biochemical role of C150 in rods and cones. Our data show that in cones lacking C150, Rds loses the ability to interact with Rom-1, whereas in rods this interaction is maintained. Consistent with the absolute requirement for higher-order Rds oligomers to support the formation of OSs, photoreceptors expressing only C150S mutant form of Rds (MOP-T/rds−/− and COP-T/rds−/−) do not support OS formation.
Comparisons between this study and our previous work highlight the phenotypic differences between mutations which interrupt intramolecular disulfide bonding compared with those which interrupt intermolecular disulfide bonding. We have previously studied an intramolecular disulfide bonding Rds mutant (C214S) and reported that the mutant protein is not stable and has a simple loss-of-function phenotype (25), which is in stark contrast to the C150S phenotype reported here, which manifests as neither a simple loss-of-function nor a simple gain-of-function phenotype. The fact that other tetraspanin proteins do not have an additional unpaired cysteine (a C150 equivalent) (29) suggests that tetraspanin proteins in retinal photoreceptors have evolved to fulfill a different role than the traditional tetraspanin proteins.
The hypothesis that Rds intermolecular disulfide bonding depends on C150 was first formulated based on the results of several lines of experimental evidence. It has been shown that WT Rds is capable of flattening isolated microsomal vesicles (mimicking an OS disc rim), but when C150S-Rds is expressed, this flattened morphology is lost (30). This transition was also induced by the addition of reducing agents to the preparation, supporting the idea that it is the ability of C150 to form disulfide bonds that results in the formation of the rim-like structure (30). These data correlate nicely with reports showing that, after transfection of COS cells, C150S-Rds protein does not form disulfide bonds. On the other hand, the exogenous protein was stable, properly folded and could form heterotetramers with Rom-1 (19). While these studies provide a fairly clear picture of the role of C150 in Rds complex formation, they provide very little insight into how that role is relevant to different OS structures of rods and cones or to the electrophysiological competence of these cells required for normal vision.
Previous work has shown that Rds has a different function in rods versus cones (13,31); however, no cellular or biochemical data has been heretofore available to explain why this should be the case. Here we report three findings that contribute to our understanding of the differential requirement of rods and cones for Rds. First, we show a difference in the pattern of Rds/Rom-1 binding in rods and cones. Our biochemical studies show that in rods, C150S-Rds can interact with Rom-1 to form tetramers, whereas in cones, C150S-Rds does not associate with Rom-1. Second, we show a difference in the phenotypic consequences of expressing an oligomerization-incompetent form of Rds (i.e. C150S-Rds) in rods versus cones. In good agreement with the Xenopus laevis study cited above (9), we show that in rods, the presence of the C150S Rds mutation does not cause a dominant degeneration (i.e. in the presence of native Rds), whereas in cones, C150S-Rds is associated with a striking, dominant negative structural and functional defect. Third, we show evidence suggesting that Rds traffics differently in rods versus cones. Consistent with studies in amphibian rods, C150S-Rds in murine rods traffics properly to the OS, but in cones is often mislocalized throughout the photoreceptor and is associated with mislocalization of cone opsins.
Studies have shown that in some cases protein mislocalization and dominant degenerations (32,33) have been associated with overexpression. It is important to note here that overexpression of C150S-Rds in cones is not the cause of the observed phenotype. Although we and others have shown that opsin overexpression causes degeneration (33,34), we have clearly demonstrated that Rds overexpression does not cause any toxic or degenerative response (26). We have previously shown in several other transgenic lines that transgene message levels are not correlated with the protein levels (35), possibly due to lower translation efficiency of the transgene message. Although we report that COP-T message levels are ∼25% of WT levels, this does not likely mean that C150S protein levels in COP-T are 25% of WT. This is supported by the lack of detection of the COP-T protein on western blots of retinal homogenates. We have shown that Rds levels much less than 25% of WT are detectable by western blot analysis (26,27). Furthermore, our preliminary characterization (biochemical and functional) of mid-expresser COP-T lines showed the same phenotype as the COP-T line presented here (data not shown).
