Abstract
Cerebral cavernous malformations (CCMs) are vascular anomalies of the central nervous system, comprising dilated blood-filled capillaries lacking structural support. The lesions are prone to rupture, resulting in seizures or hemorrhagic stroke. CCM can occur sporadically, manifesting as solitary lesions, but also in families, where multiple lesions generally occur. Familial cases follow autosomal-dominant inheritance due to mutations in one of three genes, CCM1/KRIT1, CCM2/malcavernin or CCM3/PDCD10. The difference in lesion burden between familial and sporadic CCM, combined with limited molecular data, suggests that CCM pathogenesis may follow a two-hit molecular mechanism, similar to that seen for tumor suppressor genes. In this study, we investigate the two-hit hypothesis for CCM pathogenesis. Through repeated cycles of amplification, subcloning and sequencing of multiple clones per amplicon, we identify somatic mutations that are otherwise invisible by direct sequencing of the bulk amplicon. Biallelic germline and somatic mutations were identified in CCM lesions from all three forms of inherited CCMs. The somatic mutations are found only in a subset of the endothelial cells lining the cavernous vessels and not in interstitial lesion cells. These data suggest that CCM lesion genesis requires complete loss of function for one of the CCM genes. Although widely expressed in the different cell types of the brain, these data also suggest a unique role for the CCM proteins in endothelial cell biology.
INTRODUCTION
Cerebral cavernous malformations (CCMs) are vascular anomalies of the central nervous system, comprising grossly dilated blood-filled capillaries (1). The vessels within the CCM lesions are devoid of structural support and intervening neural tissue. Consequently, CCM lesions are prone to hemorrhage, often resulting in seizures, hemorrhagic stroke and death (2,3). The onset of CCM may occur either sporadically or in an inherited autosomal-dominant form due to mutations in one of the three genes: CCM1/KRIT1 (4,5), CCM2/malcavernin (6,7) or CCM3/PDCD10 (8). The function of each of these proteins is just beginning to be elucidated. Recent data suggests that the three CCM proteins interact with one another to participate in several signaling complexes. The KRIT1 protein is involved in Rap1 and integrin-mediated signaling, whereas malcavernin acts as a scaffolding protein as part of the p38 MAP kinase signaling cascade (9–16). Disruption of one or more components of this signaling mechanism likely results in the onset of CCM.
Prior to the discovery of the CCM genes, sporadic cases of CCMs could be distinguished from inherited cases by lesion burden. In general, solitary lesions are seen in sporadic cases, whereas inherited cases usually display multiple lesions (1). Since the discovery of the CCM genes, molecular data have confirmed this trend. Indeed, many individuals displaying multiple lesions, but lacking a clear family history, have been shown to harbor a de novo germline mutation (6,8,17,18). Additionally, familial cases of CCMs also show a more aggressive phenotype, with an increased risk of hemorrhage and seizure as well as an earlier age of onset (19–21). These clinical observations are generally analogous to those seen for the inherited cancer disorder, retinoblastoma. Similar epidemiological observations for patients with retinoblastoma were used by Knudson (22) to devise the two-hit hypothesis that requires both alleles of a tumor suppressor gene to be inactivated to result in tumor formation.
We and others have hypothesized that CCM lesions might also follow a similar two-hit molecular mechanism for pathogenesis, requiring loss of both copies of one of the CCM genes for lesion genesis. Two earlier attempts to identify somatic mutations (23) or loss of heterozygosity (LOH) (24) in support of the two-hit hypothesis for CCMs have been limited and largely unsuccessful. These earlier studies used detection strategies, which relied on direct sequence analysis of bulk lesion-derived DNA. Consequently, these studies may have lacked an appropriate level of sensitivity to detect somatic mutations, which might be present in only a minor fraction of the cells comprising the bulk lesion tissue. Another study (25) has provided some supportive evidence for the two-hit mechanism, such that two distinct potential somatic mutations were identified in the CCM1 gene from a sporadic CCM lesion. However, the missense mutations that were identified are inconsistent with the allelic series observed in germline CCM1 cases, which consists exclusively of protein-truncating mutations, casting some doubt whether the sequence variants identified in the somatic tissue represent bona fide loss-of-function mutations. Furthermore, a crucial aspect of the two-hit hypothesis—the determination of allelic status of the mutations—was not investigated (25).
Mouse models of CCMs lend some support to the two-hit hypothesis. We and others have generated knockout alleles for both murine Ccm1 and Ccm2 (26,27). With few exceptions, the heterozygous animals do not develop CCM lesions. However, when these mutant alleles are crossed on background of p53 knockout, the animals develop lesions, showing all the hallmarks of CCMs (28,29). The p53 null background was chosen to increase genetic and genomic instability and create a sensitized background for somatic mutation. This sensitized animal model for CCM further supports the two-hit mechanism of pathogenesis. However, to date, somatic mutations have not been identified in lesions from the murine models of CCM.
The strongest evidence for the two-hit model of CCM pathogenesis thus far is that from a lesion from a familial case of CCM1, where a 34 bp deletion in the CCM1 gene was identified in lesion tissue and confirmed to be biallelic to the germline mutation (30). It is unclear whether this mechanism is general for all CCM1 cases and whether this mechanism applies to the other inherited forms of CCMs. In the present study we have used a robust, staged, DNA sequence-based strategy to identify biallelic somatic mutations in all forms of inherited CCMs, and we have attempted to identify the cellular component harboring the somatic mutation.
RESULTS
Somatic mutation detection strategy
We have attempted to identify biallelic somatic mutations within surgically excised CCM lesion tissue with inherited cases of CCM. Once the germline mutation had been established, we faced the more difficult task of uncovering somatic mutations that might be present in only a subpopulation of the cells isolated from the bulk lesion. Any somatic mutation present in only one cell type and only a subset of that cell type will represent substantially <50% of the sequenced alleles. Because of this, a somatic point mutation cannot be identified as a heterozygous change in a direct sequence tracing of bulk lesion-derived DNA. To identify mutations potentially present at a frequency of <50% of the alleles, we have developed a rigorous four-stage approach to identify somatic mutations that are biallelic to the germline mutations.
In the first stage, each amplified exon for the gene of interest is cloned into a plasmid, and 48 individual clones are sequenced. By sequencing multiple individual clones for each exon, we also create the potential to observe rare, PCR-induced replication errors. We have devised a staged replication/validation approach to discern whether a rare variant is a PCR-induced error or a bona fide somatic mutation present in the tissue. We often observe sequence changes that are found in only one of the 48 clones, which we term singletons. In contrast, we only rarely identify a sequence change that is observed in more than one clone. The second stage of our strategy involves a second round of amplification, cloning and sequencing of any exon containing a change seen in more than one clone. Significantly, in the second round of sequencing, any previously identified singletons are not observed, although instead new singletons are sometimes seen. Because these changes are not reproducible, we consider these singly observed changes to be PCR-induced errors.
In contrast to PCR-induced errors, in the first round of amplification, we occasionally observe a sequence variant in two or more clones. This change is invariably seen again in multiple clones in the second round. The third stage of our procedure involves a third round of amplification, cloning and sequencing, this time using a different polymerase in the amplification step. If this same change is identified again in the third stage, it is considered a bona fide somatic mutation. The fourth and final stages involve determination of the allelic status of the somatic mutation in reference to the germline mutation. The approach used to determine allelic status is specific to the nature of the particular germline and somatic mutations and is described for each case below.
Biallelic somatic mutations are identified in CCM1
Case 1: sample 2049
Germline mutation: The lesion sample for the first case is part of the cohort of de-identified archived CCM lesion samples for which matched blood was not available. Sequence analysis of lesion-derived DNA for exon 10 of CCM1 reveals that this patient carries the common Hispanic germline mutation (5), a premature stop codon in the CCM1 gene c.1363C>T, Q455X. As a constitutional mutation, the patient DNA is heterozygous for this change in all tissues, including the lesion. Thus, the germline mutation is readily apparent as a heterozygous change when the amplified product is directly sequenced (Fig. 1A).
