Abstract
Autosomal recessive retinal diseases caused by mutations in the ABCA4 gene are being considered for gene replacement therapy. All individuals with ABCA4-disease show macular degeneration, but only some are thought to progress to retina-wide blindness. It is currently not predictable if or when specific ABCA4 genotypes will show extramacular disease, and how fast it will progress thereafter. Early clinical trials of focal subretinal gene therapy will aim to arrest disease progression in the extramacular retina. In 66 individuals with known disease-causing ABCA4 alleles, we defined retina-wide disease expression by measuring rod- and cone-photoreceptor-mediated vision. Serial measurements over a mean period of 8.7 years were consistent with a model wherein a normal plateau phase of variable length was followed by initiation of retina-wide disease that progressed exponentially. Once initiated, the mean rate of disease progression was 1.1 log/decade for rods and 0.45 log/decade for cones. Spatio-temporal progression of disease could be described as the sum of two components, one with a central-to-peripheral gradient and the other with a uniform retina-wide pattern. Estimates of the age of disease initiation were used as a severity metric and contributions made by each ABCA4 allele were predicted. One-third of the non-truncating alleles were found to cause more severe disease than premature truncations supporting the existence of a pathogenic component beyond simple loss of function. Genotype-based inclusion/exclusion criteria and prediction of the age of retina-wide disease initiation will be invaluable for selecting appropriate candidates for clinical trials in ABCA4 disease.
INTRODUCTION
Human ocular gene therapy success in a rare autosomal recessive blindness (1–4) has recently paved the path to clinical trials in more common diseases of the retina such as those caused by mutations in the ABCA4 gene (5–7). ABCR protein expressed by the ABCA4 gene is localized to photoreceptors of the retina (8,9) and is hypothesized to be involved in the clearance from photoreceptor cells of a byproduct of the retinoid cycle of vision (10–14). Pre-clinical studies of gene therapy in abcr-deficient mice are accelerating now that the gene can be packaged in recombinant vectors derived from lentivirus (15) or adeno-associated virus (16).
Are there any impediments to translating this promising therapeutic direction into human trials? One major hurdle to initiating treatment is the dearth of understanding of which ABCA4-mutant humans would be suitable candidates for early trials with the goal of halting or slowing down the relentless progression to blindness. The spectrum of clinically defined ABCA4 disease is notably broad (17–19), but there currently is no way of predicting whether or when an individual will progress to retina-wide blindness and thereby warrant a possible experimental intervention. This study quantifies the initiation and progression of disease in a large cohort of patients, defines a predictive model for the natural history of disease and characterizes the relationship between the parameters of this model and disease-causing ABCA4 variants. The results provide criteria for identifying appropriate candidates at appropriate ages and with specific ABCA4 variants for upcoming human gene therapy trials in ABCA4 disease.
RESULTS
ABCA4 mutations lead to a wide spectrum of disease severity
Sixty-six individuals representing 54 families were studied (Supplementary Material, Table S1). All individuals were found to harbor two ABCA4 variants likely to cause the retinal disease (18,20–26). In 40 families (74%), independent segregation of the two alleles was demonstrated. The ages at the time of their first visit ranged from 9 to 74 years (mean = 35.9, median = 35.2 years); in the majority of individuals (36/66=55%), data were available from a second visit that occurred on average 8.7 years (range=2–20 years, median = 6.9 years) after the first visit.
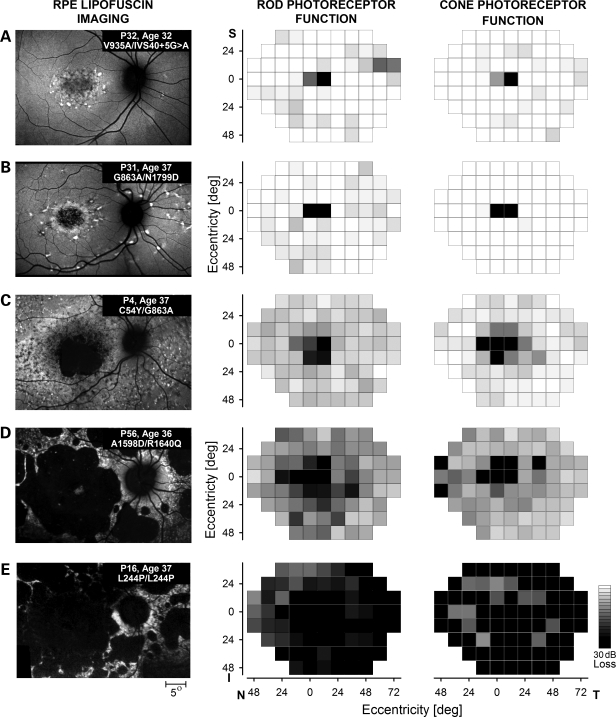
The range of disease expression in the macula is illustrated with retinal pigment epithelial (RPE) cell lipofuscin imaging (Fig. 1, left column). Retina-wide disease is shown with maps of photoreceptor-mediated function (Fig. 1, middle and right columns). The five similar-aged individuals from different families in Figure 1 are representative of the disease expressions in the cohort. Mild macular disease could include minor lipofuscin disturbance limited to the foveal and/or parafoveal region such as P32 (patient 32) carrying V935A and IVS40+5G>A alleles (Fig. 1A). P32 retained foveal vision and a visual acuity (VA) of 20/32. Intermediate stages of macular disease showing different extents of macular atrophy surrounded by flecks or speckled regions are exemplified by individuals such as P31 carrying G863A and N1799D alleles (Fig. 1B) and P4 with C54Y and G863A alleles (Fig. 1C). P31 had complete foveal degeneration and used a macular location ∼6° supero-temporal to the anatomical fovea to score a VA of 20/125. P4 also used an extramacular retinal locus and VA was 20/320. More severe macular disease consisted of a near-complete loss of the lipofuscin signal and atrophy of the macular RPE surrounded by additional smaller patches of atrophy as illustrated by P56, a compound heterozygote for A1598D and R1640Q alleles (Fig. 1D) or by P16, a homozygote for the L244P allele (Fig. 1E). P56 and P16 used extramacular preferred retinal loci at ∼16° eccentric and VAs were 20/200 and 20/1000, respectively. The VA results in these five individuals were representative of the range of acuities (20/20 to ‘hand motion’) in the entire cohort (data not shown).
Figure 1.
The wide spectrum of ABCA4 disease severity in the macula and across the retina, illustrated in unrelated individuals in their fourth decade of life. Images (left column) show the macular distribution of RPE disease by taking advantage of lipofuscin autofluorescence excited with short-wavelength light. All images are individually contrast stretched for better visibility of features. Patient number, age and disease-causing ABCA4 mutations are shown (insets). Maps (middle and right columns) show retina-wide distribution of rod- and cone-mediated sensitivity losses obtained with psychophysical testing on a uniform grid. Magnitudes of the losses compared with mean normal values are shown on a gray scale; the physiological blind spot is represented with a black square at 12° temporal visual field locus. All images and maps in this and subsequent figures are shown as equivalent right eyes to facilitate comparisons. I, inferior field; N, nasal field; S, superior field; and, T, temporal field.
