Abstract
Hematopoietic stem cells (HSCs) remain by far the most well-characterized adult stem cell population both in terms of markers for purification and assays to assess functional potential. However, despite over 40 years of research, working with HSCs in the mouse remains difficult because of the relative abundance (or lack thereof) of these cells in the bone marrow. The frequency of HSCs in bone marrow is about 0.01% of total nucleated cells and approximately 5000 can be isolated from an individual mouse depending on the age, sex and strain of mice as well as purification scheme utilized. This prohibits the study of processes in HSCs which require large amounts of starting material. Adding to the challenge is the continual reporting of new markers for HSC purification, which makes it difficult for the uninitiated in the field to know which purification strategies yield the highest proportion of long-term, multi-lineage HSCs. This report will review different hematopoietic stem and progenitor purification strategies and compare flow cytometry profiles for HSC sorting and analysis on different instruments. We will also discuss methods for rapid flow cytometric analysis of peripheral blood cell types, and novel strategies for working with rare cell populations such as HSCs in the analysis of cell cycle status by BrdU, Ki-67 and Pyronin Y staining. The purpose of this review is to provide insight into some of the recent experimental and technical advances in mouse hematopoietic stem cell biology.
INTRODUCTION
Hematopoietic stem cells have tremendous therapeutic potential and have been harnessed in the clinic for more than 40 years in the context of bone marrow transplantation. Multipotent long-term HSCs (LT-HSCs) reside in the bone marrow and through a process of asymmetric cell division, can self-renew to sustain the stem cell pool or differentiate into short-term HSCs (ST-HSCs) or lineage-restricted progenitors that undergo extensive proliferation and differentiation to produce terminally differentiated, functional hematopoietic cells. ST-HSCs or multipotent progenitors (MPPs) are only able to sustain hematopoiesis in the short term, while the LT-HSCs must persist for the lifespan of the organism to perpetually replenish the hematopoietic system. HSCs can be isolated from bone marrow or peripheral blood using enrichment (magnetic cell separation - MACS) and / or single-cell sorting (fluorescence-activated cell sorting - FACS) based on cell surface markers and / or vital dye staining. The HSC has served as the paradigm for adult stem cell populations by virtue of a well-defined differentiation cascade with distinct intermediaries connecting the differentiation of LT-HSCs into mature, functional hematopoietic cells. Each of the cell stages of HSC differentiation can be purified from the bone marrow or peripheral blood using characteristic cell surface markers which has greatly facilitated the study of hematopoietic biology and revealed important signaling molecules and molecular pathways crucial to HSC function. In this review, we will discuss a range of methods for characterizing HSCs, progenitors, and mature hematopoietic cells which can then be applied to the analysis of mutant mice or non-steady state conditions.
RESULTS
Hematopoietic Stem Cell Purification Schemes
The study of hematopoietic stem cells has been greatly facilitated in the last 20 years by advances in flow cytometric technology and monoclonal antibody availability. The identification and purification of HSCs relies on the unique cell surface molecule expression found on these cells compared to the remainder of bone marrow cells including closely related hematopoietic progenitor cell counterparts. Although there does not appear to be any single marker that segregates HSCs from other hematopoietic cell types, HSCs can be readily identified using multi-parameter flow cytometry. However several different antibody combination schemes have been developed by different laboratories to achieve this, although all have the ultimate goal of producing the highest yield of long-term, multi-lineage reconstituting HSCs. Almost all HSC purification strategies revolve around the cell surface phenotype of positive selection for the markers c-Kit and Sca-1 and negative selection for markers of mature hematopoietic cell lineages (typically B220, CD4, CD8, Gr-1, Mac-1 and Ter-119). Although this c-Kit+Lin-Sca-1+(KLS) phenotype greatly enriches for hematopoietic reconstituting activity, this bone marrow compartment contains progenitor cells in addition to long-term HSCs. In fact only approximately 10% of KLS cells are bona fide long-term HSCs, and as such the KLS compartment should be regarded as merely enriched for HSCs. A variety of strategies have been used to further enrich bone marrow for HSCs, with or without the KLS as a foundation. These strategies include identification of HSCs as KLS-CD34-Flk-2- [2], KLS-CD150+CD48- cells [3], the Hoechst-effluxing side population (SP) [1], and associated variations on that theme (e.g. CD45midLin-HoechstlowRhodaminelow [4] or SP-EPCR [5]). Here, we will review and compare the major strategies, and also show the corresponding methods to purify the various short-term HSC and committed progenitor populations. A summary of cell surface phenotypes and the hematopoietic cell types they enrich for is presented in Table 1.