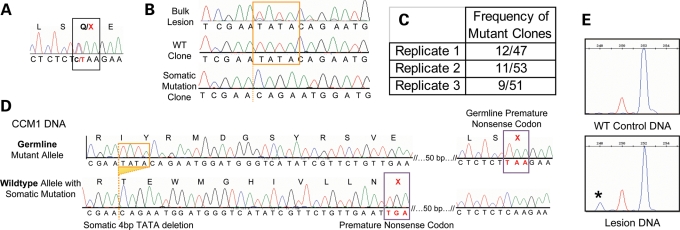
Figure 1.
A biallelic somatic mutation identified in CCM1 sample 2049. (A) Direct sequence analysis of bulk lesion tissue identified the germline mutation as the common Hispanic mutation, c.1363C>T, Q455X in exon 10 of CCM1. (B) Clonal analysis of CCM1 exon 10 reveals wild-type clones, germline mutant clones and clones with the 4 bp deletion somatic mutation, c.1270_1273DelTATA. The deleted bases are boxed in orange in the wild-type sequences, and the deletion site is marked with an orange dotted line in the somatic mutant sequence (bottom tracing). The somatic mutation is barely visible from direct sequence of bulk lesion. (C) The somatic mutation was identified in the lesion tissue multiple times in each of three rounds of PCR, cloning and sequencing. Replicates 1 and 2 used HiFi Platinum Taq Polymerase (Invitrogen) and replicate 3 used HotStar Polymerase (Qiagen). (D) The somatic and germline mutations are biallelic. Sequencing analysis of individual colonies containing both mutation loci shows three classes of clones, germline mutant alleles, wild-type alleles, and alleles carrying the somatic mutation that are wild-type for the germline mutation. (E) Fragment analysis validates the somatic mutation in the lesion. Normal control DNA shows all amplicons of predicted 252 bp length (blue peak). The red size marker is at 250 bp, and the lesion-derived DNA shows products of WT length and those from the somatic mutant allele at 248 bp. The mutant allele represents 11% of the total amplicons.
Somatic mutation: Using our somatic mutation detection strategy, we identified a second-site somatic mutation also in exon 10 of CCM1. The somatic mutation is a 4 bp deletion, c.1270_1273delTATA, resulting in a frameshift and leading to a premature stop codon. This somatic mutation is readily apparent on sequence tracings of individual clones (Fig. 1B) and was identified in multiple clones in each of the three separate amplification/cloning stages, in ∼20% (12/47, 11/53 and 9/51) of the total clones analyzed (Fig. 1C). By fragment analysis, 11% of the total amplicons display the somatic mutation (Fig. 1E). In contrast to the germline mutation, the somatic mutation is not readily apparent in the sequence tracing when the amplified product is directly sequenced. Only in retrospect, after the identification of the somatic mutation, is the signal from the somatic mutation able to be distinguished from background noise on a sequence tracing (Fig. 1B). The contrast between the readily identified germline mutation and the cryptic somatic mutation illustrates the absolute requirement for DNA sequence analysis of multiple individual clones for each exon of the appropriate gene. These data illustrate the sensitivity required for somatic mutation analysis for CCM vascular lesions.
The presence of the somatic and germline mutations in the same exon of the CCM1 gene allowed the allelic relationship of the mutations to be determined at the genomic DNA level. Further analysis of sequence derived from the clones used to identify the somatic mutation yielded three categories, each representing different haplotypes present in the lesion DNA. The first category represents the germline mutant allele. These show the germline mutation, but significantly never show with the somatic mutant allele, consistent with the two-hit hypothesis. The second category represents the wild-type allele. These clones lack both the germline and the somatic mutations and represent wild-type alleles that have not been somatically mutated. The third category represents the somatic mutation. These are always wild-type for the germline mutation, but also harbor the somatic mutation (Fig. 1D). Consistent with the two-hit hypothesis, only wild-type germline alleles show the somatic mutation. The somatic mutation is therefore biallelic to the germline mutation. Only a fraction (∼20%) of the wild-type alleles harbors the somatic mutation, suggesting that only a subset of the cells within the lesion contains the somatic mutation.
Case 2: sample 2009
Germline mutation: case 2 is that of a CCM lesion resected from another patient of the de-identified archived tissue cohort described earlier. Direct sequence analysis of amplified exons from lesion-derived DNA reveals that this sample also harbors the common Hispanic germline mutation, CCM1 c.1363C>T, Q455X (Fig. 2A).
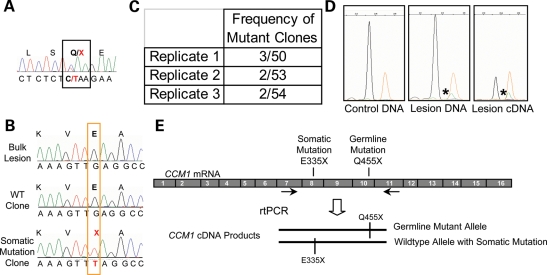
Figure 2.
A biallelic somatic mutation identified in CCM1 sample 2009. (A) Direct sequence analysis of bulk lesion-derived DNA identified the germline mutation as the common Hispanic mutation, CCM1 exon 10 c.1363C>T, Q455X. (B) Somatic mutation analysis identified a somatic mutation in CCM1 exon 8, c1003G>T, E335X. Direct sequence of lesion DNA shows only wild-type sequence. Sequence of individual clones reveals sequence for both wild-type clones and clones with the somatic mutation. (C) The somatic mutation was identified multiple times in each of the three rounds of PCR. Replicates 1 and 2 used HiFi Platinum Taq Polymerase (Invitrogen), and replicate 3 used HotStar Polymerase (Qiagen). (D) The somatic mutation is validated using the SNapShot Assay. The wild-type allele in blue (arrow) is present in all samples, whereas the somatic mutation in green (asterisk) is only present in the lesion-derived DNA or cDNA products and orange peaks are the size standard. Somatic mutant alleles represent ∼6% of the DNA-derived fragments and 24% of the cDNA-derived fragments analyzed. (E) The somatic and germline mutations are biallelic. RT–PCR products containing both somatic and germline mutant loci were amplified, cloned and sequenced. Sequencing analysis of individual colonies shows three classes of clones: germline mutant alleles, wild-type alleles and alleles carrying the somatic mutation that are wild-type for the germline mutation.
Somatic mutation: a second-site somatic mutation was identified in exon 8 of the CCM1 gene, c.1003G>T, E335X, resulting in premature termination. Once again, direct sequencing of the amplified product for exon 8 does not reveal the somatic mutation (Fig. 2B). However, the somatic mutation is identified when individual clones were sequenced for each exon. This somatic mutation is present in ∼4% (3/50, 2/50, 2/54) of the total clones analyzed for this exon (Fig. 2C). As this somatic mutation was identified near the edge of our threshold for detection (two or more of 48 clones sequenced), we sought an independent method to validate the presence of the mutation. We used a SNaPshot assay (Applied Biosystems) to determine the sequence of thousands of PCR amplicons in a single reaction (Fig. 2D). This primer extension assay interrogates the sequence at a specified variant site, with each allele identified with a different fluorescent tag. The wild-type sequence (G) is represented by a black peak and the mutant sequence (T) is shown with a green peak, with the size standard in orange. Blood-derived DNA from an unaffected individual was used as a control in this assay to determine whether the putative somatic mutation occurs merely as a result of a PCR-induced error. This assay confirms that the somatic mutation is present only in the CCM tissue and not in control blood DNA (Fig. 2D). By SNaPshot, the mutation is found in ∼6% of the amplification products from the lesion-derived DNA. Isolation of high-quality RNA for cDNA analysis was possible for this lesion sample because it was cryopreserved following surgery. Using cDNA as a PCR template, the presence of the somatic mutation is also confirmed by SNapShot, and amplicons with the somatic mutation represent 24% of the total population (Fig. 2D). From the total lesion tissue, only cells expressing the CCM1 transcript would contribute to the clones, thus increasing the percentage of amplicons showing the mutation.