How is it that identical molecules of Rds have a different ability to bind Rom-1 in different cell types (i.e. C150S-Rds can bind Rom-1 in rods, but not in cones)? To answer this question, we turn to studies from non-ocular tetraspanin proteins (e.g. CD151 and CD9), which demonstrate that such proteins function as central anchoring points, binding a variety of other proteins to form functional membrane microdomains (36–38). It has been shown that Rds interacts with several proteins in addition to Rom-1 (39,40). We hypothesize that additional interacting proteins bind with Rds/Rom-1 tetramers (in the inner segment during trafficking) or higher-order complexes (in the OS). It is likely that these binding partners are different in rods versus cones, thus allowing Rds to behave differently in these two cell types.
More importantly, what is the biological purpose of differential Rds/Rom-1 binding in rods versus cones? We showed previously that, in the WT retina, Rom-1 is found as part of the heterotetramers and intermediate oligomeric complexes but not higher-order oligomers. However, in the cone-dominant nrl−/− retina, Rom-1 is only found as part of heterotetramers (17). It is possible that cones rely on these Rds/Rom-1 tetramers to perform a yet to be elucidated role in maintaining the open part of cone rims (a function which would not be necessary in rods). In this case, inhibiting the interaction between Rds and Rom-1 in cones (e.g. in the case of the C150S mutation) would lead to a distinct dominant cone degenerative phenotype. It has been well established that Rom-1 is less important in the maintenance of rod OS structure than is Rds, but the role of Rom-1 specifically in cones has not been well studied.
In response to the second point raised above, the vast differences in the consequences of ablating Rds intermolecular disulfide bonding in rods versus cones relate to the core of what makes rods different from cones. Differences in rod versus cone OS biogenesis are poorly understood, due in part to the lack of good cone-dominant animal models; but structurally the two types of OSs are quite distinct. While we (and others) have thoroughly characterized the composition of Rds complexes in rods (16,17), little is known about the composition of Rds complexes in cones. This is particularly important, as cones do not form closed OS discs; rather, they maintain open membranous lamellae. The rim regions of these lamellae have two distinct environments: around one portion of the OS the lamellae are covered by plasma membrane, thus forming a rim-membrane microstructure indistinguishable from that found in rods; around the rest of the cone, the lamellae are open to the extracellular space, which represents an entirely different rim environment from that found in rods. Although we have recently shown (17) that the distribution of Rds complexes (oligomers, intermediate complexes and tetramers) is similar in both WT (rod-dominant) and nrl−/− (cone-dominant) mouse retinas, it is impossible to tell whether Rds complexes on the open side of cones are the same as Rds complexes on the closed side of cones or whether the structural demands on Rds complexes are the same in rods (closed discs inside the cell) versus cones (lamellae in open contact with the extracellular space).
Indeed, evidence from our current study suggests that the structural demands on Rds complexes in rods and cones are quite different. We show that the C150S-Rds is capable of partially alleviating the Rds haploinsufficiency phenotype in the rds+/− mouse. From our previous work, we know that rod structure improves as the quantity of Rds (normal or mutant) present increases. In this case, we propose that C150S can participate in higher-order oligomers (or else why would it improve rod structure?), but that as these oligomers would be held together by fewer disulfide bonds than normal (due to the presence of the C150S), they are less structurally sound than the WT oligomers. In rods, this alteration in structural stability would logically be less important than in cones. First, rod OSs are entirely contained by the plasma membrane and are of a cylindrical shape and, therefore, less likely to be adversely affected by any mechanical forces in the retina. Secondly, in rod OSs, it has been shown that Rds binds to the GARP subunit of the cyclic nucleotide-gated channel, which is expressed on the plasma membrane, thus providing an additional layer of structural stability for the OS. On the contrary, cone lamellar rims are exposed to the extracellular space, and the conical shape of cone OSs may make their lamellae more subject to perturbation. Additionally, we have shown (unpublished data) that Rds does not bind to the cone cyclic nucleotide-gated channel, thus eliminating a secondary support mechanism for cone lamellae. These observations suggest that proper formation of cone OSs relies on the presence of Rds that can make perfect, structurally resilient higher-order oligomers, whereas in rods the total amount of Rds oligomers is the primary determinant of structure. This hypothesis is supported by our past work on the R172W mutation. R172W-Rds causes a dominant cone degeneration (similar to C150S), but the only biochemical difference between R172W-Rds and WT-Rds is that R172W-Rds containing complexes are slightly more subject to tryptic digestion than WT, again suggesting that although R172W-Rds can form oligomers, those complexes are slightly less stable than normal and, therefore, are not capable of supporting cone OS structure.