The spatial distribution of retina-wide visual function mediated by rod or cone photoreceptors was sampled on a regular grid by using dark- and light-adapted chromatic perimetry. All five patients had loss of sensitivity at the fovea. Outside the fovea, P32, P31 and P4 had mostly normal rod- or cone-mediated sensitivities (Fig. 1A–C); the average sensitivity losses were 0.3, 0.7 and 4.5 dB for rods (3SD limits for normal +/−7.4 dB) and −0.6, −3 and 2 dB for cones (3SD limits for normal +/−4.4 dB), respectively. P56 and P16, on the other hand, had abnormal sensitivities retina-wide (Fig. 1D and E); average sensitivity losses were 10.8 and 29.5 dB for rods and 8.0 and 16.8 dB for cones, respectively. Data from these five representative individuals suggest that visual function may represent a method to quantify and compare the range of retina-wide disease severities observed in different ABCA4 genotypes.
Progression of retina-wide ABCA4 disease in younger and older patients
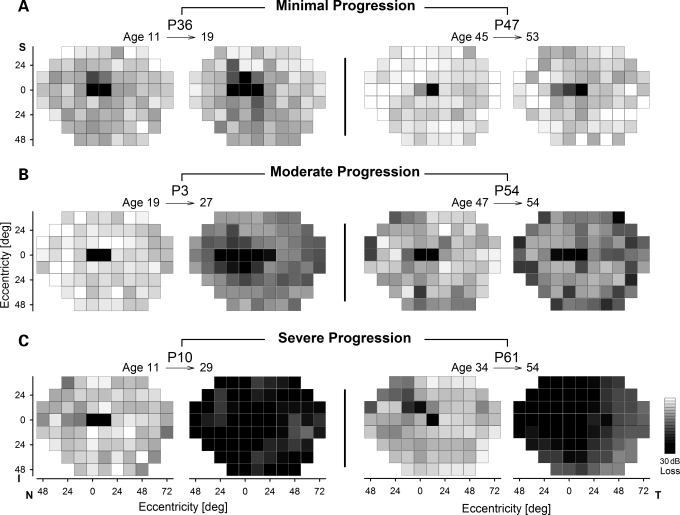
Longitudinal studies over years to decades are the ideal data set for understanding the spatio-temporal progression of disease. Figure 2 illustrates serial observations made over 7–20 years in six individuals who at their initial visit showed no significant extramacular disease. P36 and P47 (Fig. 2A) showed minimal progression of disease over an extended period of time. At age 11, P36 (A1038V;L541P/N965S) had a sensitivity loss of 5.3 dB which was within normal limits; 8 years later at age 19 there was no significant change in sensitivity loss (5.6 dB). Similarly, P47 (P1380L/G1961E) had a sensitivity loss (2.3 dB) that was within normal limits at age 45, and this increased, but not significantly, to 3.6 dB over an 8-year period. In contrast, P3 and P54 (Fig. 2B) represent moderate (and significant) progression over a similar interval. P3 (W41X/R1098C) progressed from normal sensitivity (3.1 dB loss) at age 19 to a loss of 12.5 dB at age 27. P54 (T1526M/R2030Q) progressed from a mildly abnormal loss of 7.6 dB at age 47 to a greater loss of 14.4 dB at age 54. In some individuals, longer periods of follow-up revealed severe progression of disease (Fig. 2C). For example, P10 (R152X/IVS38-10T>C) progressed from normal sensitivity (5.5 dB loss) at age 11 to a dramatically abnormal sensitivity (30.9 dB loss) at age 29. P61 (L1940P/IVS40+5G>A) progressed from normal sensitivity (6.1 dB loss) at age 34 to an abnormal sensitivity (23.8 dB loss) at age 54. It is notable that there were also individuals showing no apparent progression over 12 or more years (e.g. P42, P55), on the other hand, all individuals with significant retina-wide disease at their initial visit showed significant further progression (data not shown). Cone sensitivity maps generally followed similar trends as the rod maps but losses were of smaller magnitude (data not shown).
Figure 2.
Serial data illustrating different levels of retina-wide disease progression. Evaluations in three younger (left two panels) and three older (right two panels) individuals with ABCA4 mutations demonstrate examples of minimal (A), moderate (B) and severe (C) progression of retina-wide rod disease over intervals ranging from 7 to 20 years.
Natural history of retina-wide disease: application of a model
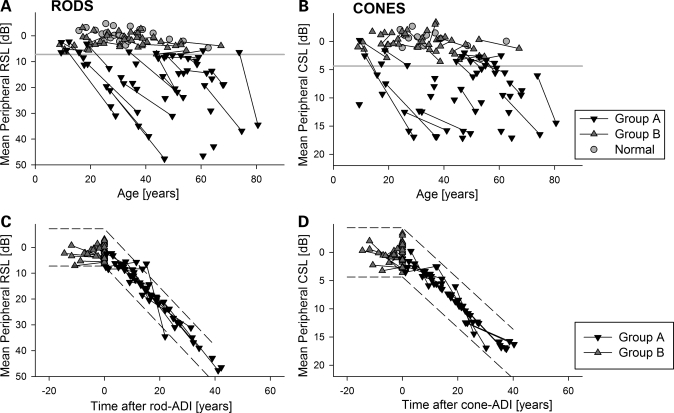
In order to decipher the wide spectrum of retina-wide ABCA4 disease and its progression, we considered the principal study population as the subset of individuals (Group A, n = 36) who had abnormal extramacular rod or cone sensitivity at one or more visits. Data from the remaining individuals with normal sensitivities (Group B, n = 30) were used as a supporting study population since it was not known if or when in the future they would demonstrate retina-wide disease. Quantitative evidence for the wide spectrum of disease can be seen from the plots of rod- and cone-sensitivity loss against age (Fig. 3A and B). Group A individuals exhibited varying extents of sensitivity loss ranging from normal to severely abnormal, and those with longitudinal data show a tendency for progression from less abnormal to more abnormal (diagonal lines). Median (and mean) progression rate of rod-mediated sensitivity loss in Group A was 1.1 log/decade (95% CI = 0.42 log/decade, n = 19). The rate of progression for cones tended to be smaller than that for rods and clustered around a median value of 0.45 log/decade (95% CI = 0.17 log/decade, n = 17). In contrast and as expected, Group B results overlapped with those of normal subjects across a wide range of ages, and individuals with longitudinal data showed a tendency for no progression (nearly horizontal lines connecting the symbols).
Figure 3.
Derivation of a quantitative severity metric based on a mathematical model of retina-wide rod and cone disease progression. (A and B) Plots of mean retina-wide rod and cone sensitivity loss (RSL, CSL) as a function of age illustrate the range of ABCA4 disease severity observed in the current cohort. Serial evaluations in the same individual are shown with a line connecting symbols. The lower limit (mean−3SD) of normal is shown (horizontal gray line). Group A individuals showed significant sensitivity loss at one or both of their evaluations. Group B individuals had normal sensitivities. (C and D) Plot of mean RSL and CSL data against time after age of disease initiation (ADI) supports a common underlying function of disease progression by reducing the scatter apparent in (A) and (B). The function of disease progression assumes that all individuals progress with an invariant exponential rate after a variable delay. Dashed lines represent the normal variability (+/−3SD) extended along the respective exponentials for rod and cone disease.