The genomic distance between these two mutation sites is prohibitively large, such that they may not be amplified as a single amplicon at the genomic DNA level. Therefore, to confirm that the somatic c.1003G>T, E335X (exon 8) mutation is biallelic to the germline c.1363C>T, Q455X (exon 10) mutation, we performed reverse transcriptase (RT)–polymerase chain reaction (PCR) to amplify the region of CCM1 from the cDNA that would encompass both mutation loci. The sequence analysis of individually cloned amplicons reveals three categories of clones: those with the germline mutation, those that are fully wild-type and those that are wild-type at the germline mutation, but showing the somatic mutation. These results demonstrate that the somatic and germline mutations are biallelic as the somatic mutation was only observed in the context of the wild-type allele (Fig. 2E).
Biallelic somatic mutation identified in CCM2
Case 3: family 307 sample 3911
Germline mutation: the lesion sample for case 3 is a CCM lesion resected from the proband of family 307 (31). The germline mutation inherited within this family is a large deletion of the CCM2 gene, encompassing nine of the ten coding exons (Fig. 3A). This germline common deletion has been repeatedly identified in our North American CCM cohort and is readily screened for by a PCR reaction spanning the deletion (31,32).
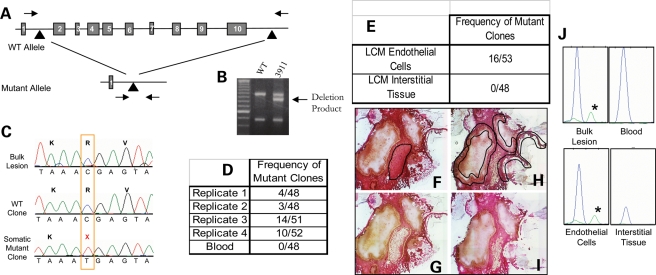
Figure 3.
A biallelic somatic mutation is identified in CCM2 sample 3911. (A) The germline mutation is identified as a large deletion of the CCM2 gene encompassing exons 2–10, which is readily screened for using primers spanning the deleted region (arrows). (B) Sample 3911 is heterozygous for the CCM2 2-10 deletion. The intermediate size band results from a PCR, which spans the 77 kb deletion (32), and is not present in wild-type controls. The upper and lower PCR products are control reactions, each requiring one of the primers used to generate the deletion product band (32). (C) Somatic mutation analysis reveals a somatic mutation, c.55C>T, R19X in exon 2 of the CCM2 gene. The somatic mutation is not readily apparent on a sequence tracing from bulk lesion. Sequence from multiple clones containing exon 2 inserts reveal both wild-type and somatic mutant clones. (D) The somatic mutation is present in multiple clones in each of three PCR replicates from bulk lesion tissue. Using template DNA derived from histology slides, the third replicate also had multiple clones with the somatic mutation. A different polymerase, Hotstar polymerase (Qiagen), was used for the final replicate. The somatic mutation is not present in clones generated from leukocyte-derived template DNA. (E) Laser capture microscopy was preformed to isolate endothelial cells surrounding the caverns as well as interstitial (non-endothelial) lesion tissue. The somatic mutation was identified using endothelial cell-derived DNA as template. The interstitial tissue-derived DNA clones were entirely wild-type. (F–H) Laser capture microscopy to isolate inter-cavernous control cells [(F) before LCM with targeted tissue outlined in black and (G) after isolation] and endothelial cells surrounding blood-filled caverns [(H) before LCM with targeted EC's outlined in black and (I) after LCM]. (J) The somatic mutation is validated using the SNapShot assay. The somatic mutation is present in bulk lesion and laser-captured endothelial cell-derived DNA (green peak with asterisks). The wild-type allele (blue) is present in all samples.
Somatic mutation: we identified a somatic nonsense mutation c.55C>T, R19X in exon 2 of CCM2 in DNA isolated from the lesion of sample 3911. Direct sequence of bulk lesion-derived DNA does not reveal the somatic mutation; however, the somatic mutation is readily identified by sequence analysis of individual clones (Fig. 3C). In four replicates of somatic mutation analysis using lesion DNA as a PCR template, 16% (4/48, 3/48, 14/51, 10/52) of the total clones displayed the somatic mutation (Fig. 3D). The analysis of 48 clones of CCM2 exon 2, from the blood-derived DNA, resulted in all clones, showing wild-type sequence at the somatic mutation site, supporting the hypothesis that somatic mutations in CCM patients are specific to the lesion tissue.
The germline deletion in this sample encompasses nine of the 10 exons of CCM2, removing nearly the entire gene of the mutant allele. Any sequence generated from these deleted exons is, by definition, derived from the wild-type CCM2 allele. Because exon 2 is deleted in the mutant germline allele, the somatic mutation identified within exon 2 of CCM2 is biallelic to the germline deletion.
This frozen lesion sample was of sufficient structural quality to enable histological examination. Using laser capture microscopy, we have further defined the scope of somatic mutations in CCM lesions. We isolated two populations of cells: cavernous endothelial cells and non-endothelial interstitial tissue located between neighboring caverns within the lesion (Fig. 3F–I). Endothelial and non-endothelial tissue isolated from five serial section histology sides was pooled according to the cell type, and the resulting DNA was used for somatic mutation analysis. The analysis of exon 2 in 48 clones from the interstitial tissue of the lesion resulted in all clones, showing wild-type sequence the somatic mutation site. Conversely, the endothelial cell-derived DNA showed the somatic mutation in 16/53 (30%) of the clones (Fig. 3E). Using the SNaPshot assay to quantify the fraction of mutant amplicons, the somatic mutation is present in 5–10% of the DNA isolated from bulk lesion. Different individual sections of the CCM lesion showed a reproducible fraction of mutant clones; however, the fraction varied from section to section. In contrast, the somatic mutation was present in 15% of the laser-captured endothelial cell-derived DNA (Fig. 3J). This suggests that the somatic mutation occurs within a subset of the endothelial cells lining the caverns within this late-stage CCM lesion.
Somatic mutation identified in CCM3
Case 4: family 283 sample 4035
Germline mutation: the sample for the fourth case is a member of a family harboring a known splice altering germline mutation in CCM3, exon 8, c.474+1G>A (17). As expected, the direct sequence analysis of bulk blood-derived DNA for the coding exons of CCM3 revealed this patient to also harbor the CCM3 germline mutation (Fig. 4A).
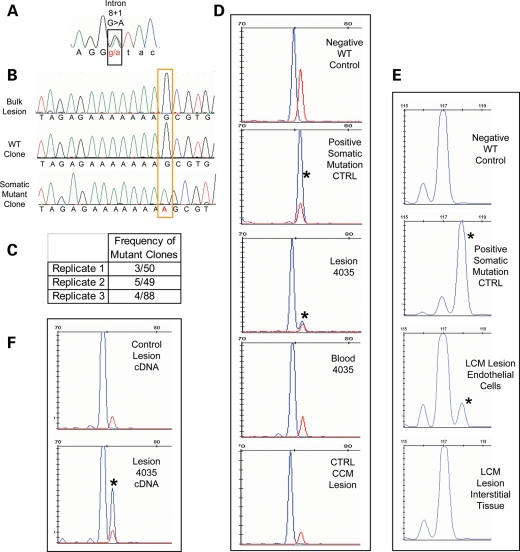
Figure 4.
