Abstract
The discovery and mapping of cis-regulatory elements is important for understanding regulation of gene transcription in mosquito vectors of human diseases. Genome sequence data are available for 3 species, Aedes aegypti, Anopheles gambiae, and Culex quinquefasciatus (Diptera: Culicidae), representing 2 subfamilies (Culicinae and Anophelinae) that are estimated to have diverged 145 to 200 million years ago. Comparative genomics tools were used to screen genomic DNA fragments located in the 5′-end flanking regions of orthologous genes. These analyses resulted in the identification of 137 sequences, designated “mosquito motifs,” 7 to 9 nucleotides in length, representing 18 families of putative cis-regulatory elements conserved significantly among the 3 species when compared to the fruit fly, Drosophila melanogaster. Forty-one of the motifs were implicated previously in experiments as sites for binding transcription factors or functioning in the regulation of mosquito gene expression. Further analyses revealed associations between specific motifs and expression profiles, particularly in those genes that show increased or decreased mRNA abundance in females following a blood meal, and those accumulating transcription products exclusively or preferentially in the midgut, fat bodies, or ovaries. These results validate the methodology and support a relationship between the discovered motifs and the conservation of hematophagy in mosquitoes.
Keywords: gene expression, hematophagy, Aedes, Anopheles, Culex
Many mosquito species are vectors of pathogens that cause widespread human diseases. This medically significant role makes these insects the center of research, the aim of which is to find ways to reduce the burden these diseases impose (1). The genomes of 3 species, Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus, were sequenced (2, 3, http://www.vectorbase.org), and the information acquired has furthered the knowledge of many aspects of their biology. For example, genome-wide studies focusing on mosquito immunity (4), olfaction (5), and insecticide resistance (6), have led to proposals for innovative alternatives for vector population management and control of disease transmission.
The feasibility of using genetics-based technologies to control transmission of vector-borne diseases, either by limiting the size of vector populations (population reduction), or altering the populations so that they do not transmit pathogens (population replacement), is a major research focus (1, 7, 8). Further knowledge of the mechanisms involved in regulation of gene expression in vector species is necessary for development of these technologies. Promoter and other cis-acting regulatory DNA fragments are needed to regulate restricted expression of selected antimosquito or antipathogen effector molecules. The possibility of designing synthetic promoters comprising well-defined cis-regulatory elements (CREs) to drive robust and tissue-specific transgene expression stimulated active research in both biotechnology and gene therapy (9), and would be beneficial for mosquito-based disease-control strategies. The availability of the 3 mosquito genomes allows comprehensive exploration of CREs for these purposes.
The search for mosquito CREs is complicated by several features of the species and their corresponding genomes. In addition to relatively long divergence times (Fig. 1), An. gambiae, Ae. aegypti and Cx. quinquefasciatus have noticeably distinct genome sizes [278, 1310, and 575 million base-pairs in length, respectively (http://www.vectorbase.org)] caused in part by variations in amounts of repetitive elements, especially near the 5′- and 3′-end untranslated regions of genes (3). The long divergence times and variability make difficult CRE discovery and analyses that require regional sequence alignments. An algorithm, motif discovery using orthologous sequences (MDOS) was developed that does not require anchoring of orthologous sequences (10). MDOS assigns a conservation z-score, which is a statistical measure of how often a specific, short-DNA sequence (7–9 nucleotides in our study) is conserved in the putative control DNA of orthologous genes. We apply it here to one-to-one orthologous genes to discover putative CREs conserved among all 3 mosquito species. We also present evidence of conservation of CREs associated with blood meal-regulated genes among mosquito species of the Culicinae and Anophelinae subfamilies.
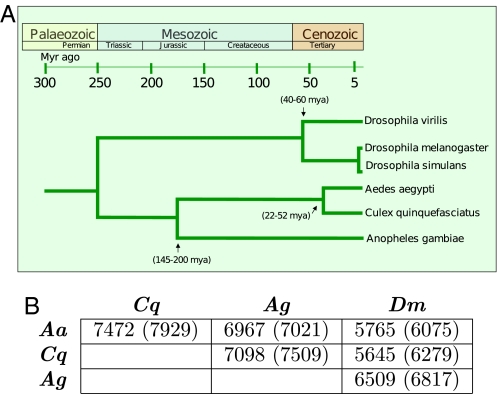
Fig. 1.
Phylogenetic relationships of 6 dipteran species and numbers of one-to-one orthologous gene pairs analyzed. (A) Schematic representation of the deduced evolutionary history of Aedes aegypti, Culex quinquefasciatus, Anopheles gambiae, Drosophila melanogaster, D. simulans, and D. virilis. The nodes and branches depicted in the tree are derived from published data (11, 22, 55). D. melanogaster and D. simulans represent the most divergent species included in the Drosophila 12 Genomes Consortium Study (57). (B) Numbers of pairs of one-to-one orthologous genes between species considered for the discovery of conserved cis-regulatory elements. Numbers in parenthesis give the gene pairs in the datasets before our screening procedures. Abbreviations: Aedes aegypti (Aa), Culex quinquefasciatus (Cq), Anopheles gambiae (Ag), Drosophila melanogaster (Dm).