Next, we evaluated a model wherein disease progresses exponentially after an initial delay; variants of this model have previously been used to describe progression of different parameters related to vision in hereditary retinopathies (27–32) as well as photoreceptor death kinetics in animal models (33,34). The model has two parameters: a time delay called age of disease initiation (ADI) and an exponential rate defining progression of disease after the ADI. Based on relatively small variability observed in progression rates of patients at different ages, we hypothesized that the ADI parameter is the dominant contributor to the observed variation in ABCA4 disease. For each Group A individual, the ADI value corresponded to the age when the x-axis intercepts the progression line of invariant slope (equal to the group median) fit to the sensitivity loss. For Group A individuals with two visits (n = 19) progression lines were fit to minimize the error. Rod and cone ADIs were separately estimated from their respective data sets. For Group B patients, the ADI was specified to be greater than the age of their most recent visit with the presumption that these individuals would show initiation of retina-wide disease at a future visit (Supplementary Material, Table S1). Replotting the data shown in Fig. 3A and B against the time after rod- and cone-ADIs demonstrated an orderly natural history applicable to all individuals over a cumulative time period ranging over four decades (Fig. 3C and D). Rod- and cone-ADI values were highly correlated (linear regression, r2 = 0.92) with a slope of near unity, and there was a tendency for the cone ADI to occur earlier than the rod ADI at younger ages (data not shown).
Electroretinograms (ERGs) were available from 55 of 66 (83%) individuals at a single visit. ERGs represent an objective measure of the mean function across the retina. Rod and cone ERG amplitudes of Group A individuals showed a wide spectrum of severity across all ages, as expected from their rod and cone sensitivity loss data. ERG results of all Group B subjects were within normal limits (Supplementary Material, Fig. S1A and B). Application of the rod- and cone-ADI parameters derived from sensitivity data to the ERG data reduced the apparent variability and resulted in natural history functions showing an orderly progression of disease (Supplementary Material, Fig. S1C and D). The psychophysical sensitivity and ERG results taken together suggest that the natural history of extramacular ABCA4 rod and cone disease is represented with a delayed exponential function to a good first approximation, and that ADI is a single parameter that can quantify the age of the delay and hence the severity of retina-wide disease.
Spatial progression of ABCA4 disease
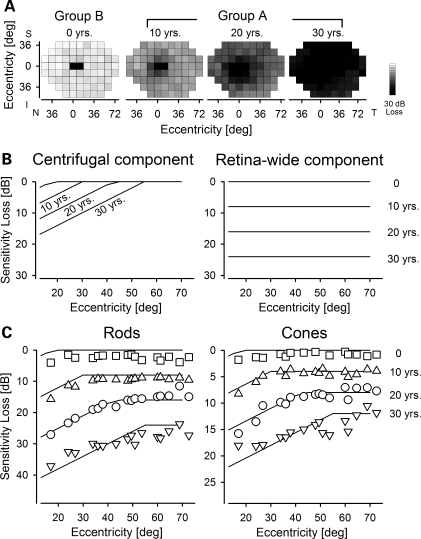
There are notable extremes of ABCA4 disease: at one end of the spectrum are individuals with mild retinopathy confined to the macula, and at the other end are those with severe retina-wide disease (Fig. 1). Spatial progression involving a centrifugal expansion of disease between the two extremes is often assumed but has never been established. We used our model of temporal disease progression to better understand spatial disease progression (Fig. 4). Group A individuals were divided into three decades based on the estimated duration of the retina-wide disease (10, 20 or 30 years, Fig. 4A) at the time of their visit. Maps of mean sensitivity loss for each decade of disease duration were compared with those individuals with no retina-wide disease (0 years, Fig. 4A), which were conservatively chosen from the subset of Group B individuals with follow-up. Mean maps showed the deepening of the severity of macular sensitivity loss, centrifugal enlargement of the central dysfunction and progression of loss in peripheral regions (Fig. 4A). The results suggest the existence of two components of disease: one showing a central-to-peripheral (centrifugal) spatial gradient of sensitivity loss with a boundary that expands over time, and the other causing a uniform progression of sensitivity loss over time. In order to quantitatively assess these two components, we used a spatio-temporal disease model based on the same delayed exponential model described earlier. The centrifugal disease component was defined by a spatially variant ADI that linearly increases with eccentricity and a spatially invariant progression rate; the uniform disease component included spatially invariant ADI and progression rates (Fig. 4B). The sum of these two components (Fig. 4C, lines) was fit as an ensemble to the mean ABCA4 data (Fig. 4C, symbols) and there was a good correspondence over an estimated disease duration of 30 years. Centrifugal component parameters included 0.8 years/deg linear expansion of ADI for rods and cones, and progression rates of 0.5 and 0.3 log/decade for rods and cones, respectively. The retina-wide component parameters included 0.8 and 0.4 log/decade progression rates for rods and cones, respectively.
Figure 4.
Spatio-temporal progression of retina-wide disease. (A) Maps of mean rod sensitivity loss in Group B individuals with no retina-wide disease (0 years, n = 16) and Group A individuals divided into three sub-groups based on their estimated duration of retina-wide disease (10 years, n = 9; 20 years, n = 13; 30 years, n = 8). (B) Two components (centrifugal and retina-wide) of a model describe spatio-temporal progression of rod disease as a function of eccentricity from the fovea. (C) Mean values of the rod and cone sensitivity losses before initiation of retina-wide disease (0 years) and over three decades of disease duration (10, 20 and 30 years) are shown as a function of eccentricity from the fovea (symbols). Spatio-temporal model of disease progression (lines) consisting of the sum of a centrifugal component and a diffuse retina-wide component are fit (by eye) as ensemble to the data.
Predicting mutant allele severity from disease severity
In our cohort of 54 families, there was considerable genetic heterogeneity with 59 different variants accounting for the disease causing alleles (Supplementary Material, Table S1). There were 6 truncations, 3 frame shifts, 4 presumed splicing defects and 46 missense mutations. Some of these variants were relatively more common and occurred in two or more families (Supplementary Material, Table S2), but most of them were found in one family only.
We hypothesized that each allele makes an independent and additive contribution to severity of the retina-wide disease, and under certain simplifying assumptions, the disease contribution of each of the 41 Group A alleles could be estimated from the severity of the individuals carrying them. First, we assumed that all eight truncations and frame shift mutations resulted in no protein product and assigned them a standard severity value of 0. Next, we defined a standard ADI resulting from two truncating mutation alleles as 10.6 years based on the average value of the ADIs estimated for siblings (P19, P20) homozygous for the Y245X truncation. Then, we calculated a severity value for six alleles (D654N, R1098C, IVS38-10T>C, G1961R, G1961E and C2150Y) occurring in trans to the truncation and frame shift mutations by subtracting the standard ADI from the ADI of each individual; if there were several individuals with the same genotype, their ADIs were averaged. For four missense mutations occurring homozygously (L244P, R220C, N965S and P1380L), we assumed that each allele contributed equally to disease severity. And for an additional eight mutations (G818E, A1038V;L541P, E1087D, R1108C, E1122K, IVS40+5G>A, L1940P and K2172R), we performed severity estimates recursively by using estimates established above. For the remaining 15 alleles occurring singly, we assumed that each allele contributes half of the severity. The resulting severity estimates for the 33 non-truncating variants in Group A individuals showed a substantial range where the contribution to delay of retina-wide disease initiation for the mildest allele was 70 years later than the most severe allele (Table 1). To predict the ADI (in years) for an individual carrying alleles from this list, the severity values of each allele are added to the standard ADI.
Table 1.