A CCM3 somatic mutation identified in sample 283-4035. (A) The germline mutation identified by sequence analysis of leukocyte-derived DNA is a splicing mutation in CCM3 exon 8, c.474+1 G>A. (B) Somatic mutation identified as an insertion of A in exon 6 of CCM3, c.205-211insA. The somatic mutation is not readily identifiable from the direct sequence of bulk lesion-derived DNA. Clonal analysis reveals both wild-type and somatic mutant clones. (C) The somatic mutation is identified multiple times in each of three replicates. The final replicate is of Phusion-derived PCR products. (D) The somatic mutation is validated by fragment analysis. Size standard marker at 75 bp is in red. Phusion-derived PCR fragments appear in blue. The wild-type fragment product is 74 bp (Negative Ctrl), and the somatic mutation product is 75 bp long (Positive Ctrl). The lesion-derived sample yields two products, both wild-type and mutant (asterisk) with the mutant allele representing 10% of the total amplicons. The larger fragment of the somatic mutation is not present in either matched blood for lesion sample 4035 or another unrelated CCM lesion sample that is WT at the site of the somatic mutation. (E) In a different fragment analysis assay, normal WT length fragments are 117 bp, and those with the somatic mutation (asterisk) are 118 bp long. From the laser-captured tissue, only the endothelial cells and not the interstitial tissue harbor the somatic mutation. Thirteen percent of the endothelial cell-derived amplicons carry the somatic mutation. (F) The somatic mutation is biallelic to the germline mutation. Fragment analysis of a phusion-derived PCR product from lesion cDNA spanning exons 6 to 9 of CCM3. The somatic mutation (asterisk) is only present in the RT–PCR product from 4035 lesion cDNA, not from a control CCM lesion.
Somatic mutation: we have identified a somatic mutation in exon 6 of CCM3 that is an insertion of a single adenosine, c.205-211insA, resulting in a frameshift mutation and premature termination. The sequence of individual clones readily identifies the presence of this somatic mutation, whereas it is undetectable by direct sequence of bulk lesion-derived PCR products (Fig. 4B). This somatic mutation was observed multiple times from each of three amplifications. In total, 7% (3/50, 5/49, 4/88) of the clones from bulk lesion-derived DNA harbored the somatic mutation (Fig. 4C). This mutation is an insertion of an adenosine in a string of seven adenosines. Repeat units such as this can be prone to mutation by polymerase error, both in vitro and in vivo. To confirm that this sequence alteration was present within the lesion tissue and not created during amplification by polymerase error, we again chose to use a fragment size-based assay to analyze thousands of amplicons in a single reaction. Using a high fidelity enzyme (PHusion™), we generated amplified products for exon 6 of CCM3 from lesion DNA and controls. The wild-type sequence for this amplicon will result in a 74 bp fragment, and the somatic mutation insertion will increase the product size to 75 bp. Only the DNA isolated from the CCM lesion displays amplicons of 75 bp. Thus, only lesion 4035 harbors this somatic mutation (Fig. 4D, middle panel). A matched blood DNA control does not display the somatic mutant 75 bp fragment. In order to determine whether the mutation was a polymerase error induced by poor DNA quality of lesion-derived DNA, we used DNA isolated from another lesion sample of similar quality (also formalin-fixed and paraffin-embedded). This sample showed only the 74 bp wild-type allele. Thus, the somatic mutation is not due to PCR error or poor quality DNA, but is a bona fide somatic mutation in CCM sample 4035.
Analogous to case 3, laser capture microscopy was performed on this sample to isolate endothelial and non-endothelial cell populations from within the lesion. By fragment analysis, only DNA derived from the endothelial cells harbored the somatic mutation, and the interstitial tissue yielded products of only wild-type size (Fig. 4E).
We determined the allelic nature of this mutation using RNA (cDNA) isolated from lesion. Because the germline mutation in this case disrupts normal splicing due to mutation of the canonical splice donor site in exon 8, the resulting mutant transcript would skip this exon. We amplified RT–PCR products from the lesion-derived cDNA to contain both mutation sites in an amplicon spanning from exons 6 to 9. Using these PCR products as a template for the fragment analysis assay, the somatic mutation is observed in only full-length spliced transcripts containing exon 8, indicating that it arose on the wild-type allele. Furthermore, it was found only in RT–PCR products from the lesion 4035 and not a different CCM lesion (Fig. 4F).
Additional samples analyzed
In addition to these previously described cases, we employed our somatic mutation detection strategy on four additional CCM2 cases and two additional CCM3 cases. In these cases, we were unable to identify a somatic mutation using our strategy. We discuss the meaning of this below.
DISCUSSION
The heterogeneous cellular nature of the CCM lesions may have inhibited previous attempts to detect somatic mutations within the vascular tissue. Direct sequencing of an amplified product effectively identifies heterozygous germline mutations. However, if a somatic mutation is present in substantially <50% of the alleles, it may not be easily detected. Our strategy for somatic mutation analysis involving sequencing of individual PCR amplicons provided the sensitivity to detect mutations that are not present in an entire cellular population and that might otherwise remain undetected by direct sequencing of the amplified product. The overall sensitivity of our approach is approximately similar to that used by Gault et al. for the same purpose (30). Denaturing high-pressure liquid chromatography also enabled screening of all the CCM1 exons, such that a biallelic somatic mutation could be identified in a CCM lesion despite its presence in only a fraction of the cells.
We have shown that somatic mutations are present in all forms of inherited CCMs, although we were unable to detect somatic mutations in each sample examined. We suggest that the architecture and microanatomy of the CCM lesions can affect the ability to detect somatic mutations. Tissue heterogeneity can cause problems with somatic mutation detection for solid tumors (33) and also pose similar problems with the CCM lesion, which comprised multiple cell and tissue types. In addition to vascular components, the bulk lesion contains intervening connective tissue. Additionally, as the lesion is itself a vascular structure, it can be difficult to distinguish the vasculature of the lesion proper from feeding and draining vessels. Any given section through the surgically resected lesion may contain different fractions of cells harboring the somatic mutation. Some sections might contain more interstitial tissues, fewer caverns, more blood or a part of the lesion consisting primarily of feeding or draining vessels. This problem is especially relevant for cases in which histology slides are the only source of tissue available, as the slides represent a single slice through the lesion. In support of this contention, we have observed that different DNA preparations isolated from different regions of the same CCM lesion result in an apparent difference in the frequency of the somatic mutation. In extreme cases, the fraction of clones exhibiting a somatic mutation might fall below the threshold for initial detection.
We chose to sequence 48 clones for each exon as two exons could be completed in a 96-well microtiter plate. Using this number of clones, power calculations showed that we have 95% power to observe a mutation in more than one clone if 20% of the alleles harbored the mutation. However, the power to detect a mutation decreases markedly as the number of alleles harboring the mutation falls below the 20% level. Although in one case we have identified a somatic mutation that was present in only 6% of the alleles sampled, it is more likely that we would miss mutations present at such low levels in the section of the lesion that was analyzed.
We have also noted that the quality of the tissue sample can have profound effects on the ability to identify a somatic mutation. Surgically resected tissue that was immediately frozen provided genomic DNA that amplified very well and, in general, showed the fewest singleton changes and other PCR-induced errors. In contrast, formalin-fixed paraffin-embedded tissue samples contained DNA that was more difficult to amplify and usually showed a higher frequency of random nucleotide changes. Both singletons and even multiple clone changes were detected in formalin-fixed tissue, none of which was validated through multiple rounds of amplification, cloning and sequencing. The fixation process can destroy the integrity of the DNA and result in sequence changes that are indistinguishable from PCR error.