Finally, the observation that some C150S-Rds is mislocalized in cones, but not rods, provides a further explanation for the severity of the C150S cone defect. Studies in amphibian rods have shown that Rds contains an OS trafficking signal in its C-terminus; so there is no immediately apparent reason why cone trafficking would be affected by C150S, unless rods and cones traffic Rds differently. This hypothesis is supported by the observation that C150S-Rds mislocalization in cones is accompanied by cone opsin mislocalization, whereas in rods substantial evidence has shown that Rds trafficking occurs by a separate pathway from rhodopsin trafficking. Certainly, mislocalization of cone opsin would contribute to the cone-dominant visual defect we observe in COP-T mice.
The data shown here represent a significant step forward in our understanding rod and cone OS biogenesis and the role of Rds in that process. We present evidence to suggest that Rds intermolecular disulfide bonding is differentially important in rods versus cones, likely due to the differential structural requirements of the closed rod discs compared with the open cone lamellae and that Rom-1 may be differentially required by rods versus cones. Finally, we show evidence suggesting that OS trafficking may be quite different in rods versus cones. These data together provide several new avenues for further investigations into the critical cellular processes required for the formation of rod and cone OSs and, thus, the mechanisms underlying vision. Furthermore, these results dramatically enhance our understanding of the role of Rds in rods versus cones and will further efforts to understand and treat the many rod- and cone-dominant blinding diseases caused by mutations in Rds.
MATERIALS AND METHODS
Generation and characterization of C150S transgenic mice
The transgene consists of the 1.6 kb full-length mouse RDS cDNA carrying the C150S mutation and the P341Q modification to enable specific detection with the mAb 3B6, and was based on our previously published normal mouse peripherin/Rds (NMP) transgene (26). The P341Q modification was included to facilitate the detection of the transgene product in the presence of WT Rds, using the mAB 3B6 (a generous gift from Dr Robert S. Molday, University of British Columbia). The transgene was expressed in rods by a 221 bp fragment of the MOP (20) and in cones by a 6.5 kb fragment of the human red/green-opsin promoter (COP) (generously shared by Dr Jeremy Nathans, John Hopkins University, MD) which has been shown to direct robust expression in all mouse cone types (21,22). Pigmented transgenic animals were generated, bred and screened as described previously (22,23,25). Twenty potential founders for the MOP transgene (MOP-T) and 15 potential founders for COP transgene (COP-T) underwent initial characterization and the highest expressing lines from each group were used to generate the data presented in this study. Real-time PCR was performed in triplicate on each cDNA sample with at least three independent animals from each group (iCycler; Bio-Rad Laboratories), and cT values were calculated against the neuronal house keeping gene hypoxanthine phosphoribosyltransferase (HPRT). Results are presented as mean±standard error of the mean (S.E.M.). All experiments and animal maintenance were approved by the local Institutional Animal Care and Use Committee (IACUC; University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA) and conformed to the guidelines on the care and use of animals adopted by the Society for Neuroscience and the Association for Research in Vision and Ophthalmology (Rockville, MD, USA).
Western blot analysis and IP
Western blot analysis and IP were performed using polyclonal antibodies specific to the C-terminal region of Rds (Rds-CT) and Rom-1 (Rom-1-CT) (17,27). Frozen retinas were homogenized on ice in solubilization buffer containing 50 mm Tris–HCl, pH 7.5, 100 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.05% SDS, 2.5% glycerol and 1.0 mm phenylmethyl-sulfonyl fluoride. Western blot (20 µg protein) and IP (100 µg protein from MOP-T and 300 µg protein from COP-T) were undertaken as previously described (17). For analysis of disulfide-linked dimers, gel electrophoresis under non-reducing condition was conducted by omitting dithiothreitol from the sample buffer.