Estimated severity of ABCA4 alleles and their properties
| ABCA4 allele | Delay of retina-wide disease initiation (years)a | In vitro or in vivo studiesb | Molecular structural localizationc |
|---|---|---|---|
| C2150Y | −25.8 | NBD-2 | |
| A1038V;L541P | −14.0 | 35, 38 | ECD-1/NBD-1 |
| IVS38-10 T>C | −11.1 | ||
| L244P | −5.7 | ECD-1 | |
| E1122K | −3.5 | NBD-1 | |
| C54Y | −2.1 | 35 | ECD-1 |
| IVS35+2 T>C | −2.1 | ||
| R602W | −1.8 | 38 | ECD-1 |
| V1896D | −1.8 | TM12 | |
| L1940P | −1.4 | NBD-2 | |
| Truncation mutationsd | 0.0 | ||
| E1087D | 2.8 | NBD-1 | |
| R220C | 3.9 | ECD-1 | |
| A1598D | 3.9 | ECD-2 | |
| R1640Q | 3.9 | ECD-2 | |
| R1098C | 4.9 | NBD-1 | |
| P1380L | 7.4 | 35 | TM7 |
| N965S | 7.6 | 35 | NBD-1 |
| V1433I | 8.6 | ECD-2 | |
| R1108C | 10.4 | 35 | NBD-1 |
| T1526M | 14.5 | 35 | ECD-2 |
| R2030Q | 14.5 | NBD-2 | |
| L2027F | 15.1 | 35,37 | NBD-2 |
| G818E | 17.3 | 35 | TM5/TM6 |
| S100P | 18.2 | ECD-1 | |
| L1201R | 18.2 | NBD-1 | |
| R18W | 18.5 | Nt | |
| D600E | 18.5 | ECD-1 | |
| L11P | 21.7 | Nt | |
| D654N | 25.3 | 36 | ECD-1 |
| K2172R | 27.9 | NBD-2 | |
| IVS40+5 G>A | 28.1 | ||
| G1961E | 37.9 | 35 | NBD-2 |
| G1961R | 44.0 | NBD-2 |
aDelay of retina-wide disease initiation relative to the standard of age 10.6 years.
bReferences to publications reporting in vitro protein expression, ATP binding and in vivo mislocalization of the protein to the rod inner segments.
cNt, amino-terminal domain; ECD-1 and ECD-2, exocytoplasmic domains 1 and 2; NBD-1 and NBD-2, nucleotide binding domains 1 and 2; TM5, TM6, TM7, TM12, within, near or between transmembrane helices 5, 6, 7 and 12.
dW41X, R152X, Y245X, Y362X, W663X, M669del2tccAT, R681X and A1739 del11gcTGGGCTGGTGG.
Reliability of the allele severity estimates were evaluated from multiple members of the same family. Generally, ADI estimates showed a consistency better than a decade (range 1–13 years) even though the ages of the individuals at the time of the evaluation could be more than three decades apart (Supplementary Material, Table S1). Predictability of the allele severity estimates was tested in 9 Group B individuals who were not included in the calculations above but who carried two alleles with available estimates. Unrelated individuals P11 and P12 had the same pair of mutant alleles, R152X and G1961E. At ages 21 and 19, respectively, these individuals had normal extramacular function which is consistent with the predicted ADI of 48.5 years (=10.6 + 0.0 + 37.9 years, Table 1) for this allele pairing. P7 with C54Y and G1961E alleles had normal extramacular vision at age 40 consistent with a predicted ADI of 46.4 years (=10.6 – 2.1 + 37.9 years, Table 1). Other individuals showing such consistency included P47 and P60 with normal extramacular vision at ages 52 and 30 with predicted ADIs of 56 and 27 years. P37 had nearly-normal results at age 19 but the ADI prediction from the genotype was age 4. In three individuals, there was qualitative consistency but quantitative differences. In P42, P55 and P58, the genotype estimates suggested late-onset retina-wide disease with ADIs ranging from 40 to 49 years. These individuals indeed had late-onset disease but at least 11–18 years later than predicted.
Were there clues from previous in vitro or in vivo studies to explain the differences in severity between mutations? In vitro biochemical analyses evaluating defects in protein yield, azido-ATP binding, basal and retinal-stimulated ATP hydrolysis (35–37) do not provide criteria to differentiate between two of the severe mutations and seven of the milder mutations studied (Table 1). In vivo studies in photoreceptors of transgenic Xenopus laevis tadpoles have shown mislocalization of several mutants in the inner segment due to severe misfolding of the proteins (38). Curiously, two of these misfolding mutants are alleles predicted to be more severe than truncation mutations (Table 1). In order to evaluate possible relationships between severity estimates and molecular structural aberrations, we used a topological model of the ABCR molecule and three dimensional homology models of its two nucleotide binding domains (Supplementary Material, Fig. S2). Missense mutations were distributed throughout the different domains of the ABCR protein (Table 1). Possible biochemical abnormalities and molecular structural aberrations could be postulated based on the observed phenotypic severity of mutants (Supplementary Material, Text) but phenotypic severity could not be predicted from molecular considerations.
DISCUSSION
The universal phenotypic feature of ABCA4 disease is progressive degeneration of photoreceptors and RPE in the macula (18,24,39) commonly leading to difficulty in performing everyday tasks such as reading small print (40). When ABCA4 disease progresses from macula-only degeneration to retina-wide blindness, the quality of life deteriorates dramatically with loss of mobility and independence. Ideally, treatments should anticipate disease onset and cure the genetic abnormality before any vision is lost. More realistic, however, are clinical trials of gene therapy that will enroll individuals with existing macular degeneration. The best result for such therapy would be either to delay the initiation of retinal degeneration in genetically abnormal but phenotypically uninvolved extramacular retina or to arrest progression of disease in already affected areas. To enroll candidates for such a trial, there should be a way to predict reliably which individuals are destined for retina-wide disease progression within the 3–5-year time frame of a clinical trial. The current study took an early step towards preparation for such therapeutic strategies by defining a model of disease initiation and progression, and applying this model to predict the natural history of retina-wide disease for individuals with different genotypes.
There is agreement in the literature that retinal disease caused by mutations in ABCA4 covers a wide spectrum of severity (17–19). Extremes of phenotype are easily identifiable. But in most individuals with ABCA4 disease, the phenotype is intermediate between the extremes, and there are no well-accepted metrics for comparing severities. Clinical criteria have been commonly used as a method of comparison. A diagnosis of STGD is considered to represent less severe disease than CRD or atypical RP (6,7,41,42) because of the retina-wide involvement implicit in the latter diagnoses. However, many individuals with STGD can also show retina-wide involvement, and wide variation in clinical severity is observed within each diagnostic category (20,23,43–50). Furthermore, rare longitudinal studies or cross-sectional studies in individuals with the same genotype provide evidence that ABCA4 disease can progress from a maculopathy consistent with STGD to a severe retina-wide degeneration (48,50–54). Therefore, it is unlikely that limited diagnostic criteria established in the pre-molecular era can reliably and reproducibly represent the spectrum of ABCA4 disease severity observed.
There have been previous attempts to quantify ABCA4 disease severity. For example, patient-reported age of disease onset has been used as a measure of disease severity (55,56), but it is not known how this metric relates to macular or retina-wide disease. ERG is a well-established objective metric of retina-wide visual function and studies using ERG have demonstrated a wide range of disease severity in ABCA4 mutations (53,57). In the current study, we used a psychophysical method of assessing rod and cone vision that sampled the whole visual field at regular intervals. Our previous work has shown that photoreceptors are the dominant contributors to loss of psychophysical sensitivity in ABCA4 disease (18). Sampling of sensitivities across the visual field afforded us the advantage of determining spatial distribution of disease from the macula to the far periphery. Using a combination of longitudinal and cross-sectional data on sensitivity losses, we defined a simple model of disease progression based on exponential rates of rod and cone sensitivity loss. The only free parameter of the current model—the age of retina-wide disease initiation, ADI—can be thought to represent an absolute metric of disease severity that can be used to compare individuals within or across studies.