Finally, our somatic mutation identification strategy was specifically designed to identify mutations that are readily apparent by DNA sequencing: single nucleotide changes and small insertions or deletions. Our approach would miss altogether other types of second hits. A classic mechanism for a second genetic hit is LOH, resulting from large deletions, unbalanced translocations or mitotic recombination. The identification of this type of second hit requires the ability to observe the loss of one allele of a linked heterozygous marker. In a relatively homogeneous lesion sample, the loss of one allele is readily apparent, either as complete loss of one allele (LOH) or, more commonly, as a statistically significant imbalance in the ratio of the two alleles. However, it becomes increasingly difficult to observe LOH with increasing amounts of cellular heterogeneity in the lesion tissue (33). Given the complex microanatomy and mixed cellular composition of CCM lesions, LOH would be very difficult if not impossible to detect. Epigenetic effects such as promoter methylation may also represent a mechanism by which the wild-type allele may be silenced. Again, these types of mutations are not readily identifiable by our strategy. Samples that did not exhibit somatic mutations as simple sequence changes might be explained by any of the previously described technical caveats or by different classes of second hit mutations.
Another potential two-hit mechanism would be the development of a somatic mutation in a gene other than that which harbors the germline mutation. For CCMs, one of the other CCM genes might make attractive candidates for a second hit in samples, where we did not identify a somatic mutation. The concept of trans-heterozygosity as a two-hit mechanism has precedence in autosomal-dominant polycystic kidney disease (PKD). PKD has been shown to follow the two-hit mechanism for pathogenesis. PKD cysts may arise due to inheritance of a mutated copy of PKD1 or PKD2, followed by a second hit to the germline mutation (34,35). The cysts sometimes instead show trans-heterozygous germline and somatic mutations, one in each of the two PKD genes (34,35). However, our mouse models of CCM argue against trans-heterozygosity. We have crossed the Ccm1 and Ccm2 lines to create mice that are doubly heterozygous for both germline mutations. These animals are trans-heterozygous in all cells and tissues and yet they do not develop CCM lesions. In light of this observation, we do not favor trans-heterozygosity as a mechanism for the second genetic hit for CCMs.
The two-hit mechanism for disease pathogenesis was originally described for tumor suppressor genes and inherited cancers. However, in recent years, the two-hit mechanism underlying disease pathogenesis has also emerged for non-malignant diseases. PKD is a clear example of a non-malignant phenotype that follows the two-hit mechanism (34,35). The two-hit genetic mechanism has been implicated as the underlying mechanism for the development of venous and glomuvenous malformations (36,37). Somatic mutations have been identified in a population of non-proliferating T cells in autoimmune lymphoproliferative syndrome (ALPS). This rare population of cells is believed to promote the proliferation of non-mutated lymphocytes and to indirectly cause the onset of ALPS (38). CCM may follow a similar mechanism of pathogenesis. The second hit of the wild-type allele might initiate both the proliferative and remodeling capabilities of neighboring endothelial cells to result in the genesis of multicavernous CCM lesions.
All three CCM genes have been shown by in situ and immunohistochemical studies to be expressed in a variety of cell types within the brain, including neurons, astrocytes and vascular endothelium. Although these studies have shown some inconsistencies in the expression pattern, all agree that all three CCM transcripts/proteins are robustly expressed in the neurons (26,39–44). These collective observations suggested that CCM pathogenesis might be primarily due to a neural defect. However, we have shown here that the somatic mutations in the CCM tissue are present only in the endothelial cells lining the caverns of the lesion. Regardless of the role of these proteins in other cell types, this observation strongly supports an important role of the CCM proteins in the vascular endothelium for the maintenance of normal vessel integrity. A critical role for the CCM proteins in the vascular endothelium is also supported by biochemical studies, which demonstrate that KRIT1 is localized to endothelial cell junctions and is responsible for maintenance and stabilization of the integrity of tight junctions (14). Additionally, a significant role of the CCM1 and CCM2 gene products in the endothelial cell is supported by developmental studies of the null phenotypes in mice (CCM1) (27) and zebrafish (santa and valentine, CCM1 and CCM2, respectively) (45,46), which exhibit primarily vascular (or cardiovascular) phenotypes.
Our data also support existing clinical and histological data, showing that CCM lesions exhibit characteristics of both vascular tumors and vascular malformations. These lesions appear to grow as a result of both remodeling of the existing vasculature and proliferation of the lesion endothelial cells. The three forms of CCM follow the two-hit mechanism of pathogenesis, suggesting that the CCM genes also show characteristics of tumor suppressor genes. Loss of both copies of the gene may explain some of the proliferative capacity in the EC component (47–49). However, loss of both copies also appears to initiate other pathogenic mechanisms, including vascular remodeling (20,21) and changes in vascular permeability due to loss of tight junctions (3,50). Further analysis of these genes and gene products in normal vascular growth will shed further light on the distinct roles of the CCM genes in vascular biology and in the pathogenesis of CCM.
MATERIALS AND METHODS
Study subjects and germline mutation analysis
This study used the genetic material of 10 distinct CCM lesion tissues from 10 individual patients (Supplementary Material, Table S1).
CCM lesion samples with germline mutation in CCM1
From a cohort of 52 de-identified archived lesion samples, we prepared DNA from each lesion. Using standard direct sequencing techniques to sequence amplified exon products for CCM1 exon 10, 30 lesion samples were specifically screened for the common Hispanic mutation (CCM1 c.1363C>T, Q455X). Eight of the lesions harbored this germline mutation. We performed somatic mutation analysis on two lesions, 2009 and 2049, each with the germline mutation in the CCM1 gene. These two samples were flash frozen following surgery. No matched blood samples were available for these tissues.
The lesion samples with germline mutations in both CCM2 and CCM3 were obtained after receipt of informed consent, and DNA was prepared from these samples through use of standard methods.
CCM lesion samples with germline mutation in CCM2
Sample 362-4286 was received as paraffin-embedded formalin-fixed slides, and no matched blood was available. Samples 4296, 4297 and 4312 were obtained as frozen samples. DNA was prepared directly from these lesions, and we identified the germline mutation to be a common deletion in CCM2 encompassing exons 2 through 10. Thus, mutation is readily identified with a PCR reaction to amplify across the deleted region (32). Sample 307-3911 is also a frozen lesion sample that also carried the common CCM2 2–10 deletion as the germline mutation. This tissue sample was collected with matched blood, and the germline mutation was identified using leukocyte-derived DNA (31).
CCM lesion samples with germline mutation in CCM3
Blood and lesion tissue was available for all of the CCM3 samples. We acquired formalin-fixed paraffin-embedded slides for lesion samples 180-3933 (17) and 283-4035, whereas sample 162-4309 (17) is a frozen lesion sample. The germline mutation for each of the CCM3 cases was determined by sequence analysis of leukocyte-derived DNA. The germline mutation for 180-3933 is CCM3 c.608T>G, L203X, for 283-4035 is CCM3 c.474+1G>A and for 162-4309 is CCM3 c.474+5G>A.
Somatic mutation analysis
Because two of the 10 CCM lesion samples used in this study were de-identified archived surgical specimens, we first had to identify whether the lesion derives from an inherited case, defined the presence of a heterozygous germline mutation. However, for other lesions, blood was not available. In these cases (2009 and 2049), we identified the germline mutation directly from the lesion tissue. Some germline mutations will be present in any tissue in the appropriate Mendelian frequency for autosomal-dominant disease, e.g. 50% of the total alleles.