Anautogeny, the requirement for a blood meal to promote egg development, is conserved in the clades represented by An. gambiae (Anophelinae) and Ae. aegypti/Cx. quinquefasciatus (Culicinae) over divergence times estimated to be 145 to 200 Mya (11). Hematophagy in mosquitoes stimulates a series of events characterized by the induction and repression of specific genes (12–14). The temporal-, tissue-, and sex-specific expression of groups of these genes is hypothesized to be under some form of coordinate regulation (15). Furthermore, it is proposed that this coordinate regulation is achieved by the presence of common CREs in control DNA in analogy to what is observed for hormone-, heat shock-, or immune-modulated genes in insects (16–18). Our findings support the conclusion that the shared life history of hematophagy in mosquitoes is a selective force in the conservation of CREs.
Results
Identification of Conserved Putative CREs in Mosquitoes.
A separate study produced the set of orthologous genes found between any 2 of the 3 mosquito species, An. gambiae, Ae. Aegypti, and Cx. quinquefasciatus, analyzed here (http://www.vectorbase.org/Other/ComparativeAnalyses). We focused on unique orthologous genes between pairs of species (one-to-one orthologues) because selective pressures and drift may result in changes in the sequences of control DNA in paralogous genes (19–21). Gene pairs whose predicted assembly and primary structure were ambiguous or uninformative were removed from this dataset, resulting in 21,537 available combinations (see Fig. 1). Of these, 18,873 (87.6%) were present in all 3 mosquito species.
MDOS analyses identified a total of 1,001 motifs (391 7-mers, 432 8-mers, and 178 9-mers) between species pairs that were conserved significantly (conservation z-scores ≥3) within DNA fragments up to 2 kb in length located at the 5′-end gene boundaries defined by VectorBase [supporting information (SI) Table S1]. More conserved motifs (including reverse complement sequences) were found between Ae. aegypti and Cx. quinquefasciatus (n = 723) than between Cx. quinquefasciatus and An. gambiae (n = 454), or Ae. aegypti and An. gambiae (n = 371). In addition, comparisons between Ae. aegypti and Cx. quinquefasciatus generally produced higher conservation z-scores. These results are not surprising, given the more recent proposed phylogenetic divergence between these 2 species (22). Uninformative “N” designations at the 5′- or 3′-ends were removed in subsequent analyses and displays resulting in some motifs having lengths of 6 nucleotides.
Of the 1,001 motifs, 153 were determined to be conserved significantly (conservation z-scores ≥3) among all 3 mosquito species (Table S1). MDOS comparisons also were made between Drosophila melanogaster and each mosquito species to assess conservation of the discovered motifs in a more evolutionarily distant (≈250 Mya) and nonblood feeding Dipteran. Sixteen of the 153 motifs had conservation z-scores ≥2 among all four Dipterans, and 4 of these (CGATCG, GATCGG, YGATCG, and RCGATCR) were present with z-scores ≥3 in all mosquito-fruit fly combinations. The remaining 137 show no consistently significant conservation within 5′-end flanking regions of D. melanogaster gene orthologues and these were designated “mosquito motifs.” Twenty (14.5%) of the 137 mosquito motifs received conservation z-scores ≥3 in only 1 pairwise D. melanogaster/mosquito comparison, 6 (4.3%) received conservation z-scores ≥3 in two D. melanogaster/mosquito pairwise comparisons, and none received a conservation z-score ≥3 in all 3 D. melanogaster/mosquito comparisons. Mosquito motifs with higher conservation z-scores among mosquitoes were in general those receiving the lowest conservation z-scores in D. melanogaster/mosquito comparisons (Fig. 2). The TTTGACAG motif and variations are associated with the highest conservation z-scores in mosquitoes (Aa/Ag = 9.7, Aa/Cq = 11.4, Cq/Ag = 12.1), and have consistently negative conservation z-scores for D. melanogaster/mosquito comparisons (Dm/Aa = –2.1, Dm/Ag = –5.3, Dm/Cq = –0.4) (see Table S1).
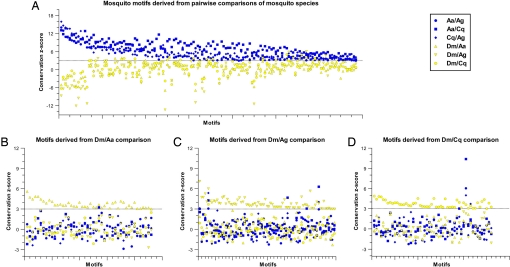
Fig. 2.