Velocity sedimentation analysis
Non-reducing velocity sedimentation was performed on 200 µg whole retinal extract as described previously (17,27). Continuous density gradients of 5–20% sucrose were prepared by sequentially layering 0.5 ml each of 20, 15, 10 and 5% (w/v) sucrose solutions in phosphate-buffered saline containing 0.1% Triton X-100 and 10 mm N-ethylmaleimide (NEM). Gradients were allowed to become continuous by diffusion at room temperature for 1 h and then chilled on ice for at least 30 min prior to sample loading. On average, 12 fractions were collected for each sample and labeled 1–12, where #1 is the heaviest fraction (from the bottom of the tube) and #12 to the lightest fraction (from the top of the tube). The samples were then analyzed by SDS–PAGE and western blotting.
Electroretinography
Rod and cone full-field electroretinographies (ERGs) were performed as previously described (13,24,27). Measurement of scotopic a-wave amplitude was made from the pre-stimulus baseline to the a-wave trough, and the b-wave amplitude was made from the trough of the a-wave to the crest of b-wave. To evaluate photopic response, animals were light adapted for 5 min prior to recording and measurement of photopic b-wave amplitude was made from the trough of the a-wave to the crest of the b-wave.
Immunofluorescence labeling
Tissue fixation and sectioning were performed as previously described (17). Primary antibodies were anti-M-opsin (1:1000), anti-S-opsin (1:200), Rds-CT (1:1000) and mAb 3B6 (1:100) (26). Anti-S-opsin and anti-Rds-CT were generated in house (31) and anti-M-opsin was generously provided by Dr Cheryl Craft (University of Southern California) (41). Antigens were visualized after incubation with Alexa Fluor® 488 or 555-conjugated goat IgGs (Molecular Probes). Cone number was quantified by counting the number of cone OSs (stained with either M- or S-opsin) in the 400 µm immediately superior to (S-cones) or inferior to (M-cones) the optic nerve (N = 3–5). Fluorescent images were captured using either a 20× (air, 0.75 NA) or 60× (oil, 1.42 NA) objective with an Olympus BX-62 microscope equipped with a spinning disc confocal unit. Images were stored and deconvolved (no neighbors paradigm) using Slidebook® version 4.2 and are either epifluorescent (Fig. 6) or single slices of a confocal stack (Fig. 7). Figure assembly was done in Adobe Photoshop CS.
Light/electron microscopy and immunogold labeling
The methods employed for tissue collection and processing for plastic-embedment light and electron microscopy and immunogold labeling were as previously described (13,25). Tissue sections were obtained with a Reichert–Jung Ultracut E microtome using glass or diamond knives. Thin (600–800 Å) sections were collected on copper 75/300 mesh grids for EM analysis and stained with 2% (w/v) uranyl acetate and Reynolds’ lead citrate. Sections for immunogold were collected on nickel 75/300 mesh grids; primary antibodies (anti-M-opsin and anti-S-opsin) were used at 1:10 dilution; secondary antibodies (AuroProbe® 10 nm gold-conjugated goat anti-rabbit IgG; Amersham) were used at 1:50 dilution. Sections were viewed with a JEOL 100CX electron microscope at an accelerating voltage of 60 keV.
FUNDING
This study was supported by grants from the National Institutes of Health [EY10609 (M.I.N.), EY018656 (M.I.N.) and EY007361 (S.J.F.)], Core Grant for Vision Research EY12190 (M.I.N.), the Foundation Fighting Blindness (M.I.N.) and the Knights Templar Eye Research Foundation (D.C.). Dr Naash is the recipient of a Research to Prevent Blindness James S. Adams Scholar Award. Dr Fliesler is the recipient of a Research to Prevent Blindness Senior Scientific Investigator Award.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs Molday and Craft for sharing their antibodies and to Dr Nathans for sharing his promoter. We thank Alexander Quiambao, Mark Ballard, Jeff S. Skaggs, Barbara A. Nagel and Carla Hansens for providing excellent technical assistance.