Following identification of the germline mutation, we looked for somatic mutations in the CCM gene that harbors the germline mutation. In accordance with the two-hit hypothesis we sought to identify somatic mutations that are biallelic to the germline mutation (for primer sequences, see Supplementary Material, Table S1). For cases of CCM1 and CCM3, we looked for somatic mutations in each coding exon. For CCM2, we looked for somatic mutations in exons 2 through 10 and alternatively spliced exon 1B, as all of these exons are deleted as part of the germline deletion mutation. Any sequence from these exons would, by definition, be derived from the wild-type allele. The strategy used standard sequencing techniques modified by an intermediate cloning step. Briefly, we used a high fidelity Taq-derived polymerase to amplify each of the exons for the gene of interest from lesion-derived DNA. The resulting product was subsequently ligated into a T-A cloning vector, p-Gem T-Easy (Promega), following manufacturer's protocol, overnight at 4°C. To generate PCR products that contained minimal polymerase-induced errors and that also allowed for non-template-directed A-tailing of products, we used Platinum Taq Hi-Fidelity (Invitrogen) or Hotstar polymerase (Qiagen) for exon amplification. The first two replicates for any given exon were amplified with the Invitrogen Taq polymerase, and the third confirmatory amplification used the Qiagen polymerase to control for polymerase-specific errors. Amplification products generated by either of these polymerases were directly ligated into the p-Gem vector. In one case, for the third replicate for 4035 CCM3 exon 6, we amplified using Phusion™ High-Fidelity DNA Polymerase (Finnzymes), which generated blunt-ended products. These PCR fragments were purified using the High Pure PCR Product Purification Kit (Roche) and then were A-tailed prior to ligation by Taq polymerase during a 30 min incubation with 10 mm dATP, 1× PCR buffer and 5 U Taq polymerase.
Ligated plasmids were transformed into competent DH5α Escherichia coli and plated overnight on Luria–Bertani plates with ampicillin, X-Gal and IPTG. Using blue–white screening, we picked 48 white insert-positive colonies for use as template in a colony-PCR reaction. Each sequenced clone represents a single allele, enabling identification of sequence changes that may be in low abundance within the lesion tissue. We attempted to sequence the exon product of 48 insert-positive colonies, each representing a single PCR amplicon and to be used as a template in a colony PCR. For the colony PCR, we used vector-specific primers SP6 5′-ATTTAGGTGACACTATAG and T7 5′-TAATACGACTCACTATAGGG. Again, a high fidelity polymerase, Platinum Taq HiFidelity (Invitrogen), was used and each PCR product was sequenced using BigDye® Terminator v1.1 cycle sequencing kit (Applied Biosystems) and an ABI 3730 DNA Sequencer. Sequencing of 48 clones for each exon provides us the power to detect any somatic mutation, as a sequence variant in two or more clones, which is present in at least 20% of the amplicons, with 95% confidence.
Due to cloning or sequencing failures, sometimes, the number of sequenced clones was less than 48. When this number was substantially less than 48, we sequenced additional clones from the same amplification. At this point, we included enough clones to ensure that we would hit at least 48 clones. Thus, the final number for any given analysis stage did not always equal 48 and, in some cases, was slightly more.
Three rounds of amplification, cloning and sequencing were performed for any sequence variant, which was observed in more than one clone. In the second round of amplification, we used the same polymerase as the first round, and if the sequence change was observed again in multiple clones, we proceed to a third round of amplification, which used a different high fidelity polymerase. This change of polymerase was performed to validate that the somatic mutation is a bona fide mutation, and not due to a polymerase-specific error. Only bona fide somatic mutations were observed in all three rounds of PCR. Polymerase errors were typically seen as single clone changes and were not replicated in multiple rounds of amplification.
Snapshot assay
For confirmation of the somatic mutation in samples 2009 and 307–3911, we used a single base extension assay, the SNaPshot Assay (Applied Biosystems). For sample 2009, we amplified exon 8 of CCM1 from the lesion DNA and exons 7 to 11 from the lesion cDNA as described below, and for 307–3911, we amplified CCM2 exon 2. PCR products were treated with 1 U exonuclease I (New England Biolabs) and 1 U of shrimp alkaline phosphatase (SAP) (Promega). The somatic mutation was labeled green in both cases, and the wild-type allele was labeled black for 2009 and blue for 307-3911 following the SNaPshot reaction, which included the treated PCR product, 2.5 µl of the pre-mix from the ABI PRISM® SNaPshot® Multiplex Kit, 2 µM primer 5′-TAACAATATGCGAGTGGCCT (sample 2009) or 5′-CTCTTTTCACCTTTTAGGAATACTC (sample 307-3911), 1× PCR buffer, 0.5 mm MgCl2. Cycling conditions were as follows: 40 cycles of 95°C for 10 s, 50°C for 5 s and 60°C for 30 s. These products were phosphatase-treated again with 1 U of SAP, prior to running on an ABI 3130 sequencer and analysis using GeneMapper® Software Version 4.0.
Fragment analysis
The presence of the 4 bp deletion somatic mutation in sample 2049 was validated by a fragment analysis assay. Using primers 5′-6FAM-TTCGTTACGTAAGTTATCGTTTCTTG and 5′-CTACCAACCCACTCCCAAAA, we amplified a 252 bp fragment of CCM1 containing the somatic mutation site. The presence of the somatic mutation shifts that fragment size to 248 bp, detected as described below.
The presence of a single base insertion somatic mutation in sample 4035 was validated by a fragment analysis assay. For the fragment analysis, PCR products for exon 6 of CCM3 were amplified using one fluorescently labeled primer 6-FAM-CTCACACAAGACATCATTATG and one unlabeled primer 5′-CCATACGAAGAAGGGACTCC or 5′-AAACAAGGTTCTTCTGTCCGTTA. The resulting Phusion™ PCR products were resuspended in formamide (Applied Biosystems) with Rox 350 size standards (Applied Biosystems) and were characterized on an ABI 3130 Sequencer. Subsequent analysis of fragment sizes was performed using GeneMapper® Software Version 4.0. Positive and negative controls were colony PCR products from clones, with the sequence showing either wild-type (negative control) or the somatic mutant (positive control) sequence.
cDNA synthesis and RT–PCR
For RT–PCR reactions, total RNA was isolated from frozen lesions using the TRIzol® reagent (Invitrogen) and manufacturer's protocol. From formalin-fixed paraffin-embedded tissues, the total RNA was isolated using the PureLink™ FFPE Total RNA Isolation Kit (Invitrogen) and manufacturer's protocol. First-strand cDNA synthesis was performed using iScript reverse transcriptase (BioRad), and the resulting products were used as the template for RT–PCR. The PCR product containing CCM1 exons 7 to 11 was generated with the following primers: exon 7 RT forward 5′-CAGAATTACTAAGCCGTCTTCTCA and exon 11 RT reverse 5′-GGATCCAGATTAGTCAATTCAGC.
The following primers were used to amplify exons 6 to 9 of CCM3: exon 6 RT forward 5′-TCCAGGTCTCACACAAGACATC and exon 9 RT reverse 5′-CTTTCTTTTGGTGTTCAAGTGC.
Laser capture microscopy
Endothelial cells and interstitial control tissue were isolated from CCM pathology slides by laser capture microscopy. Cellular populations were isolated from five 17 µM frozen sections for sample 307-3911 and from three thin paraffin sections for sample 283-4035, using the infrared laser on a Veritas Microdissection system by Arcturus. On average, laser settings were as follows: 70 mW power, pulse time 5500 s, intensity 180 mV and spot size 33.6 µm. Tissue samples were collected on Arcturus CapSure Macro LCM Caps, and DNA was prepared using the QIAamp DNA Micro Kit (Qiagen). Endothelial cell and control interstitial cell populations from each slide were pooled for DNA isolation. Following manufacturer's protocol for the QIAmp DNA Micro Kit, LCM DNA was purified and resuspended in 20 µl elution solution. An aliquot of 5 µl of the DNA solution was used for subsequent PCR reactions to amplify CCM2 exon 2 prior to clonal analysis for case 3 or CCM3 exon 6 for fragment analysis in case 4.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the American Heart Association (grant 0715225U to A.L.A.) and the National Institutes of Health (Grant 1F31NS061468-01A1 to A.L.A., and grant RO1NS43543 to D.A.M.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the patients and their families for tissue donation and their continued support for our research. We thank Tracey Leedom and Connie Lee with the Angioma Alliance Tissue Bank for their help with tissue acquisition. We thank Rich Roberts, Adam Foye, Stephanie Li and Kenneth Igbalode for technical assistance.