Evolutionary conservation of motifs as evaluated using MDOS within 5′ flanking regions of orthologous genes. (A) Conservation z-scores for 137 mosquito enriched motifs derived through species pair-wise comparisons of the 5′ flanking regions of one-to-one orthologous genes shared between Aedes aegypti (Aa), Anopheles gambiae (Ag), and Culex quinquefasciatus (Cq), evaluated in species pair-wise comparisons of orthologous genes of Aa, Ag, Cq and Drosophila melanogaster (Dm). (B–D) Conservation z-scores for the 177 Dmel/mosquito 8-mers derived from species pairwise comparisons of either Dm and (B) Aa, (C) Ag, or (D) Cq and evaluated in species pairwise comparisons of orthologous genes of Aa, Ag, Cq, and Dm. The dotted lines denote a conservation z-score equal to 3.
A reciprocal MDOS analysis was applied to test whether the results of the mosquito analyses were biased by the order in which they were discovered. Drosophila melanogaster and mosquito orthologous genes were screened for conserved 8-mers (Dmel-mosquito 8-mers) using the same criteria applied to the mosquito pair analyses. Despite the discovery of 177 nonredundant Dmel-mosquito 8-mers, none had a conservation z-score ≥3 in all three D. melanogaster/mosquito pairwise comparisons (see Fig. 2 B–D; Table S1). Two motifs (ATCTWAATC and CGATCKT) received conservation z-scores ≥3 in all mosquito/mosquito combinations, and were designated previously as mosquito motifs (see Table S1). One, GTGGAAKT, received a conservation z-score ≥2 in all 3 D. melanogaster/mosquito comparisons and its biological function is currently unknown.
Mosquito Motif Enrichment Within Temporally and Spatially Defined Gene Clusters.
The 137 mosquito motifs were classified into 18 families (a–r) based on sequence similarity (Fig. S1 and Table S1). Although sequence-based clustering defines putative CRE families, each of the 137 mosquito motifs was tested individually in subsequent analyses for association with genes that display similar expression profiles, because different members of a motif family may act as either activators or repressors of gene expression during reproductive development in mosquitoes (23, 24).
Expression data derived from Marinotti et al. (13, 14) on 8,661 An. gambiae genes were screened and used to cluster 4,067 of these according to the time course (TC) of their mRNA abundance profiles following a blood meal (103 TC clusters) or to their exclusive or preferential accumulation in a specific tissue (9 clusters) (Table S2). A total of 624 associations comprising 122 of the 137 mosquito motifs (89%) were found within the 5′-end flanking regions of genes whose mRNA abundance varied significantly (P-value ≤0.01) in response to a blood meal (Fig. S2, Table S3). Notable examples include the association of GATA-containing motifs in the g family with genes up-regulated at 3 h after blood meal (hPBM) (Fig. 3). Motif families a, b, c, and e also showed significant association with genes whose mRNAs increased in abundance following a blood meal. Sixty-four motif-cluster associations identified 35 (26%) of the 137 mosquito motifs as enriched significantly (P-value ≤0.01) within 5′-end flanking regions of tissue-specific/enhanced gene clusters, especially in those expressed within the midgut (23) (17%) (see Fig. 3; Table S3). Of the mosquito motifs identified, 23 are enriched significantly (P-value ≤0.01) in putative regulatory regions of genes induced in the midgut of Ae. aegypti after a blood meal (12) (Fig. S3; Table S4). Nine of the 23 are shared among genes expressed in the midguts of Ae. aegypti and An. gambiae.
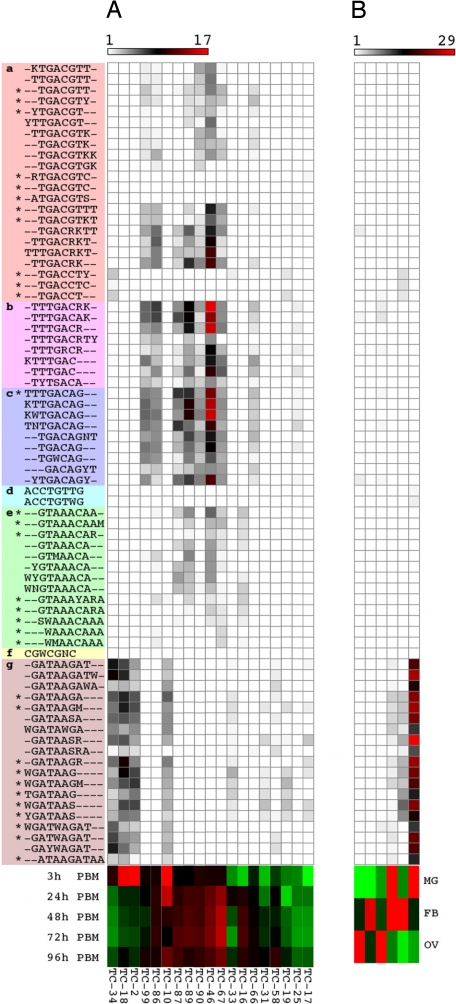
Fig. 3.