Conflict of Interest statement. None of the authors has any conflict of interest.
REFERENCES
- 1.Corless J.M., Fetter R.D., Costello M.J. Structural features of the terminal loop region of frog retinal rod outer segment disk membranes. I. Organization of lipid components. J. Comp. Neurol. 1987;257:1–8. doi: 10.1002/cne.902570102. [DOI] [PubMed] [Google Scholar]
- 2.Eckmiller M.S. Cone outer segment morphogenesis: taper change and distal invaginations. J. Cell Biol. 1987;105:2267–2277. doi: 10.1083/jcb.105.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roof D.J., Heuser J.E. Surfaces of rod photoreceptor disk membranes: integral membrane components. J. Cell Biol. 1982;95:487–500. doi: 10.1083/jcb.95.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang J.Z., Zhao Y., Sung C.H. SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell. 2007;130:535–547. doi: 10.1016/j.cell.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arikawa K., Molday L.L., Molday R.S., Williams D.S. Localization of peripherin/Rds in the disk membranes of cone and rod photoreceptors: relationship to disk membrane morphogenesis and retinal degeneration. J. Cell Biol. 1992;116:659–667. doi: 10.1083/jcb.116.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molday R.S., Hicks D., Molday L. Peripherin: a rim-specific membrane protein of rod outer segment discs. Invest. Ophthalmol. Vis. Sci. 1987;28:50–61. [PubMed] [Google Scholar]
- 7.Connell G., Bascom R., Molday L., Reid D., McInnes R.R., Molday R.S. Photoreceptor peripherin is the normal product of the gene responsible for retinal degeneration in the Rds mouse. Proc. Natl Acad. Sci. USA. 1991;88:723–726. doi: 10.1073/pnas.88.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J., Norton J.C., Allen A.C., Burns J.B., Hasel K.W., Burns J.L., Sutcliffe J.G., Travis G.H. Retinal degeneration slow (Rds) in mouse results from simple insertion of a t-haplotype-specific element into protein-coding exon II. Genomics. 1995;28:212–219. doi: 10.1006/geno.1995.1133. [DOI] [PubMed] [Google Scholar]
- 9.Loewen C.J., Moritz O.L., Tam B.M., Papermaster D.S., Molday R.S. The role of subunit assembly in peripherin-2 targeting to rod photoreceptor disk membranes and retinitis pigmentosa. Mol. Biol. Cell. 2003;14:3400–3413. doi: 10.1091/mbc.E03-02-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kajiwara K., Hahn L.B., Mukai S., Travis G.H., Berson E.L., Dryja T.P. Mutations in the human retinal degeneration slow gene in autosomal dominant retinitis pigmentosa. Nature. 1991;354:480–483. doi: 10.1038/354480a0. [DOI] [PubMed] [Google Scholar]
- 11.Wells J., Wroblewski J., Keen J., Inglehearn C., Jubb C., Eckstein A., Jay M., Arden G., Bhattacharya S., Fitzke F., et al. Mutations in the human retinal degeneration slow (RDS) gene can cause either retinitis pigmentosa or macular dystrophy. Nat. Genet. 1993;3:213–218. doi: 10.1038/ng0393-213. [DOI] [PubMed] [Google Scholar]
- 12.van Soest S., Westerveld A., de Jong P.T., Bleeker-Wagemakers E.M., Bergen A.A. Retinitis pigmentosa: defined from a molecular point of view. Surv. Ophthalmol. 1999;43:321–334. doi: 10.1016/s0039-6257(98)00046-0. [DOI] [PubMed] [Google Scholar]
- 13.Farjo R., Skaggs J.S., Nagel B.A., Quiambao A.B., Nash Z.A., Fliesler S.J., Naash M.I. Retention of function without normal disc morphogenesis occurs in cone but not rod photoreceptors. J. Cell Biol. 2006;173:59–68. doi: 10.1083/jcb.200509036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mears A.J., Kondo M., Swain P.K., Takada Y., Bush R.A., Saunders T.L., Sieving P.A., Swaroop A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg A.F., Moritz O.L., Molday R.S. Heterologous expression of photoreceptor peripherin/Rds and Rom-1 in COS-1 cells: assembly, interactions, and localization of multisubunit complexes. Biochemistry. 1995;34:14213–14219. doi: 10.1021/bi00043a028. [DOI] [PubMed] [Google Scholar]