The relationship between ABCA4 genotype and resulting severity of retinal disease phenotype has been of longstanding interest (5–7). According to the commonly held residual ABCR function hypothesis, each ABCA4 allele produces either no protein or an abnormally functioning protein. The sum total of the residual function from the pair of ABCA4 alleles can range from normal to non-existent. The clinical severity of ABCA4 disease is thought to be inversely related to the residual ABCR function (7,41,42). Indeed, individuals with truncation mutations in both of their alleles likely represent complete loss ABCR function and the corresponding phenotype reported has been among the more severe diseases (6,50–52,58,59). Consistent with this hypothesis were siblings in the current study (P19, P20) homozygous for Y245X truncation showing retina-wide disease within the first decade. However, for the majority of individuals carrying non-truncating (missense and splicing) mutations, the predictions of the residual ABCR function hypothesis are more tenuous. Our demonstration of individuals with two non-truncating mutations (such as P15, P16, P17, P18, P25 and P38) showing greater severity of disease than individuals with two truncating mutations are not consistent with the hypothesis relating ABCA4 disease severity solely to the extent of residual ABCR function. It could be argued that environmental and genetic modifiers contribute to the phenotypic variation, but until proven otherwise, we will assume that the dominant contributor to severity of this Mendelian disease is the underlying pathophysiology caused by the two mutant alleles.
ABCA4 disease pathogenesis could potentially include at least two pathways. One of these pathways is loss-of-function. Partial loss-of-function may result from reduced activity of mutant proteins (35–37) properly synthesized, folded and transported to the rims of outer segment discs. Complete loss-of-function may result from degradation of misfolded or truncated proteins (60,61) and there is in vivo evidence for the existence of misfolding alleles (38). Loss of ABCR function is thought to result in retinal disease due to accumulation of toxic substances within photoreceptors and/or the neighboring RPE cells (10,13,14,38,62–64). It is important to note that misfolding alleles could exacerbate the loss-of-function pathology by a second pathway. Normally, the cellular system for degradation and clearance of misfolded proteins is highly efficient (60,65). But persistently elevated photoreceptor stress caused by atRAL accumulation (14) may overwhelm the protein turnover (66–68) causing ER stress and initiation of the unfolded-protein response (UPR) (69). Failure of the UPR to establish cellular homeostasis can result in apoptotic signaling (70,71). Our observations of a substantial subset of non-truncating mutations causing greater disease than truncating mutations could be explained by the exacerbation of the primary loss-of-function pathogenesis by toxic gain-of-function and ER stress (72). This subset of severe non-truncating mutations would not be expected to benefit from gene replacement therapy.
Accurate predictions of disease effects in the future will not only require a better understanding of the underlying pathophysiology, taking into account constantly evolving hypotheses about the exact function of ABCR, but also better biochemical assays of mutant alleles that are quantitatively validated against the disease severity they cause in humans. Resolving the crystal structure of the ABCR molecule also is required to understand more precisely how various mutations may affect its function. In the meantime, prospective studies of the natural history of vision loss in ABCA4 patients to provide further proof for the hypotheses raised in the current work, are valuable prerequisites in preparation for clinical trials of gene therapy in ABCA4-associated diseases.
MATERIALS AND METHODS
Human subjects
The study population consisted of 66 patients (from 54 families) with known ABCA4 genotypes (Supplementary Material, Table S1). Informed consent was obtained; procedures followed the Declaration of Helsinki and had institutional review board approval.
Visual function
A complete eye examination was performed in all subjects including best-corrected ETDRS VA and Goldmann kinetic perimetry with V-4e and I-4e targets. Preferred locus of fixation was documented by using fundus imaging (39) and all psychophysical tests were adjusted to each individual’s preferred fixation locus. Sensitivities were measured by using a modified automated perimeter across the full extent of the visual field at 71 loci placed on a 12° square grid (18,73). Chromatic (500 nm, blue, and 650 nm, red) stimuli (1.7° diameter, 200 ms duration) were used under dark-adapted conditions to estimate rod-mediated vision. Light-adapted testing with an orange (600 nm) target ensured mediation by the cone system. Sensitivity losses were calculated at each locus by comparison to locus-specific mean normal values. Mean retina-wide loss of sensitivity was calculated by considering loci at ≥30° eccentricity from the fovea (18). In a subset of visits, macular imaging was performed with a confocal scanning laser ophthalmoscope (HRA1 or HRA2, Heidelberg Engineering, Dossenheim, Germany) as previously described (18,24,39).
Molecular modeling of ABCR
Topological model of ABCR was based on currently available experimental evidence (12,74). Initial homology models of NBD-1 and NBD-2 domains of the ABCR molecule were generated with the SWISS-MODEL server (75). Three models were created for each NBD. The templates for NBD-1 were the crystal structures of the ABC transporter ATP-binding protein from Thermotoga maritima (PDB ID 1VPL, 33.3% sequence identity), the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus (PDB ID 1OXT, (76), 30.3% sequence identity) and PH0203 protein from Pyrococcus horikoshii (PDB ID 2IT1, 34.7% sequence identity). The templates for NBD-2 consisted of crystal structures of the ABC transporter ATP-binding protein from Thermotoga maritima (PDB ID 1VPL, 31.5% sequence identity), the ATPase subunit CysA of the putative sulfate ABC transporter from Alicyclobacillus acidocaldarius (PDB ID 1Z47, (77), 31.3% sequence identity) and the ATP-binding protein of the sugar ABC transporter from Thermotoga maritima (PDB ID 2YYZ, 30.2% sequence identity). The models were refined by energy minimization in NAMD Molecular Dynamics Simulator (78) using the CHARM22 force field. RAMPAGE software (79) implemented in the CCP4 package (80) was used for validation of the structures. No residues were found in disallowed regions of Ramachandran plots. Structural superpositions of the models showed close similarity with respect to location and general environment of the mutated residues. Therefore, models of NBD-1 and NBD-2 based on the 1VPL structure were chosen for further considerations. Figures were prepared using Pymol (http://pymol.sourceforge.net/).