Associations of mosquito motifs with gene expression profiles in An. gambiae. Motif enrichment within (A) 5′-end flanking regions of genes in clusters responsive to blood meal ingestion, and in (B) 5′-end flanking regions of genes in clusters enriched in selected tissues. The significance of motif enrichment is indicated by pseudocolor of -log10 (P-value) determined through hypergeometric statistics, and the median expression profile of each gene cluster is shown below each respective column. Red and green colors represent higher and lower relative mRNA accumulation, respectively. Asterisks (*) indicate a match to a previously described mosquito TFBS (Table S5). Heatmaps were created with Matrix2png (58). FB, fat body; hPBM, hours post blood meal; MG, midgut; OV, ovaries; TC, time course clusters (Table S2).
The motif/cluster associations were validated by shuffling the nucleotides within each mosquito motif to produce random permutations followed by expression-cluster enrichment analyses. Permutations that did not maintain nucleotide composition resulted in only 1 motif with associated P-value ≤0.001 (data not shown). Permutations constrained to maintain nucleotide composition resulted in few shuffled motifs enriched (P-value ≤0.001) within the 5′-end flanking regions of genes whose mRNA increased in abundance following a blood meal (11 motifs), or enriched in genes expressed in the midgut, fat body, or ovaries (2 motifs) (Fig. S4). These results support the conclusion that the previously established associations are not determined by nucleotide composition (for example, AT richness).
The sequences of 41 of the 137 conserved mosquito motifs align with transcription factor binding sites (TFBS) identified previously in 8 genes from mosquitoes using experimental approaches, such as electrophoretic mobility shift assays, DNase I footprinting, and deletion/mutational analysis (Table S5). Nine motifs (GATAAGA, GATAAGM, GATAAGR, WGATAAG, WGATAAGM, TGATAAG, WGATAAS, ATAAGATAA, and YGATAAS) align perfectly or with 1 mismatch to GATA-factor binding sites characterized for the promoter of the Ae. aegypti vitellogenin-encoding genes [VgA1, L41842; VgB, AY380797; VgC, AY373377 (25)] or the vitellogenin receptor-encoding gene [VgR, L77800 (26)]. The TGATAAG motif also is found in the putative cis-regulatory regions of vitellogenin genes of Cx. quinquefasciatus, An. gambiae, An. stephensi, and An. albimanus [CPIJ001358, AF281078, DQ442990, AY691327, respectively; present study, (27)]. GATA-binding factors are both positive and negative regulators of vitellogenesis (23, 28), and discernible mRNA accumulation patterns are associated with distinct members of the GATA-motif family. For example, GATAAGAT and WGATWAGAT are enriched in An. gambiae gene clusters whose mRNAs increase in abundance following a blood meal, and WGATAAS is associated with both increases and decreases (see Fig. 3). Three other conserved motifs (TGACCTY, TGACCTC, TGACCT) align to known mosquito TFBS of the ecdysone receptor and ultraspiracle complex (25, 29), and RTGACGTC aligns with a recognition sequence in a gene encoding vitellogenin binding protein (30). These TFBS are associated with the regulation of vitellogenesis (31), and the enrichment of the vitellogenin binding protein and ecdysone receptor and ultraspiracle TFBS within specific gene-expression clusters is consistent with this function (gene clusters induced after a blood meal and those enriched within fat body). Finally, 6 motifs (YGATCKT, TTTGACAG, TATCAGY, YTATCAGY, TWATCAGY, and TTTTATAC) aligned to putative trypsin response elements (PTRE) or coordinating elements located directly 5′ to the PTRE. The PTRE and its associated elements have been implicated in the regulation of early and late trypsin genes in response to the blood meal in anopheline mosquitoes (32).
Discussion
The role CREs play in regulating gene expression during development is well-established (33), and the development of tools for their identification is an active area of research following the publication of genome sequence and associated genome-wide expression datasets. However, the discovery in silico of CREs is challenging because typically they are short, degenerate, and contained within vast amounts of intergenic genomic DNA. Despite these limitations, various computational approaches have been developed for their discovery (34–37). Comparative genomics represents a powerful extension to CRE discovery that diminishes these effects. Functional gene regulatory elements, including CREs, are proposed to diverge at much lower rates compared to neutral sequences because of selective pressures, and therefore may stand out from surrounding neutral DNA by virtue of their greater levels of conservation among orthologous sequences. Previous work has demonstrated the utility of this concept (38–40) and comparative genomics of insects has been applied successfully to map putative CREs in the genomes of relatively closely related Drosophila species, [divergence times estimated ≤50 Mya (39)], in which orthologous intergenic sequences are aligned more easily. The work presented in the present study includes mosquito species with phylogenetic relationships spanning 20 to 200 Mya with genomic comparisons complicated by large amounts of dispersed repetitive elements (3).