Table 1.
Cell surface phenotypes of various hematopoietic stem and progenitor cell populations.
| Marker Phenotype | Cell Type | Reference |
|---|---|---|
| KLS | Hematopoietic stem and progenitor cells | [19] |
| SPKLS | Long-term HSCs (LT-HSC) | [1] |
| Flk-2-CD34-KLS | Long-term HSCs | [2] |
| CD150+CD48-CD41-KLS | Long-term HSCs | [3] |
| CD45midLin-RhodaminelowSP | Long-term HSCs | [4] |
| Flk-2+CD34+KLS | Short-term HSC (ST-HSC) and multipotent progenitors (MPP) | [20] |
| Lin-I17rα+c-Kit+Sca-1+ | Common lymphoid progenitors (CLP) | [15] |
| Lin-I17rα-c-Kit+Sca-1- | Myeloid progenitors | [16] |
| Lin-I17rα-c-Kit+Sca-1-CD34+CD16/32- | Common myeloid progenitor (CMP) | [16] |
| Lin-I17rα-c-Kit+Sca-1-CD34-CD16/32- | Megakaryocyte-erythrocyte (MEP) | [16] |
| Lin-I17rα-c-Kit+Sca-1-CD34+CD16/32+ | Granulocyte-macrophage progenitors (GMP) | [16] |
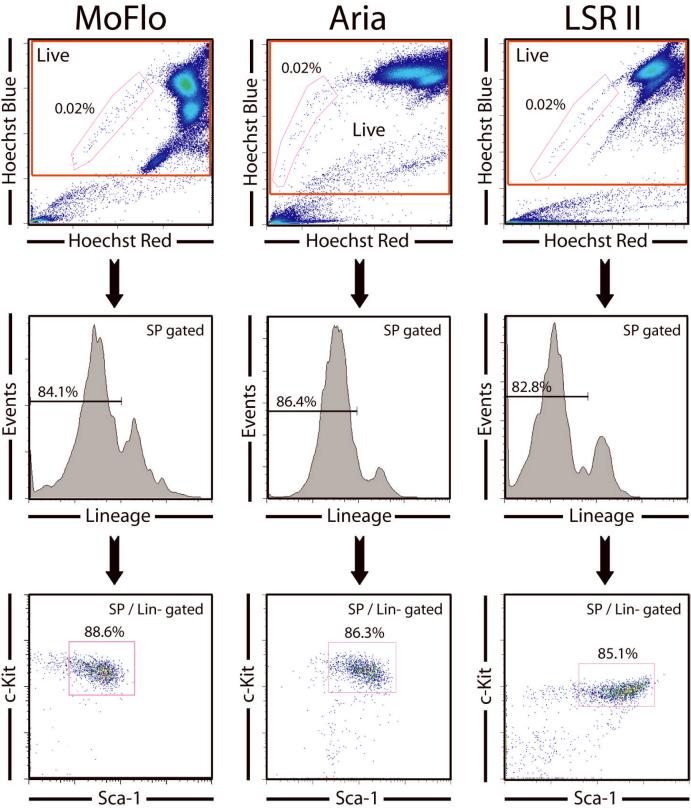
Our laboratory typically uses Hoechst 33342 staining to identify HSCs, the so called side population, or SP. The Hoechst dye is retained at low levels in HSCs due to their ability to efflux the dye via membrane transport pumps which are highly active in these cells compared to other bone marrow cell types. The distinctive staining pattern of HSCs, easily observed when Hoechst fluorescence is displayed at two different wavelengths, results in their presence at the side of the Hoechst fluorescence profile, hence the “SP”. Several studies have shown that even without other markers such as KLS, the SP is remarkably enriched for HSC activity, and most of the SP cells bear other surface markers of HSC, such as KLS [6]. In addition, virtually all of the long-term HSC activity is contained within this SP fraction [1]. However, one drawback of SP staining is that it is highly sensitive to slight modifications in preparation techniques, so KLS antibody staining is often used to complement SP staining to ensure a highly pure HSC population is obtained by the phenotype of SP+c-Kit+Lineage-Sca-1+ which we termed SPKLS (or “SParKLS”). When the Hoechst staining is performed correctly on completely unfractionated mouse bone marrow, the SP fraction should represent between 0.01-0.03% of whole bone marrow and approximately 85% of SP cells should be positive for c-Kit and Sca-1 and negative for lineage markers. These SPKLS cells are additionally homogeneously CD34-/low, Flk-2-, and CD48- [6, 