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health [EY13203, EY09339], Macula Vision Research Foundation, Foundation Fighting Blindness, Hope for Vision and Research to Prevent Blindness.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Elaine Smilko, Louisa Affatigato and Jean Andorf for their critical help.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bainbridge J.W., Smith A.J., Barker S.S., Robbie S., Henderson R., Balaggan K., Viswanathan A., Holder G.E., Stockman A., Tyler N., et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 2.Maguire A.M., Simonelli F., Pierce E.A., Pugh E.N., Jr, Mingozzi F., Bennicelli J., Banfi S., Marshall K.A., Testa F., Surace E.M., et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauswirth W.W., Aleman T.S., Kaushal S., Cideciyan A.V., Schwartz S.B., Wang L., Conlon T.J., Boye S.L., Flotte T.R., Byrne B.J., et al. Treatment of Leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Human Gene Therapy. 2008;19:979–990. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cideciyan A.V., Aleman T.S., Boye S.L., Schwartz S.B., Kaushal S., Roman A.J., Pang J-j., Sumaroka A., Windsor E.A.M., Wilson J.M., et al. Human gene therapy for RPE65-isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc. Natl Acad. Sci. USA. 2008;105:15112–15117. doi: 10.1073/pnas.0807027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allikmets R., Singh N., Sun H., Shroyer N.F., Hutchinson A., Chidambaram A., Gerrard B., Baird L., Stauffer D., Peiffer A., et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat. Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Mir A., Paloma E., Allikmets R., Ayuso C., del Rio T., Dean M., Vilageliu L., Gonzalez-Duarte R., Balcells S. Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat. Genet. 1998;18:11–12. doi: 10.1038/ng0198-11. [DOI] [PubMed] [Google Scholar]
- 7.Cremers F.P., van de Pol D.J., van Driel M., den Hollander A.I., van Haren F.J., Knoers N.V., Tijmes N., Bergen A.A., Rohrschneider K., Blankenagel A., et al. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene ABCR. Hum. Mol. Genet. 1998;7:355–362. doi: 10.1093/hmg/7.3.355. [DOI] [PubMed] [Google Scholar]
- 8.Sun H., Nathans J. Stargardt’s ABCR is localized to the disc membrane of retinal rod outer segments. Nat. Genet. 1997;17:15–16. doi: 10.1038/ng0997-15. [DOI] [PubMed] [Google Scholar]
- 9.Molday L.L., Rabin A.R., Molday R.S. ABCR expression in foveal cone photoreceptors and its role in Stargardt macular dystrophy. Nat. Genet. 2000;25:257–258. doi: 10.1038/77004. [DOI] [PubMed] [Google Scholar]
- 10.Weng J., Mata N.L., Azarian S.M., Tzekov R.T., Birch D.G., Travis G.H. Insights into the function of Rim protein in photoreceptors and etiology of Stargardt’s disease from the phenotype in abcr knockout mice. Cell. 1999;98:13–23. doi: 10.1016/S0092-8674(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 11.Sun H., Molday R.S., Nathans J. Retinal stimulates ATP hydrolysis by purified and reconstituted ABCR, the photoreceptor-specific ATP-binding cassette transporter responsible for Stargardt disease. J. Biol. Chem. 1999;274:8269–8281. doi: 10.1074/jbc.274.12.8269. [DOI] [PubMed] [Google Scholar]
- 12.Molday R.S. ATP-binding cassette transporter ABCA4: molecular properties and role in vision and macular degeneration. J. Bioenerg. Biomem. 2007;39:507–517. doi: 10.1007/s10863-007-9118-6. [DOI] [PubMed] [Google Scholar]
- 13.Pawar A.S., Qtaishat N.M., Little D.M., Pepperberg D.R. Recovery of rod photoresponses in ABCR-deficient mice. Invest. Ophthalmol. Vis. Sci. 2008;49:2743–2755. doi: 10.1167/iovs.07-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda A., Maeda T., Golczak M., Palczewski K. Retinopathy in mice induced by disrupted all-trans-retinal clearance. J. Biol. Chem. 2008;283:26684–26693. doi: 10.1074/jbc.M804505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong J., Kim S.R., Binley K., Pata I., Doi K., Mannik J., Zernant-Rajang J., Kan O., Iqball S., Naylor S., et al. Correction of the disease phenotype in the mouse model of Stargardt disease by lentiviral gene therapy. Gene Ther. 2008;15:1311–1320. doi: 10.1038/gt.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allocca M., Doria M., Petrillo M., Colella P., Garcia-Hoyos M., Gibbs D., Kim S.R., Maguire A., Rex T.S., Di Vicino U., et al. Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice. J. Clin. Invest. 2008;118:1955–1964. doi: 10.1172/JCI34316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozet J.M., Gerber S., Souied E., Ducroq D., Perrault I., Ghazi I., Soubrane G., Coscas G., Dufier J.L., Munnich A., et al. The ABCR gene: a major disease gene in macular and peripheral retinal degenerations with onset from early childhood to the elderly. Mol. Genet. Metab. 1999;68:310–315. doi: 10.1006/mgme.1999.2925. [DOI] [PubMed] [Google Scholar]
- 18.Cideciyan A.V., Aleman T.S., Swider M., Schwartz S.B., Steinberg J.D., Brucker A.J., Maguire A.M., Bennett J., Stone E.M., Jacobson S.G. Mutations in ABCA4 result in accumulation of lipofuscin before slowing of the retinoid cycle: a reappraisal of the human disease sequence. Hum. Mol. Genet. 2004;13:525–534. doi: 10.1093/hmg/ddh048. [DOI] [PubMed] [Google Scholar]
- 19.Klevering B.J., Deutman A.F., Maugeri A., Cremers F.P., Hoyng C.B. The spectrum of retinal phenotypes caused by mutations in the ABCA4 gene. Graefes Arch. Clin. Exp. Ophthalmol. 2005;243:90–100. doi: 10.1007/s00417-004-1079-4. [DOI] [PubMed] [Google Scholar]
- 20.Fishman G.A., Stone E.M., Grover S., Derlacki D.J., Haines H.L., Hockey R.R. Variation of clinical expression in patients with Stargardt dystrophy and sequence variations in the ABCR gene. Arch. Ophthalmol. 1999;117:504–510. doi: 10.1001/archopht.117.4.504. [DOI] [PubMed] [Google Scholar]
- 21.Webster A.R., Héon E., Lotery A.J., Vandenburgh K., Casavant T.L., Oh K.T., Beck G., Fishman G.A., Lam B.L., Levin A., et al. An analysis of allelic variation in the ABCA4 gene. Invest. Ophthalmol. Vis. Sci. 2001;42:1179–1189. [PubMed] [Google Scholar]
- 22.Stone E.M. Finding and interpreting genetic variations that are important to ophthalmologists. Trans. Am. Ophthalmol. Soc. 2003;101:437–484. [PMC free article] [PubMed] [Google Scholar]
- 23.Fishman G.A., Stone E.M., Eliason D.A., Taylor C.M., Lindeman M., Derlacki D.J. ABCA4 gene sequence variations in patients with autosomal recessive cone-rod dystrophy. Arch. Ophthalmol. 2003;121:851–855. doi: 10.1001/archopht.121.6.851. [DOI] [PubMed] [Google Scholar]