The MDOS algorithm (10) was designed to mitigate these problems by not requiring alignment of orthologous sequences, and incorporating features that account for the greater probability for the co-occurrence of a motif because of shared ancestry in orthologous sequences. The application of MDOS for computational discovery of putative CREs shared among mosquitoes resulted in the identification of mosquito-specific or enriched motifs. These motifs demonstrate that MDOS can identify related DNA sequences in diverged mosquitoes and may be useful in similar scenarios in which genome sequences are being analyzed for species that have no other closely related genomes available. Furthermore, the identification in silico of GATA-factor and other TFBS that are identical to those whose function was established experimentally (25, 26, 28–30, 32, 41, 42) serves as a powerful positive control for these analyses. The 5′-end regions of immune-related genes from An. gambiae and D. melanogaster were screened for conserved motifs, and AT-rich domains were found to associate with NFκB response elements (43). These sequences were not identified in our analyses because the restrictions on the dataset we used eliminated many of the immune-related genes where one-to-one orthology may be difficult to establish. In addition, most AT-rich domains discovered in their study were ≤6 nucleotides in length, and our analysis only addressed those recovered with lengths of 7 to 9 nucleotides.
The 3 mosquito species included in this study require a blood meal for successful reproduction. Although this trait is not unique to this group of arthropods, acquiring the necessary nutrients for egg development from ingested blood is a specialized adaptation in insects. There is debate on when and how hematophagy arose in mosquito evolution (44), but it is hypothesized that the occurrence of this trait in the larger clade is monophyletic (45, 46). Most of the discovered mosquito motifs are associated with blood meal-regulated genes. Thus, these data support a common hematophagous ancestor for all mosquitoes, and indicate that hematophagy acts as a selective force for conservation of CREs.
The functionality of some of the discovered motifs was supported further in An. gambiae and Ae. aegypti by their association with genes displaying enriched transcription product accumulation within tissues responsible for blood meal digestion and reproduction. These findings bolster the conclusion that these mosquitoes share a regulatory code controlling expression levels for some genes regulated after hematophagy. The availability of additional genome-wide studies of gene expression for all 3 mosquito species will facilitate discovery of other associations of the motifs with specific gene-regulation patterns.
The present study and similar genome-wide approaches to identify putative CRE in mosquitoes (43) are furthering our understanding of the mechanisms involved in gene regulation in this group of vector insects. An expanded set of putative mosquito CREs will allow the definition of genome-wide motif-association maps and the identification of cis-regulatory modules comprising multiple, linked CREs that convey specific patterns of gene expression. Experimental validation of the functionality of each discovered motif and regulatory module is necessary and will provide support for the development of mosquito synthetic promoters that deliver desired and predetermined expression patterns in transgenic mosquitoes. Promoters that direct expression of transgenes specifically to the germ cells would be useful for the development of gene-drive mechanisms for spreading a desired (pathogen-resistance) trait in a mosquito population (47). Gene-specific knockdown or robust expression of exogenous odorant receptors or odorant binding proteins in the antenna of anthropophilic mosquitoes could redirect their preferences toward other animals (48). Targeted expression of antipathogen effector genes in the midguts, salivary glands, and hemolymph (via fat body-specific control DNA), 3 sites of interaction of most pathogens with their insect vectors, could reduce mean intensities of infection to zero, preventing pathogen transmission and disease (49). The availability of defined synthetic mosquito promoters that direct controlled, local gene expression in response to pathogens also would be a major advance. These promoters will allow engineering of mosquitoes with increased parasite or virus resistance. These and similar envisioned applications for mosquito control and the control of mosquito-borne disease transmission will benefit greatly from a better understanding of gene regulation mechanisms in these insects.
Materials and Methods
Sequence Datasets.
Orthologous gene pairs among Culex quinquefasciatus (genebuild CpiJ1.2), Aedes aegypti (genebuild AaegL1.1), Anopheles gambiae (genebuild AgamP3.4), and Drosophila melanogaster (genebuild BDGP4.3) were determined using the Ensemble Compara pipeline (50). This pipeline is based on maximum likelihood phylogenetic gene trees built from the gene transcripts and representing the evolutionary history of gene families. Duplication or speciation events are differentiated by comparing the gene trees to the species tree. This method is analogous to the reciprocal best-hit approach in the simple case of unique orthologous genes (one-to-one orthologues). The resulting lists of genes are available from Vectorbase at http://www.vectorbase.org/Other/ComparativeAnalyses.
All mosquito gene coordinates were obtained from VectorBase and D. melanogaster data were from Ensembl API (Ae. aegypti: Ensembl 49 genebuild AaeL1.1; An. gambiae: Ensembl 49, genebuild Agam3.4; Cx. quinquefasciatus: VectorBase, genebuild CpiJ1.2; D. melanogaster: Ensemble 49, genebuild BDGP4.3). Repeat-masked Cx. quinquefasciatus sequences were obtained from VectorBase and all other genome sequences were retrieved premasked using the Ensembl perl API (http://www.vectorbase.org/Help/Help:Does_VectorBase_provides_masked_sequences). The one-to-one mosquito orthologous datasets were evaluated further before using in the MDOS analyses. The pronounced intron elongation in 5′- and 3′-end UTRs resulting from the insertion of repetitive elements within these regions (3) and the presence of coding sequence incorrectly included in annotated UTRs were mitigated by only using sequences found within fragments 2 kb in length at the 5′-end of the annotated gene boundaries. Overlaps of these DNA sequences with adjacent genes were determined through use of fjoin (51) and the sequences truncated accordingly. Only sequences with a final size ≥10 base (bp) were analyzed. Pairwise comparisons were conducted with MDOS limits set for the discovery of 7-, 8-, and 9-mers.