7]. We standardly use SPKLS to purify HSCs, and have performed it on multiple instrument setups for cell sorting and analysis including a MoFlo (Dako), Aria (BD) and LSRII (BD). While it is possible to observe an SP with a violet laser, we obtain the best results using UV lasers. A comparison of staining profiles on different machines is presented in Figure 1; particular attention should be paid to the Hoechst profile when setting up this template on any new instrument (details of instrument parameters can be found in Table 2). The important point is that the frequency of SP cells as a proportion of a gate excluding dead cells and red blood cells should always remain within 0.01-0.03%, and ~85% of SP should be KLS.
Figure 1.
Comparison of SPKLS profiles on different flow cytometers, demonstrating how key parameters used for hematopoietic stem cell identification and isolation by flow cytometry appears visually on different machines. No FSC / SSC gate is needed as dead cells and erythrocytes are excluded by the SP gate. Hoechst Red and Hoechst Blue parameters are examined first, and the voltages adjusted to place the majority of the cells in the upper right quadrant, allowing the SP cells to be central to the plot. The Hoechst red parameter also reveals propidium iodide staining, so dead cells can be excluded on this plot (they line up against the right side), as well as red blood cells (no Hoechst stain, so lower left corner), by drawing a rectangular gate. The SP can then be identified, as gated. In normal mouse bone marrow, an appropriately stained and gated SP will comprise around 0.01 to 0.3% of the live cell gate. The SP cells are then displayed for their lineage-marker profile in a histogram (the lineages markers being a cocktail of lineage-specific antibodies). The lineage-negative cells (usually around 85% of the SP) are then gated to a Sca-1 versus c-Kit plot with the double-positive population here taken to finally identify HSCs with the phenotype SP+ / Lineage- / Sca-1+ / c-Kit+ or SPKLS. If the SP has only a much lower percentage of Lineage-c-Kit+Sca-1+ cells than shown here, the Hoechst staining is likely to be poor.
Table 2.
Comparison of instrument details for analyzing and sorting Hoechst 33342 side population with multiple parameters on different instruments. Other lasers and configurations are possible - these are the ones we routinely use.