- 24.Cideciyan A.V., Swider M., Aleman T.S., Roman M.I., Sumaroka A., Schwartz S.B., Stone E.M., Jacobson S.G. Reduced-illuminance autofluorescence imaging in ABCA4-associated retinal degenerations. J. Opt. Soc. Am. A. Opt. Image. Sci. Vis. 2007;24:1457–1467. doi: 10.1364/josaa.24.001457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aleman T.S., Cideciyan A.V., Windsor E.A., Schwartz S.B., Swider M., Chico J.D., Sumaroka A., Pantelyat A.Y., Duncan K.G., Gardner L.M., et al. Macular pigment and lutein supplementation in ABCA4-associated retinal degenerations. Invest. Ophthalmol. Vis. Sci. 2007;48:1319–1329. doi: 10.1167/iovs.06-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitiratschky V.B., Grau T., Bernd A., Zrenner E., Jägle H., Renner A.B., Kellner U., Rudolph G., Jacobson S.G., Cideciyan A.V., et al. ABCA4 gene analysis in patients with autosomal recessive cone and cone rod dystrophies. Eur. J. Hum. Genet. 2008;16:812–819. doi: 10.1038/ejhg.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berson E.L., Sandberg M.A., Rosner B., Birch D.G., Hanson A.H. Natural course of retinitis pigmentosa over a three-year interval. Am. J. Ophthalmol. 1985;99:240–251. doi: 10.1016/0002-9394(85)90351-4. [DOI] [PubMed] [Google Scholar]
- 28.Massof R.W., Finkelstein D. A two-stage hypothesis for the natural course of retinitis pigmentosa. In: Zrenner E., Krastel H., Goebel H.-H., editors. Research in Retinitis Pigmentosa. Advances in the Biosciences. Vol. 62. Oxford: Pergamon Press; 1987. pp. 29–58. [Google Scholar]
- 29.Birch D.G., Anderson J.L., Fish G.E. Yearly rates of rod and cone functional loss in retinitis pigmentosa and cone-rod dystrophy. Ophthalmology. 1999;106:258–268. doi: 10.1016/S0161-6420(99)90064-7. [DOI] [PubMed] [Google Scholar]
- 30.Iannaccone A., Kritchevsky S.B., Ciccarelli M.L., Tedesco S.A., Macaluso C., Kimberling W.J., Somes G.W. Kinetics of visual field loss in Usher syndrome Type II. Invest. Ophthalmol. Vis. Sci. 2004;45:784–792. doi: 10.1167/iovs.03-0906. [DOI] [PubMed] [Google Scholar]
- 31.Herrera W., Aleman T.S., Cideciyan A.V., Roman A.J., Banin E., Ben-Yosef T., Gardner L.M., Sumaroka A., Windsor E.A., Schwartz S.B., et al. Retinal disease in Usher syndrome III caused by mutations in the clarin-1 gene. Invest. Ophthalmol. Vis. Sci. 2008;49:2651–2660. doi: 10.1167/iovs.07-1505. [DOI] [PubMed] [Google Scholar]
- 32.Nagy D., Schönfisch B., Zrenner E., Jägle H. Long-term follow-up of retinitis pigmentosa patients with multifocal electroretinography. Invest. Ophthalmol. Vis. Sci. 2008;49:4664–4671. doi: 10.1167/iovs.07-1360. [DOI] [PubMed] [Google Scholar]
- 33.Clarke G., Collins R.A., Leavitt B.R., Andrews D.F., Hayden M.R., Lumsden C.J., McInnes R.R. A one-hit model of cell death in inherited neuronal degenerations. Nature. 2000;406:195–199. doi: 10.1038/35018098. [DOI] [PubMed] [Google Scholar]
- 34.Clarke G., Lumsden C.J. Heterogeneous cellular environments modulate one-hit neuronal death kinetics. Brain Res. Bull. 2005;65:59–67. doi: 10.1016/j.brainresbull.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Sun H., Smallwood P.M., Nathans J. Biochemical defects in ABCR protein variants associated with human retinopathies. Nat. Genet. 2000;26:242–246. doi: 10.1038/79994. [DOI] [PubMed] [Google Scholar]
- 36.Shroyer N.F., Lewis R.A., Yatsenko A.N., Wensel T.G., Lupski J.R. Cosegregation and functional analysis of mutant ABCR (ABCA4) alleles in families that manifest both Stargardt disease and age-related macular degeneration. Hum. Mol. Genet. 2001;10:2671–2678. doi: 10.1093/hmg/10.23.2671. [DOI] [PubMed] [Google Scholar]
- 37.Biswas-Fiss E.E. Functional analysis of genetic mutations in nucleotide binding domain 2 of the human retina specific ABC transporter. Biochemistry. 2003;42:10683–10696. doi: 10.1021/bi034481l. [DOI] [PubMed] [Google Scholar]
- 38.Wiszniewski W., Zaremba C.M., Yatsenko A.N., Jamrich M., Wensel T.G., Lewis R.A., Lupski J.R. ABCA4 mutations causing mislocalization are found frequently in patients with severe retinal dystrophies. Hum. Mol. Genet. 2005;14:2769–2778. doi: 10.1093/hmg/ddi310. [DOI] [PubMed] [Google Scholar]
- 39.Cideciyan A.V., Swider M., Aleman T.S., Sumaroka A., Schwartz S.B., Roman M.I., Milam A.H., Bennett J., Stone E.M., Jacobson S.G. ABCA4-associated retinal degenerations spare structure and function of the human parapapillary retina. Invest. Ophthalmol. Vis. Sci. 2005;46:4739–4746. doi: 10.1167/iovs.05-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szlyk J.P., Fishman G.A., Aslan R.J., Grover S. Legal blindness and employment in patients with juvenile-onset macular dystrophies or achromatopsia. Retina. 1998;18:360–367. doi: 10.1097/00006982-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 41.van Driel M.A., Maugeri A., Klevering B.J., Hoyng C.B., Cremers F.P. ABCR unites what ophthalmologists divide(s) Ophthalmic Genet. 1998;19:117–122. doi: 10.1076/opge.19.3.117.2187. [DOI] [PubMed] [Google Scholar]
- 42.Shroyer N.F., Lewis R.A., Allikmets R., Singh N., Dean M., Leppert M., Lupski J.R. The rod photoreceptor ATP-binding cassette transporter gene, ABCR, and retinal disease: from monogenic to multifactorial. Vision Res. 1999;39:2537–2544. doi: 10.1016/s0042-6989(99)00037-1. [DOI] [PubMed] [Google Scholar]
- 43.Aaberg T.M. Stargardt’s disease and fundus flavimaculatus: evaluation of morphologic progression and intrafamilial co-existence. Trans. Am. Ophthalmol. Soc. 1986;84:453–487. [PMC free article] [PubMed] [Google Scholar]
- 44.Yagasaki K., Jacobson S.G. Cone-rod dystrophy. Phenotypic diversity by retinal function testing. Arch. Ophthalmol. 1989;107:701–708. doi: 10.1001/archopht.1989.01070010719034. [DOI] [PubMed] [Google Scholar]
- 45.Itabashi R., Katsumi O., Mehta M.C., Wajima R., Tamai M., Hirose T. Stargardt’s disease/fundus flavimaculatus: psychophysical and electrophysiologic results. Graefes Arch. Clin. Exp. Ophthalmol. 1993;231:555–562. doi: 10.1007/BF00936518. [DOI] [PubMed] [Google Scholar]
- 46.Armstrong J.D., Meyer D., Xu S., Elfervig J.L. Long-term follow-up of Stargardt’s disease and fundus flavimaculatus. Ophthalmology. 1998;105:448–457. doi: 10.1016/S0161-6420(98)93026-3. [DOI] [PubMed] [Google Scholar]
- 47.Gerth C., Andrassi-Darida M., Bock M., Preising M.N., Weber B.H., Lorenz B. Phenotypes of 16 Stargardt macular dystrophy/fundus flavimaculatus patients with known ABCA4 mutations and evaluation of genotype-phenotype correlation. Graefes Arch. Clin. Exp. Ophthalmol. 2002;240:628–638. doi: 10.1007/s00417-002-0502-y. [DOI] [PubMed] [Google Scholar]
- 48.Klevering B.J., Blankenagel A., Maugeri A., Cremers F.P., Hoyng C.B., Rohrschneider K. Phenotypic spectrum of autosomal recessive cone-rod dystrophies caused by mutations in the ABCA4 (ABCR) gene. Invest. Ophthalmol. Vis. Sci. 2002;43:1980–1985. [PubMed] [Google Scholar]