Discovery of Evolutionarily Conserved Putative CREs Among Mosquitoes.
Motifs receiving a conservation z-score ≥3 in all 3 mosquito pairwise comparisons were combined into a nonredundant list. To discover motifs with greater exclusivity within the 3 mosquitoes, the conservation z-scores for each motif in 2-kb 5′-end flanking regions of shared D. melanogaster orthologues also were determined. A reciprocal analysis was conducted in which 8-mers conserved in 5′-end flanking regions of one-to-one orthologs of D. melanogaster and each mosquito species also were determined (conservation z-score ≥3), followed by conservation z-scores determination of these motifs in the other 2 mosquito species. This analysis addresses the effect of the order of motif discovery, and whether the discovery process was biased by first discovering conserved motifs in mosquitoes followed by assessment in D. melanogaster or vice versa.
The discovered motifs were grouped by a “Familial Binding Profile” construction through use of the STAMP program (52, 53), using default settings (Metric = PCC, Alignment = SWU, Gap-open = 1,000, Gap-extend = 1,000, nonoverlap-align Multiple Alignment = IR, Tree = UPGMA). Putative identifications of the discovered motifs were determined using STAMP through comparisons to mosquito TFBS reported in the literature, with acceptable matches defined as those with E-values <1 × 10−5 and no more than 1 mismatched nucleotide.
Clustering of Temporal- and Spatially Regulated An. gambiae Genes.
Preexisting microarray data (13, 14) were used to identify groups of genes with specific temporal- or spatial-mRNA accumulation profiles. Alignments of probe sequences to the An. gambiae genome (Ensembl 49) were provided by Nathan Johnson (Ensembl group, EBI). Probe-sets aligning to multiple genes or with ≥2 probes with more than 1 mismatch were not included. One-way ANOVA was performed to identify probe-sets (genes) with significant changes in expression with a conservative false discovery rate of 0.001 (54), followed by k-means clustering with Euclidean distance separation using open-source software (MeV MultiExperiment Viewer v4.1.01, TM4 [55]). The probe sets showing significant dynamic expression patterns following a blood meal were clustered into distinct TC groups. To further refine the expression gene/cluster assignments, probe-sets that align to the same gene were required to have a Pearson's Correlation Coefficient ≥0.9; otherwise, the respective gene was removed from further analysis.
Expression values from 4 samples (whole-body females, midguts, fat body, and ovaries, all processed at 24 hPBM) were analyzed by one-way ANOVA. Probe-sets (genes) from each sample displaying ≥3-fold enrichment over the remaining samples as well as having a P-value ≤0.05 (Tukey honest significant difference) were considered to be enriched within the respective sample.
Determination of Association of Mosquito Motifs Within Expression Clusters.
The 5′-end flanking sequences of genes within each An. gambiae expression cluster were scanned for the occurrence of the mosquito motifs, and their enrichment scored using the hypergeometric distribution. The number of genes containing a particular motif in their 5′-end flanking sequences is designated K, and those occurring within a specific expression cluster, k. If the total number of 5′-end sequences analyzed is N, and the number of genes in that particular cluster is represented as n, all sequences without the motif (“negative set”) would be N – K and those within the sample n – k. The probability of observing by chance at least k matches within the cluster n can be calculated through the equation:
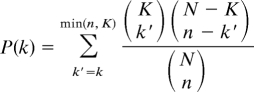 |
Distributions of P-values obtained from mosquito-motif associations with expression clusters were compared with those derived from alternative sequences. To generate alternative sequences, mosquito motifs were shuffled following 2 different procedures: the first used a translation key (A = G; G = T; T = C; C = A) to substitute the nucleotides at each position; the second produced random permutations by shuffling the order of motif constituents, maintaining the nucleotide composition.
Supplementary Material
Acknowledgments.
We thank VectorBase, the Broad Institute, and the J. Craig Venter Institute (http://www.broad.mit.edu/annotation/genome/culex_pipiens.4/Info.html/) for providing access to data before publication, and Mike Sweredoski for assistance in writing programming scripts that allowed for data acquisition and processing. Lynn Olson assisted in preparing the manuscript. This work was supported in part by a postdoctoral biomedical informatics training Fellowship National Instututes of Health (NIH)/National Library of Medicine 5 T15 LM07443 (to D.H.S.), by NIH/National Institutes of Allergy and Infectious Diseases (NIAID) Grant AI29746 (to W.A.D., O.M., and A.A.J.), and by NIH/NIAID Contract HHSN2662004000039C (to K.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813264106/DCSupplemental.