Conflict of Interest statement. We declare no conflicts of interest.
REFERENCES
- 1.Rigamonti D., Hadley M.N., Drayer B.P., Johnson P.C., Hoenig-Rigamonti K., Knight J.T., Spetzler R.F. Cerebral cavernous malformations. Incidence and familial occurrence. N. Engl. J. Med. 1988;319:343–347. doi: 10.1056/NEJM198808113190605. [DOI] [PubMed] [Google Scholar]
- 2.Gault J., Sarin H., Awadallah N.A., Shenkar R., Awad I.A. Pathobiology of human cerebrovascular malformations: basic mechanisms and clinical relevance. Neurosurgery. 2004;55:1–16. discussion 16–17. [PubMed] [Google Scholar]
- 3.Clatterbuck R.E., Eberhart C.G., Crain B.J., Rigamonti D. Ultrastructural and immunocytochemical evidence that an incompetent blood-brain barrier is related to the pathophysiology of cavernous malformations. J. Neurol. Neurosurg. Psychiatry. 2001;71:188–192. doi: 10.1136/jnnp.71.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laberge-le Couteulx S., Jung H.H., Labauge P., Houtteville J.P., Lescoat C., Cecillon M., Marechal E., Joutel A., Bach J.F., Tournier-Lasserve E. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat. Genet. 1999;23:189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- 5.Sahoo T., Johnson E.W., Thomas J.W., Kuehl P.M., Jones T.L., Dokken C.G., Touchman J.W., Gallione C.J., Lee-Lin S.Q., Kosofsky B., et al. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1) Hum. Mol. Genet. 1999;8:2325–2333. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- 6.Liquori C.L., Berg M.J., Siegel A.M., Huang E., Zawistowski J.S., Stoffer T., Verlaan D., Balogun F., Hughes L., Leedom T.P., et al. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am. J. Hum. Genet. 2003;73:1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denier C., Goutagny S., Labauge P., Krivosic V., Arnoult M., Cousin A., Benabid A.L., Comoy J., Frerebeau P., Gilbert B., et al. Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am. J. Hum. Genet. 2004;74:326–337. doi: 10.1086/381718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergametti F., Denier C., Labauge P., Arnoult M., Boetto S., Clanet M., Coubes P., Echenne B., Ibrahim R., Irthum B., et al. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am. J. Hum. Genet. 2005;76:42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zawistowski J.S., Serebriiskii I.G., Lee M.F., Golemis E.A., Marchuk D.A. KRIT1 association with the integrin-binding protein ICAP-1: a new direction in the elucidation of cerebral cavernous malformations (CCM1) pathogenesis. Hum. Mol. Genet. 2002;11:389–396. doi: 10.1093/hmg/11.4.389. [DOI] [PubMed] [Google Scholar]
- 10.Zawistowski J.S., Stalheim L., Uhlik M.T., Abell A.N., Ancrile B.B., Johnson G.L., Marchuk D.A. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum. Mol. Genet. 2005;14:2521–2531. doi: 10.1093/hmg/ddi256. [DOI] [PubMed] [Google Scholar]
- 11.Hilder T.L., Malone M.H., Bencharit S., Colicelli J., Haystead T.A., Johnson G.L., Wu C.C. Proteomic identification of the cerebral cavernous malformation signaling complex. J. Proteome. Res. 2007;11:4343–4355. doi: 10.1021/pr0704276. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J., Rigamonti D., Dietz H.C., Clatterbuck R.E. Interaction between krit1 and malcavernin: implications for the pathogenesis of cerebral cavernous malformations. Neurosurgery. 2007;60:353–359. doi: 10.1227/01.NEU.0000249268.11074.83. discussion 359. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X.A., Hemler M.E. Interaction of the integrin beta1 cytoplasmic domain with ICAP-1 protein. J. Biol. Chem. 1999;274:11–19. doi: 10.1074/jbc.274.1.11. [DOI] [PubMed] [Google Scholar]
- 14.Glading A., Han J., Stockton R.A., Ginsberg M.H. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell–cell junctions. J. Cell. Biol. 2007;179:247–254. doi: 10.1083/jcb.200705175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhlik M.T., Abell A.N., Johnson N.L., Sun W., Cuevas B.D., Lobel-Rice K.E., Horne E.A., Dell'Acqua M.L., Johnson G.L. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat. Cell. Biol. 2003;5:1104–1110. doi: 10.1038/ncb1071. [DOI] [PubMed] [Google Scholar]
- 16.Voss K., Stahl S., Schleider E., Ullrich S., Nickel J., Mueller T.D., Felbor U. CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics. 2007;8:249–256. doi: 10.1007/s10048-007-0098-9. [DOI] [PubMed] [Google Scholar]
- 17.Liquori C.L., Berg M.J., Squitieri F., Ottenbacher M., Sorlie M., Leedom T.P., Cannella M., Maglione V., Ptacek L., Johnson E.W., et al. Low frequency of PDCD10 mutations in a panel of CCM3 probands: potential for a fourth CCM locus. Hum. Mutat. 2005;27:118. doi: 10.1002/humu.9389. [DOI] [PubMed] [Google Scholar]
- 18.Davenport W.J., Siegel A.M., Dichgans J., Drigo P., Mammi I., Pereda P., Wood N.W., Rouleau G.A. CCM1 gene mutations in families segregating cerebral cavernous malformations. Neurology. 2001;56:540–543. doi: 10.1212/wnl.56.4.540. [DOI] [PubMed] [Google Scholar]
- 19.Labauge P., Brunereau L., Levy C., Laberge S., Houtteville J.P. The natural history of familial cerebral cavernomas: a retrospective MRI study of 40 patients. Neuroradiology. 2000;42:327–332. doi: 10.1007/s002340050893. [DOI] [PubMed] [Google Scholar]
- 20.Del Curling O., Jr, Kelly D.L., Jr, Elster A.D., Craven T.E. An analysis of the natural history of cavernous angiomas. J. Neurosurg. 1991;75:702–708. doi: 10.3171/jns.1991.75.5.0702. [DOI] [PubMed] [Google Scholar]
- 21.Robinson J.R., Awad I.A., Little J.R. Natural history of the cavernous angioma. J. Neurosurg. 1991;75:709–714. doi: 10.3171/jns.1991.75.5.0709. [DOI] [PubMed] [Google Scholar]
- 22.Knudson A.G., Jr Mutation and cancer: statistical study of retinoblastoma. Proc. Natl Acad. Sci. USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reich P., Winkler J., Straube A., Steiger H.J., Peraud A. Molecular genetic investigations in the CCM1 gene in sporadic cerebral cavernomas. Neurology. 2003;60:1135–1138. doi: 10.1212/01.wnl.0000055470.62265.44. [DOI] [PubMed] [Google Scholar]
- 24.Marini V., Ferrera L., Pigatto F., Origone P., Garre C., Dorcaratto A., Viale G., Alberti F., Mareni C. Search for loss of heterozygosity and mutation analysis of KRIT1 gene in CCM patients. Am. J. Med. Genet. A. 2004;130A:98–101. doi: 10.1002/ajmg.a.30122. [DOI] [PubMed] [Google Scholar]
- 25.Kehrer-Sawatzki H., Wilda M., Braun V.M., Richter H.P., Hameister H. Mutation and expression analysis of the KRIT1 gene associated with cerebral cavernous malformations (CCM1) Acta. Neuropathol. (Berl) 2002;104:231–240. doi: 10.1007/s00401-002-0552-6. [DOI] [PubMed] [Google Scholar]