- 16.Loewen C.J., Molday R.S. Disulfide-mediated oligomerization of peripherin/Rds and Rom-1 in photoreceptor disk membranes: implications for photoreceptor outer segment morphogenesis and degeneration. J. Biol. Chem. 2000;275:5370–5378. doi: 10.1074/jbc.275.8.5370. [DOI] [PubMed] [Google Scholar]
- 17.Chakraborty D., Ding X.Q., Fliesler S.J., Naash M.I. Outer segment oligomerization of Rds: evidence from mouse models and subcellular fractionation. Biochemistry. 2008;47:1144–1156. doi: 10.1021/bi701807c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke G., Goldberg A.F., Vidgen D., Collins L., Ploder L., Schwarz L., Molday L.L., Rossant J., Szel A., Molday R.S., et al. Rom-1 is required for rod photoreceptor viability and the regulation of disk morphogenesis. Nat. Genet. 2000;25:67–73. doi: 10.1038/75621. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg A.F., Loewen C.J., Molday R.S. Cysteine residues of photoreceptor peripherin/Rds: role in subunit assembly and autosomal dominant retinitis pigmentosa. Biochemistry. 1998;37:680–685. doi: 10.1021/bi972036i. [DOI] [PubMed] [Google Scholar]
- 20.Quiambao A.B., Peachey N.S., Mangini N.J., Rohlich P., Hollyfield J.G., al-Ubaidi M.R. A 221-bp fragment of the mouse opsin promoter directs expression specifically to the rod photoreceptors of transgenic mice. Vis. Neurosci. 1997;14:617–625. doi: 10.1017/s095252380001258x. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y., Smallwood P.M., Cowan M., Blesh D., Lawler A., Nathans J. Mutually exclusive expression of human red and green visual pigment-reporter transgenes occurs at high frequency in murine cone photoreceptors. Proc. Natl Acad. Sci. USA. 1999;96:5251–5256. doi: 10.1073/pnas.96.9.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fei Y., Hughes T.E. Transgenic expression of the jellyfish green fluorescent protein in the cone photoreceptors of the mouse. Vis. Neurosci. 2001;18:615–623. doi: 10.1017/s0952523801184117. [DOI] [PubMed] [Google Scholar]
- 23.Naash M.I., Hollyfield J.G., al-Ubaidi M.R., Baehr W. Simulation of human autosomal dominant retinitis pigmentosa in transgenic mice expressing a mutated murine opsin gene. Proc. Natl Acad. Sci. USA. 1993;90:5499–5503. doi: 10.1073/pnas.90.12.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng T., Peachey N.S., Li S., Goto Y., Cao Y., Naash M.I. The effect of peripherin/Rds haploinsufficiency on rod and cone photoreceptors. J. Neurosci. 1997;17:8118–8128. doi: 10.1523/JNEUROSCI.17-21-08118.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stricker H.M., Ding X.Q., Quiambao A., Fliesler S.J., Naash M.I. The Cys214→Ser mutation in peripherin/Rds causes a loss-of-function phenotype in transgenic mice. Biochem. J. 2005;388:605–613. doi: 10.1042/BJ20041960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nour M., Ding X.Q., Stricker H., Fliesler S.J., Naash M.I. Modulating expression of peripherin/Rds in transgenic mice: critical levels and the effect of overexpression. Invest. Ophthalmol. Vis. Sci. 2004;45:2514–2521. doi: 10.1167/iovs.04-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding X.Q., Nour M., Ritter L.M., Goldberg A.F., Fliesler S.J., Naash M.I. The R172W mutation in peripherin/Rds causes a cone–rod dystrophy in transgenic mice. Hum. Mol. Genet. 2004;13:2075–2087. doi: 10.1093/hmg/ddh211. [DOI] [PubMed] [Google Scholar]