- 49.Oh K.T., Weleber R.G., Oh D.M., Billingslea A.M., Rosenow J., Stone E.M. Clinical phenotype as a prognostic factor in Stargardt disease. Retina. 2004;24:254–262. doi: 10.1097/00006982-200404000-00011. [DOI] [PubMed] [Google Scholar]
- 50.Simonelli F., Testa F., Zernant J., Nesti A., Rossi S., Rinaldi E., Allikmets R. Association of a homozygous nonsense mutation in the ABCA4 (ABCR) gene with cone-rod dystrophy phenotype in an Italian family. Ophthalmic. Res. 2004;36:82–88. doi: 10.1159/000076886. [DOI] [PubMed] [Google Scholar]
- 51.Fukui T., Yamamoto S., Nakano K., Tsujikawa M., Morimura H., Nishida K., Ohguro N., Fujikado T., Irifune M., Kuniyoshi K., et al. ABCA4 gene mutations in Japanese patients with Stargardt disease and retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 2002;43:2819–2824. [PubMed] [Google Scholar]
- 52.Fukui T., Fujikado T., Tsujikawa M., Okada M., Yamamoto S., Tano Y. Null ABCA4 gene mutations found in Japanese patients with panretinal degeneration. Jpn. J. Ophthalmol. 2006;50:179–181. doi: 10.1007/s10384-005-0279-8. [DOI] [PubMed] [Google Scholar]
- 53.Beit-Ya’acov A., Mizrahi-Meissonnier L., Obolensky A., Landau C., Blumenfeld A., Rosenmann A., Banin E., Sharon D. Homozygosity for a novel ABCA4 founder splicing mutation is associated with progressive and severe Stargardt-like disease. Invest. Ophthalmol. Vis. Sci. 2007;48:4308–4314. doi: 10.1167/iovs.07-0244. [DOI] [PubMed] [Google Scholar]
- 54.Rosenberg T., Klie F., Garred P., Schwartz M. N965S is a common ABCA4 variant in Stargardt-related retinopathies in the Danish population. Mol. Vis. 2007;13:1962–1969. [PubMed] [Google Scholar]
- 55.Lewis R.A., Shroyer N.F., Singh N., Allikmets R., Hutchinson A., Li Y., Lupski J.R., Leppert M., Dean M. Genotype/Phenotype analysis of a photoreceptor-specific ATP-binding cassette transporter gene, ABCR, in Stargardt disease. Am. J. Hum. Genet. 1999;64:422–434. doi: 10.1086/302251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yatsenko A.N., Shroyer N.F., Lewis R.A., Lupski J.R. Late-onset Stargardt disease is associated with missense mutations that map outside known functional regions of ABCR (ABCA4) Hum. Genet. 2001;108:346–355. doi: 10.1007/s004390100493. [DOI] [PubMed] [Google Scholar]
- 57.Oh K.T., Weleber R.G., Stone E.M., Oh D.M., Rosenow J., Billingslea A.M. Electroretinographic findings in patients with Stargardt disease and fundus flavimaculatus. Retina. 2004;24:920–928. doi: 10.1097/00006982-200412000-00013. [DOI] [PubMed] [Google Scholar]
- 58.Briggs C.E., Rucinski D., Rosenfeld P.J., Hirose T., Berson E.L., Dryja T.P. Mutations in ABCR (ABCA4) in patients with Stargardt macular degeneration or cone-rod degeneration. Invest. Ophthalmol. Vis. Sci. 2001;42:2229–2236. [PubMed] [Google Scholar]
- 59.Singh H.P., Jalali S., Hejtmancik J.F., Kannabiran C. Homozygous null mutations in the ABCA4 gene in two families with autosomal recessive retinal dystrophy. Am. J. Ophthalmol. 2006;141:906–913. doi: 10.1016/j.ajo.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 60.Römisch K. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- 61.Kuzmiak H.A., Maquat L.E. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol. Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 62.Birnbach C.D., Järveläinen M., Possin D.E., Milam A.H. Histopathology and immunocytochemistry of the neurosensory retina in fundus flavimaculatus. Ophthalmology. 1994;101:1211–1219. doi: 10.1016/s0161-6420(13)31725-4. [DOI] [PubMed] [Google Scholar]
- 63.Mata N.L., Weng J., Travis G.H. Biosynthesis of a major lipofuscin fluorophore in mice and humans with ABCR-mediated retinal and macular degeneration. Proc. Natl Acad. Sci. USA. 2000;97:7154–7159. doi: 10.1073/pnas.130110497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radu R.A., Mata N.L., Bagla A., Travis G.H. Light exposure stimulates formation of A2E oxiranes in a mouse model of Stargardt’s macular degeneration. Proc. Natl Acad. Sci. USA. 2004;101:5928–5933. doi: 10.1073/pnas.0308302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schubert U., Antón L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 66.Lomas D.A., Parfrey H. Alpha1-antitrypsin deficiency. 4: Molecular pathophysiology. Thorax. 2004;59:529–535. doi: 10.1136/thx.2003.006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Görlach A., Klappa P., Kietzmann T. The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxid. Redox. Signal. 2006;8:1391–1418. doi: 10.1089/ars.2006.8.1391. [DOI] [PubMed] [Google Scholar]
- 68.Malhotra J.D., Kaufman R.J. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid. Redox. Signal. 2007;9:2277–2293. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- 69.Schröder M., Kaufman R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 70.Rutkowski D.T., Kaufman R.J. A trip to the ER: coping with stress. Trends Cell. Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 71.Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gregersen N., Bross P., Vang S., Christensen J.H. Protein misfolding and human disease. Annu. Rev. Genomics Hum. Genet. 2006;7:103–124. doi: 10.1146/annurev.genom.7.080505.115737. [DOI] [PubMed] [Google Scholar]
- 73.Jacobson S.G., Voigt W.J., Parel J.M., Apáthy P.P., Nghiem-Phu L., Myers S.W., Patella V.M. Automated light- and dark-adapted perimetry for evaluating retinitis pigmentosa. Ophthalmology. 1986;93:1604–1611. doi: 10.1016/s0161-6420(86)33522-x. [DOI] [PubMed] [Google Scholar]
- 74.Bungert S., Molday L.L., Molday R.S. Membrane topology of the ATP binding cassette transporter ABCR and its relationship to ABC1 and related ABCA transporters: identification of N-linked glycosylation sites. J. Biol. Chem. 2001;276:23539–23546. doi: 10.1074/jbc.M101902200. [DOI] [PubMed] [Google Scholar]
- 75.Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Verdon G., Albers S.V., Dijkstra B.W., Driessen A.J., Thunnissen A.M. Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations. J. Mol. Biol. 2003;330:343–358. doi: 10.1016/s0022-2836(03)00575-8. [DOI] [PubMed] [Google Scholar]
- 77.Scheffel F., Demmer U., Warkentin E., Schneider E., Ermler U. Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius. FEBS Lett. 2005;579:2953–2958. doi: 10.1016/j.febslet.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 78.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lovell S.C., Davis I.W., Arendall W.B., III, de Bakker P.I., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C. Structure validation by Calpha geometry: phi, psi and C beta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 80.Potterton E., McNicholas S., Krissinel E., Cowtan K., Noble M. The CCP4 molecular-graphics project. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:1955–1957. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.