- 26.Plummer N.W., Squire T.L., Srinivasan S., Huang E., Zawistowski J.S., Matsunami H., Hale L.P., Marchuk D.A. Neuronal expression of the Ccm2 gene in a new mouse model of cerebral cavernous malformations. Mamm. Genome. 2006;17:119–128. doi: 10.1007/s00335-005-0098-8. [DOI] [PubMed] [Google Scholar]
- 27.Whitehead K.J., Plummer N.W., Adams J.A., Marchuk D.A., Li D.Y. Ccm1 is required for arterial morphogenesis: implications for the etiology of human cavernous malformations. Development. 2004;131:1437–1448. doi: 10.1242/dev.01036. [DOI] [PubMed] [Google Scholar]
- 28.Plummer N.W., Gallione C.J., Srinivasan S., Zawistowski J.S., Louis D.N., Marchuk D.A. Loss of p53 sensitizes mice with a mutation in Ccm1 (KRIT1) to development of cerebral vascular malformations. Am. J. Pathol. 2004;165:1509–1518. doi: 10.1016/S0002-9440(10)63409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shenkar R., Venkatasubaramanian P.N., Wyriwicz A.M., Zhao J., Shi C., Akers A., Marchuk D.A., Awad I.A. Advanced magnetic resonance imaging of cerebral cavernous malformations: II imaging of lesions in murine models. Neurosurgery. 2008;63(4):790–797. doi: 10.1227/01.NEU.0000315862.24920.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gault J., Shenkar R., Recksiek P., Awad I.A. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke. 2005;36:872–874. doi: 10.1161/01.STR.0000157586.20479.fd. [DOI] [PubMed] [Google Scholar]
- 31.Liquori C.L., Berg M.J., Squitieri F., Leedom T.P., Ptacek L., Johnson E.W., Marchuk D.A. Deletions in CCM2 are a common cause of cerebral cavernous malformations. Am. J. Hum. Genet. 2007;80:69–75. doi: 10.1086/510439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liquori C.L., Penco S., Gault J., Leedom T.P., Tassi L., Esposito T., Awad I.A., Frati L., Johnson E.W., Squitieri F., et al. Different spectra of genomic deletions within the CCM genes between Italian and American CCM patient cohorts. Neurogenetics. 2008;9:25–31. doi: 10.1007/s10048-007-0109-x. [DOI] [PubMed] [Google Scholar]
- 33.Devilee P., Cleton-Jansen A.M., Cornelisse C.J. Ever since Knudson. Trends Genet. 2001;17:569–573. doi: 10.1016/s0168-9525(01)02416-7. [DOI] [PubMed] [Google Scholar]
- 34.Qian F., Watnick T.J., Onuchic L.F., Germino G.G. The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell. 1996;87:979–987. doi: 10.1016/s0092-8674(00)81793-6. [DOI] [PubMed] [Google Scholar]
- 35.Koptides M., Mean R., Demetriou K., Pierides A., Deltas C.C. Genetic evidence for a trans-heterozygous model for cystogenesis in autosomal dominant polycystic kidney disease. Hum. Mol. Genet. 2000;9:447–452. doi: 10.1093/hmg/9.3.447. [DOI] [PubMed] [Google Scholar]
- 36.Boon L.M., Mulliken J.B., Vikkula M., Watkins H., Seidman J., Olsen B.R., Warman M.L. Assignment of a locus for dominantly inherited venous malformations to chromosome 9p. Hum. Mol. Genet. 1994;3:1583–1587. doi: 10.1093/hmg/3.9.1583. [DOI] [PubMed] [Google Scholar]
- 37.Brouillard P., Boon L.M., Mulliken J.B., Enjolras O., Ghassibe M., Warman M.L., Tan O.T., Olsen B.R., Vikkula M. Mutations in a novel factor, glomulin, are responsible for glomuvenous malformations (‘glomangiomas’) Am. J. Hum. Genet. 2002;70:866–874. doi: 10.1086/339492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puck J.M., Straus S.E. Somatic mutations—not just for cancer anymore. N. Engl. J. Med. 2004;351:1388–1390. doi: 10.1056/NEJMp048116. [DOI] [PubMed] [Google Scholar]
- 39.Gunel M., Laurans M.S., Shin D., DiLuna M.L., Voorhees J., Choate K., Nelson-Williams C., Lifton R.P. KRIT1, a gene mutated in cerebral cavernous malformation, encodes a microtubule-associated protein. Proc. Natl Acad. Sci. USA. 2002;99:10677–10682. doi: 10.1073/pnas.122354499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guzeloglu-Kayisli O., Amankulor N.M., Voorhees J., Luleci G., Lifton R.P., Gunel M. KRIT1/cerebral cavernous malformation 1 protein localizes to vascular endothelium, astrocytes, and pyramidal cells of the adult human cerebral cortex. Neurosurgery. 2004;54:943–949. doi: 10.1227/01.neu.0000114512.59624.a5. discussion 949. [DOI] [PubMed] [Google Scholar]
- 41.Guzeloglu-Kayisli O., Kayisli U.A., Amankulor N.M., Voorhees J.R., Gokce O., DiLuna M.L., Laurans M.S., Luleci G., Gunel M. Krev1 interaction trapped-1/cerebral cavernous malformation-1 protein expression during early angiogenesis. J. Neurosurg. 2004;100:481–487. doi: 10.3171/ped.2004.100.5.0481. [DOI] [PubMed] [Google Scholar]
- 42.Seker A., Pricola K.L., Guclu B., Ozturk A.K., Louvi A., Gunel M. CCM2 expression parallels that of CCM1. Stroke. 2005;37:518–523. doi: 10.1161/01.STR.0000198835.49387.25. [DOI] [PubMed] [Google Scholar]
- 43.Denier C., Gasc J.M., Chapon F., Domenga V., Lescoat C., Joutel A., Tournier-Lasserve E. Krit1/cerebral cavernous malformation 1 mRNA is preferentially expressed in neurons and epithelial cells in embryo and adult. Mech. Dev. 2002;117:363–367. doi: 10.1016/s0925-4773(02)00209-5. [DOI] [PubMed] [Google Scholar]
- 44.Petit N., Blecon A., Denier C., Tournier-Lasserve E. Patterns of expression of the three cerebral cavernous malformation (CCM) genes during embryonic and postnatal brain development. Gene Expr. Patterns. 2006;6:495–503. doi: 10.1016/j.modgep.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Mably J.D., Chuang L.P., Serluca F.C., Mohideen M.A., Chen J.N., Fishman M.C. santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133:3139–3146. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- 46.Hogan B.M., Bussmann J., Wolburg H., Schulte-Merker S. ccm1 cell autonomously regulates endothelial cellular morphogenesis and vascular tubulogenesis in zebrafish. Hum. Mol. Genet. 2008;17:2424–2432. doi: 10.1093/hmg/ddn142. [DOI] [PubMed] [Google Scholar]
- 47.Uranishi R., Baev N.I., Ng P.Y., Kim J.H., Awad I.A. Expression of endothelial cell angiogenesis receptors in human cerebrovascular malformations. Neurosurgery. 2001;48:359–367. doi: 10.1097/00006123-200102000-00024. discussion 367–368. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y., Tan Y.Z., Zhou L.F., Wang H.J., Mao Y. Morphological observation and in vitro angiogenesis assay of endothelial cells isolated from human cerebral cavernous malformations. Stroke. 2007;38:1313–1319. doi: 10.1161/01.STR.0000259914.21997.89. [DOI] [PubMed] [Google Scholar]
- 49.Sure U., Butz N., Schlegel J., Siegel A.M., Wakat J.P., Mennel H.D., Bien S., Bertalanffy H. Endothelial proliferation, neoangiogenesis, and potential de novo generation of cerebrovascular malformations. J. Neurosurg. 2001;94:972–977. doi: 10.3171/jns.2001.94.6.0972. [DOI] [PubMed] [Google Scholar]
- 50.Wong J.H., Awad I.A., Kim J.H. Ultrastructural pathological features of cerebrovascular malformations: a preliminary report. Neurosurgery. 2000;46:1454–1459. doi: 10.1097/00006123-200006000-00027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.