- 28.Hemler M.E. Specific tetraspanin functions. J. Cell Biol. 2001;155:1103–1107. doi: 10.1083/jcb.200108061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemler M.E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 30.Wrigley J.D., Ahmed T., Nevett C.L., Findlay J.B. Peripherin/Rds influences membrane vesicle morphology: implications for retinopathies. J. Biol. Chem. 2000;275:13191–13194. doi: 10.1074/jbc.c900853199. [DOI] [PubMed] [Google Scholar]
- 31.Farjo R., Fliesler S.J., Naash M.I. Effect of Rds abundance on cone outer segment morphogenesis, photoreceptor gene expression, and outer limiting membrane integrity. J. Comp. Neurol. 2007;504:619–630. doi: 10.1002/cne.21476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stieger K., Mendes-Madeira A., Meur G.L., Weber M., Deschamps J.Y., Nivard D., Provost N., Moullier P., Rolling F. Oral administration of doxycycline allows tight control of transgene expression: a key step towards gene therapy of retinal diseases. Gene Ther. 2007;14:1668–1673. doi: 10.1038/sj.gt.3303034. [DOI] [PubMed] [Google Scholar]
- 33.Olsson J.E., Gordon J.W., Pawlyk B.S., Roof D., Hayes A., Molday R.S., Mukai S., Cowley G.S., Berson E.L., Dryja T.P. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- 34.Tan E., Wang Q., Quiambao A.B., Xu X., Qtaishat N.M., Peachey N.S., Lem J., Fliesler S.J., Pepperberg D.R., Naash M.I., et al. The relationship between opsin overexpression and photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 2001;42:589–600. [PubMed] [Google Scholar]
- 35.Naash M.I., Wu T.H., Chakraborty D., Fliesler S.J., Ding X.Q., Nour M., Peachey N.S., Lem J., Qtaishat N., al-Ubaidi M.R., et al. Retinal abnormalities associated with the G90D mutation in opsin. J. Comp. Neurol. 2004;478:149–163. doi: 10.1002/cne.20283. [DOI] [PubMed] [Google Scholar]
- 36.Serru V., Le Naour F., Billard M., Azorsa D.O., Lanza F., Boucheix C., Rubinstein E. Selective tetraspan–integrin complexes (CD81/alpha4beta1, CD151/alpha3beta1, CD151/alpha6beta1) under conditions disrupting tetraspan interactions. Biochem. J. 1999;340:103–111. [PMC free article] [PubMed] [Google Scholar]
- 37.Wu X.R., Medina J.J., Sun T.T. Selective interactions of UPIa and UPIb, two members of the transmembrane 4 superfamily, with distinct single transmembrane-domained proteins in differentiated urothelial cells. J. Biol. Chem. 1995;270:29752–29759. doi: 10.1074/jbc.270.50.29752. [DOI] [PubMed] [Google Scholar]
- 38.Yauch R.L., Berditchevski F., Harler M.B., Reichner J., Hemler M.E. Highly stoichiometric, stable, and specific association of integrin alpha3beta1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol. Biol. Cell. 1998;9:2751–2765. doi: 10.1091/mbc.9.10.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boesze-Battaglia K., Song H., Sokolov M., Lillo C., Pankoski-Walker L., Gretzula C., Gallagher B., Rachel R.A., Jenkins N.A., Copeland N.G., et al. The tetraspanin protein peripherin-2 forms a complex with melanoregulin, a putative membrane fusion regulator. Biochemistry. 2007;46:1256–1272. doi: 10.1021/bi061466i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poetsch A., Molday L.L., Molday R.S. The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J. Biol. Chem. 2001;276:48009–48016. doi: 10.1074/jbc.M108941200. [DOI] [PubMed] [Google Scholar]
- 41.Zhu X., Brown B., Li A., Mears A.J., Swaroop A., Craft C.M. GRK1-dependent phosphorylation of S and M opsins and their binding to cone arrestin during cone phototransduction in the mouse retina. J. Neurosci. 2003;23:6152–6160. doi: 10.1523/JNEUROSCI.23-14-06152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.