| BD LSR II | ||||
|---|---|---|---|---|
| Laser | Detector | Dichroic | Bandpass | Fluorochrome |
| Coherent Sapphire Blue(488) 20mW |
A | 735 | 780/60 | Pe-Cy7 |
| B | 685 | 695/40 | PI | |
| C | 635 | 660/20 | ||
| D | 595 | 610/20 | ||
| E | 550 | 575/26 | PE | |
| F | 505 | 530/30 | FITC | |
| G | blank | 488/10 | SSC | |
| H | blank | blank | ||
|
| ||||
| JDS Uniphase 1344 Red(633) 17mW |
A | 755 | 780/60 | APC-Alexa750 Alexa700 APC |
| B | 710 | 730/45 | ||
| C | blank | 660/20 | ||
|
| ||||
| Coherent Vioflame Violet(405) 60mW |
A | 635 | 660/20 | |
| B | 545 | 560/20 | ||
| C | 505 | 525/20 | ||
| D | blank | 450/50 | Pacific Blue | |
| E | blank | blank | ||
| F | blank | blank | ||
| G | blank | blank | ||
| H | blank | blank | ||
|
| ||||
| Lightwave Xcite UV(355) 20mW |
A | 635 | 652EFLP | Hoechst Red |
| B | blank | 450/50 | Hoechst Blue | |
| C | blank | 405/20 | ||
| BD Aria | ||||
|---|---|---|---|---|
| Laser | Detector | Dichroic | Bandpass | Fluorochrome |
| Coherent Sapphire Blue(488) 100mW |
A | 755 | 780/60 | Pe-Cy7 |
| B | 685 | 695/40 | PI | |
| C | 655 | 660/20 | ||
| D | 600 | 610/20 | ||
| E | 550 | 575/26 | PE | |
| F | 505 | 530/30 | FITC | |
| G | blank | 488/10 | SSC | |
| H | blank | blank | ||
|
| ||||
| JDS Uniphase 1344 Red(633) 17mW |
A | 755 | 780/60 | APC-Alexa750 |
| B | 710 | 730/45 | Alexa700 | |
| C | blank | 660/20 | APC | |
|
| ||||
| Lightwave Xcite UV(355) 20mW |
A | 635 | 652EFLP | Hoechst Red |
| B | blank | 450/50 | Hoechst Blue | |
| C | blank | 405/20 | ||
| Dako MoFlo | ||||
|---|---|---|---|---|
| Laser | Detector | Dichroic | Bandpass | Fluorochrome |
| Argon Blue(488) 100mW |
FL1 | 605SP | ||
| 555LP | ||||
| 505SP | 530/40 | FITC | ||
| FL2 | 605SP | |||
| 555LP | 580/30 | PE | ||
| FL4 | 605SP | |||
| 718SP | ||||
| 650SP | 670/30 | Pe-Cy5 | ||
| FL5 | 605SP | |||
| 718SP | ||||
| 740LP | Pe-Cy7 | |||
|
| ||||
| HeNe Red(633) 60mW |
FL7 | 650LP | 670/40 | APC |
|
| ||||
| Spectra Physics UV(350) 150mW |
FL8 | 440LP | 405/30 | Hoechst Blue |
| FL9 | 440LP | 630/40 | Hoechst Red | |
When establishing the SP method, inclusion of a Verapamil (50 μM) control can be used to ensure the correct population is being identified [1]. Verapamil is a drug that blocks the activity of the membrane pumps that efflux the Hoechst dye and the SP fraction is lost when this drug is included in the Hoechst staining and washing buffers. This control is useful for the uninitiated to feel confident they are identifying true SP cells, but we do not find it necessary once the method has been routinely established. Another problem for relative novices of the SP method is deciding where to draw the SP gate, particularly how far towards the top of the SP can you go and still find true HSCs. We tend to use a conservative SP gate, while attempting to maximize cell number yield and minimize contamination from non-HSCs. A good internal quality control for SP gate positioning is the KLS staining. If the SP gate contains >25% lineage+ cells or a high proportion of Sca-1- / c-Kit - cells, this is indicative that the SP gate is too generous and the gate should be restricted more.