References
- 1.Schneider DS, James AA. Bridging the gaps in vector biology. Workshop on the molecular and population biology of mosquitoes and other disease vectors. EMBO Rep. 2006;7:259–262. doi: 10.1038/sj.embor.7400643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt RA, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 3.Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterhouse RM, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou JJ, He HL, Pickett JA, Field LM. Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect MolBiol. 2008;17:47–63. doi: 10.1111/j.1365-2583.2007.00789.x. [DOI] [PubMed] [Google Scholar]
- 6.Strode C, et al. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem Mol Biol. 2008;38:113–123. doi: 10.1016/j.ibmb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Phuc HK, et al. Late-acting dominant lethal genetic systems and mosquito control. BMC Biol. 2007;5:11. doi: 10.1186/1741-7007-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terenius O, et al. Molecular genetic manipulation of vector mosquitoes. Cell Host & Microbe. 2008;4:417–423. doi: 10.1016/j.chom.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber W, Fussenegger M. Pharmacologic transgene control systems for gene therapy. J Gene Med. 2006;8:535–556. doi: 10.1002/jgm.903. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Sieglaff DH, Gervin J, Xie XS. Discovering regulatory motifs in the Plasmodium genome using comparative genomics. Bioinformatics. 2008;24:1843–1849. doi: 10.1093/bioinformatics/btn348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phylogenet Evol. 2006;39:417–423. doi: 10.1016/j.ympev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Sanders HR, Evans AM, Ross LS, Gill SS. Blood meal induces global changes in midgut gene expression in the disease vector, Aedes aegypti. Insect Biochem Mol Biol. 2003;33:1105–1122. doi: 10.1016/s0965-1748(03)00124-3. [DOI] [PubMed] [Google Scholar]
- 13.Marinotti O, Nguyen QK, Calvo E, James AA, Ribeiro JM. Microarray analysis of genes showing variable expression following a blood meal in Anopheles gambiae. Insect Mol Biol. 2005;14:365–373. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- 14.Marinotti O, et al. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol Biol. 2006;15:1–12. doi: 10.1111/j.1365-2583.2006.00610.x. [DOI] [PubMed] [Google Scholar]
- 15.Dissanayake SN, Marinotti O, Ribeiro JM, James AA. angaGEDUCI: Anopheles gambiae gene expression database with integrated comparative algorithms for identifying conserved DNA motifs in promoter sequences. BMC Genomics. 2006;7:116. doi: 10.1186/1471-2164-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farkas G, Leibovitch BA, Elgin SC. Chromatin organization and transcriptional control of gene expression in Drosophila. Gene. 2000;253:117–136. doi: 10.1016/s0378-1119(00)00240-7. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Jhang Z, Robinson GE, Palli S-R. Identification and characterization of a juvenile hormone response element and its binding proteins. J Biol Chem. 2007;282:37605–37617. doi: 10.1074/jbc.M704595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aggarwal K, Silverman N. Positive and negative regulation of the Drosophila immune response. BMB Rep. 2008;41:267–277. doi: 10.5483/bmbrep.2008.41.4.267. [DOI] [PubMed] [Google Scholar]
- 19.Castillo-Davis CI, Hartl DL, Achaz G. cis-Regulatory and protein evolution in orthologous and duplicate genes. Genome Res. 2004;14:1530–1536. doi: 10.1101/gr.2662504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown CD, Johnson DS, Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science. 2007;317:1557–1560. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- 21.Singh LN, Hannenhalli S. Functional diversification of paralogous transcription factors via divergence in DNA binding site motif and in expression. PLoS ONE. 2008;3:e2345. doi: 10.1371/journal.pone.0002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foley DH, Bryan DH, Yeates D, Saul A. Evolution and systematics of Anopheles: insights from a molecular phylogeny of Australian mosquitoes. Mol Phylogenet Evol. 1998;9:262–275. doi: 10.1006/mpev.1997.0457. [DOI] [PubMed] [Google Scholar]
- 23.Martin D, Piulachs M, Raikhel AS. A novel GATA factor transcriptionally represses yolk protein precursor genes in the mosquito Aedes aegypti via interaction with the CtBP corepressor. Mol Cell Biol. 2001;21:164–174. doi: 10.1128/MCB.21.1.164-174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JH, Attardo GM, Hansen IA, Raikhel AS. GATA factor translation is the final downstream step in the amino acid/target-of-rapamycin-mediated vitellogenin gene expression in the anautogenous mosquito Aedes aegypti. J Biol Chem. 2006;281:11167–11176. doi: 10.1074/jbc.M601517200. [DOI] [PubMed] [Google Scholar]
- 25.Kokoza VA, et al. Transcriptional regulation of the mosquito vitellogenin gene via a blood meal-triggered cascade. Gene. 2001;274:47–65. doi: 10.1016/s0378-1119(01)00602-3. [DOI] [PubMed] [Google Scholar]
- 26.Cho KH, et al. Regulatory region of the vitellogenin receptor gene sufficient for high-level, germ line cell-specific ovarian expression in transgenic Aedes aegypti mosquitoes. Insect Biochem Mol Biol. 2006;36:273–281. doi: 10.1016/j.ibmb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, et al. The Anopheles gambiae vitellogenin gene (VGT2) promoter directs persistent accumulation of a reporter gene product in transgenic Anopheles stephensi following multiple blood meals. Am J Trop Med Hygiene. 2007;76:1118–1124. [PubMed] [Google Scholar]