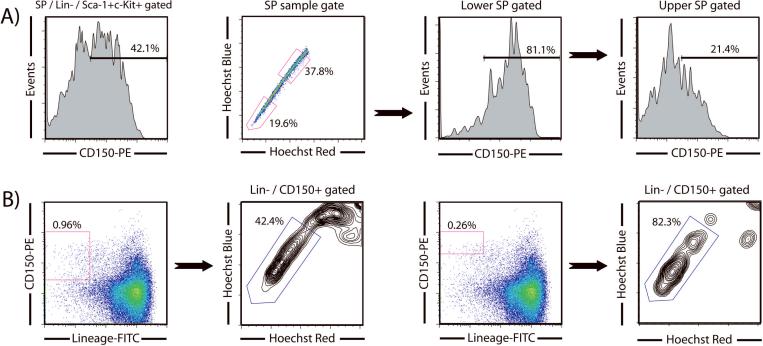
As the molecular characteristics of HSCs have become better elucidated, particularly by various global gene expression profiling studies, a continually growing arsenal of markers is becoming available to stem cell biologists to purify their populations of interest. But as each new HSC marker is published, each laboratory should validate that it works in their hands before proceeding with large scale experiments or abandoning more traditional isolation strategies. A recently proposed staining scheme based on the signaling lymphocytic activation molecule (SLAM) family of cell surface glycoproteins used a profile of CD150+CD244-CD48- to show that approximately 50% of single cells with this phenotype could reconstitute lethally irradiated mice and could be used to identify HSCs in histological sections by immunostaining [8]. We have extensively compared the SLAM purification strategy with the SPKLS method [7]. We found that when using SLAM in combination with SPKLS staining, the SP was surprisingly heterogeneous for CD150 staining, despite the nearly uniform staining of virtually every other marker we have tried. CD150+ cells are more prevalent in cells towards the tip of the SP, whereas CD150- cells were more prevalent towards the top of the SP; this appears to be the first marker that subdivides the SP and may identify different functional sub-sets of HSCs (Figure 2). We also showed that both the CD150+ and CD150- subsets of SPKLS had substantial functional HSC activity, as defined by long-term multilineage engraftment [7], although the CD150- cells have somewhat diminished potential, relative to the CD150+ subset. We have previously shown that cells from the bottom of the SP show better long-term engraftment than cells from the top of the SP [9], and so this distribution of HSC activity relative to CD150 staining is consistent with the observation that there appears to be a gradient of HSC activity along the length of the SP, perhaps representing a continuum of HSCs with slightly different functional properties.
Figure 2.
CD150 as a hematopoietic stem cell marker and overlap with SPKLS staining. (A) Gating of cells SPKLS to a CD150 plot shows that approximately half of all SP cells express CD150. Heterogeneous expression of CD150 is seen in the SP with more CD150+ cells appearing lower in the tail of the SP. This can be further demonstrated by fractionating the SP into lower and upper SP showing that approximately 80% and 40% of these cells respectively are CD150+. (B) Backgating to determine where CD150+ cells fall in the SP. If a more generous CD150 gate is used, only approximately 45% of Lineage-CD150+ cells are SP cells, but if a more stringent CD150 gate is applied, this proportion increases to over 80%.
We also examined how well the CD150 (SLAM) strategy alone identified SP-associated HSCs [7]. By backgating CD150+Lin- cells to a Hoechst plot, the proportion of these cells that are HSCs can be determined by assessing the proportion that falls into the SP gate. Using a gate defined by isotype controls, approximately 45% of CD150+Lin- cells are SP cells. However, if a more stringent CD150 gate is applied and only the brightest cells are selected, then the frequency of HSC increases to almost 83%. This indicates that the positioning of the CD150+ gate is critical when purifying HSCs. We suggest only the CD150high cells should be sorted as HSCs when using the SLAM system, or other canonical markers (such as SP or KLS) must be used in conjunction with SLAM as has been employed by the original authors in subsequent studies [3].
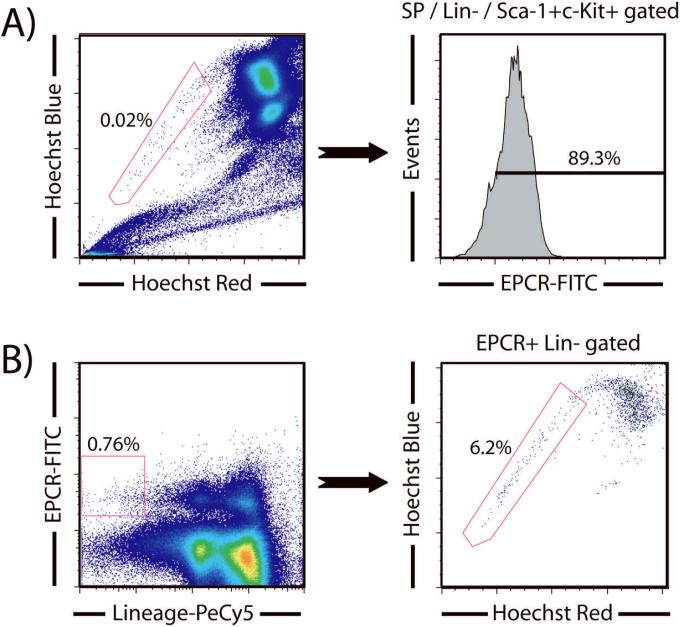
Another recent study has proposed that endothelial protein C receptor (EPCR; CD201) explicitly identifies HSCs from bone marrow [5]. Using SPKLS gating, we show that approximately 90% of these cells are also EPCR+ (Figure 3), providing excellent evidence that EPCR marks HSCs. Unlike CD150, we did not observe any heterogeneous expression of EPCR throughout the SP, it was more uniformly expressed. Using a similar backgating analysis to determine if EPCR alone marks HSCs, backgating EPCR+Lin- cells to Hoechst shows that only ~6% of these cells fall in the SP gate. Applying a more stringent gate to take only the brightest EPCR cells marginally increases the HSC frequency to ~8% (data not shown). This suggests that the intensity of EPCR expression is not critical when sorting for HSCs as is the case with CD150, and also that EPCR by itself is not a particularly useful HSC marker. It is also possible that different fluorochrome conjugates for this antibody, or combinations with different markers, may improve its utility.
Figure 3.
EPCR as a hematopoietic stem cell marker and overlap with SPKLS staining. (A) Co-staining shows that almost all SPKLS cells are also EPCR+. (B) Backgating to determine where EPCR+ cells fall in the SP. The intensity of EPCR staining does not effect SP distribution. If a generous Lineage-EPCR+ gate is used, ~6% of these cells are SP cells, but if a more stringent EPCR gate is applied, the proportion of SP cells only increases mildly to approximately 8% (data not shown).
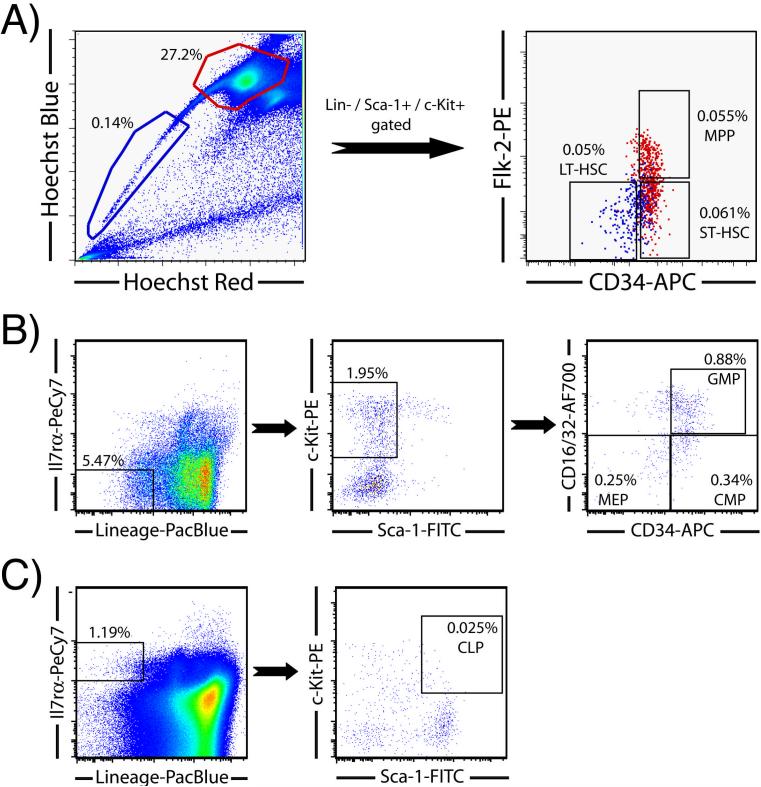
As mentioned previously, the markers CD34 and Flk-2 are commonly used to separate the KSL compartment into long-term HSCs (KLS-CD34-Flk-2-), short-term HSCs (KLS-CD34+Flk-2-) and multipotent progenitor (MPP) cells (KLS-CD34+Flk-2+). The distribution of these antigens on hematopoietic cell populations can also be demonstrated by labeling of Hoechst-stained bone marrow with these antibodies (Figure 4). Gating to a CD34 / Flk-2 plot shows that the SPKLS cells are negative for both these markers, but the non-SPKLS population contains both short-term HSCs (CD34+Flk-2-) and multipotent progenitors (CD34+Flk-2+). Thus, the Hoechst-stained population can be used to simultaneously isolate long-term and short-term HSCs as well as MPPs.