- 28.Attardo GM, Higgs S, Klinger KA, Vanlandingham DL, Raikhel AS. RNA interference-mediated knockdown of a GATA factor reveals a link to anautogeny in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2003;100:13374–13379. doi: 10.1073/pnas.2235649100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed A, et al. Genomic structure and ecdysone regulation of the prophenoloxidase 1 gene in the malaria vector Anopheles gambiae. Proc Natl Acad Sci USA. 1999;96:14795–14800. doi: 10.1073/pnas.96.26.14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pham DQ, Douglass PL, Chavez CA, Shaffer JJ. Regulation of the ferritin heavy-chain homologue gene in the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2005;14:223–236. doi: 10.1111/j.1365-2583.2004.00550.x. [DOI] [PubMed] [Google Scholar]
- 31.Attardo GM, Hansen IA, Raikhel AS. Nutritional regulation of vitellogenesis in mosquitoes: implications for anautogeny. Insect Biochem Mol Biol. 2005;35:661–675. doi: 10.1016/j.ibmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Giannoni F, et al. Nuclear factors bind to a conserved DNA element that modulates transcription of Anopheles gambiae trypsin genes. J Biol Chem. 2001;276:700–707. doi: 10.1074/jbc.M005540200. [DOI] [PubMed] [Google Scholar]
- 33.Davidson EH. The Regulatory Genome: gene regulatory networks in development and evolution. Netherlands: Academic: Amsterdam; 2006. [Google Scholar]
- 34.Das MK, Dai HK. A survey of DNA motif finding algorithms. BMC Bioinformatics. 2007;7:S21. doi: 10.1186/1471-2105-8-S7-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu J, Li B, Kihara D. Limitations and potentials of current motif discovery algorithms. Nucleic Acids Res. 2005;33:4899–4913. doi: 10.1093/nar/gki791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tompa M, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 37.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 38.Elemento O, Tavazoie S. Fast and systematic genome-wide discovery of conserved regulatory elements using a non-alignment based approach. Genome Biol. 2005;6:R18. doi: 10.1186/gb-2005-6-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dittmer NT, et al. CREB isoform represses yolk protein gene expression in the mosquito fat body. Mol Cell Endocrinol. 2003;210:39–49. doi: 10.1016/j.mce.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Meredith JM, et al. A novel association between clustered NF-kappaB and C/EBP binding sites is required for immune regulation of mosquito Defensin genes. Insect Mol Biol. 2006;15:393–401. doi: 10.1111/j.1365-2583.2006.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez-Romano J, et al. Immunity related genes in dipterans share common enrichment of AT-rich motifs in their 5′ regulatory regions that are potentially involved in nucleosome formation. BMC Genomics. 2008;9:326. doi: 10.1186/1471-2164-9-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rai K-S, Black WC., IV . In: Advances in Genetics. Mosquito Genomes: structure, organization, and evolution. Hall JC, et al., editors. Vol. 41. San Diego: Academic; 1999. pp. 1–33. [DOI] [PubMed] [Google Scholar]
- 45.Borkent A, Grimaldi DA. The earliest fossil mosquito (Diptera: Culicidae), in Mid-Cretaceous Burmese amber. Ann Entomol Soc Am. 2004;97:882–888. [Google Scholar]
- 46.Calvo E, Mans BJ, Anderson JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J Biol Chem. 2006;281:1935–1942. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- 47.Adelman ZN, et al. nanos gene control DNA mediates developmentally regulated transposition in the yellow fever mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2007;104:9970–9975. doi: 10.1073/pnas.0701515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu P, Atkinson R, Jones DN, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 49.Jasinskiene N, et al. Genetic control of malaria parasite transmission: threshold levels for infection in an avian model system. Am J Trop Med Hyg. 2007;76:1072–1078. [PubMed] [Google Scholar]
- 50.Vilella AJ, et al. EnsemblCompara GeneTrees: Analysis of complete, duplication aware phylogenetic trees in vertebrates. Genome Res. 2008 doi: 10.1101/gr.073585.107. 2008 Nov 24. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richardson JE. fjoin: simple and efficient computation of feature overlaps. J Comput Biol. 2006;13:1457–1464. doi: 10.1089/cmb.2006.13.1457. [DOI] [PubMed] [Google Scholar]
- 52.Mahony S, Benos PV. STAMP: a Web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahony S, Auron PE, Benos PV. DNA familial binding profiles made easy: comparison of various motif alignment and clustering strategies. PLoS Comput Biol. 2007;3:e61. doi: 10.1371/journal.pcbi.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 55.Saeed AI, et al. TM4: A free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 56.Porcelli D, Barsanti P, Pesole G, Caggese C. The nuclear OXPHOS genes in insecta: a common evolutionary origin, a common cis-regulatory motif, a common destiny for gene duplicates. BMC Evol Biol. 2007;7:215. doi: 10.1186/1471-2148-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drosophila 12 Genomes Consortium. Clark A-G, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 58.Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.