Figure 4.
Separation of hematopoietic progenitor populations by flow cytometry. (A) LT-HSC, ST-HSC, and MPP gating scheme. SP cells are gated to KLS as described for Figure 1, and shown here displayed in red on a CD34 / Flk-2 plot; the SPKLS are negative for both of these markers (the Hoechst-stained cells here have been previously magnetically enriched for Sca-1 to increase the overall proportion to 0.14%). The non-SP population, also shown gated on the Hoechst plot, is also taken through a KLS selection (not shown), then displayed for CD34 and Flk2. The KLS-Flk2+CD34+ cells are multi-potential progenitors (MPP), and the Flk2-CD34+ cells are considered short-term (ST) HSC. Thus, all three of these populations can be readily sorted from one sample. (B) Gating scheme for the common myeloid progenitor (CMP), megakaryocyte-erythrocyte progenitors (MEPs) and granulocyte-macrophage progenitors (GMPs). (C) Gating scheme for the common lymphoid progenitor (CLP).
In our extensive testing of multiple HSC purification protocols from other labs, we have found substantial overlap in the populations resulting from the strategies. Each method has its advantages and drawbacks, however, with regard to staining requirements, or cytometer set up in terms of available lasers and fluorochromes. It is our belief that the more popular HSC purification strategies we compared here, when used optimally in practiced hands, results in very similar purified HSC populations, which should thus be largely comparable functionally and molecularly. However, all of them are also subject to potential problems incurred due to poor staining conditions (e.g. a dim marker paired with a dim fluorochrome on an non-optimally set-up cytometer), resulting in populations with low purity and activity. Thus, we encourage investigators to perfect the HSC purification strategy they wish to follow, and to benchmark it against other published reports using both surface marker staining and functional tests.
Differences Between Mouse and Human HSC Purification
The mouse is the most widely used animal model for studying mammalian disease and many molecules show a high degree of conservation between the two species. While many aspects of HSC biology are shared between mouse and man, the purification strategies used differ slightly for experimental isolation of mouse HSCs and purification of human HSCs for therapeutic applications. While the mouse bone marrow SP represents remarkable enrichment for HSC activity, no long-term reconstitution has been reported from the human cord blood or adult bone marrow SP [10]. Another difference is that human HSCs are typically isolated by the cell surface marker phenotype CD34+CD38- [11], whereas mouse long-term HSCs have been conclusively shown to express only low levels of CD34 [9]. Despite these differences, clinical hematology has been greatly facilitated by advances in flow cytometry. The distributed nature of the hematopoietic system makes it readily amenable to flow cytometric analysis and many surface proteins and glycoproteins on stem cells, erythrocytes, leukocytes, and platelets have been studied in great detail [12,13]. Moreover, many leukemias can be identified by abnormal CD45 expression and side scatter properties with co-expression of other molecules used to discriminate the phenotype, for example CD34+CD13+ for acute myeloid leukemias [14].
Hematopoietic Progenitor Cell Analysis
As mature hematopoietic cells are lost in the peripheral blood, they must be replaced. While they are ultimately replaced from the HSC, they are more directly generated from committed progenitors, and homeostasis affects the number of these progenitors, as well as the balance of HSCs that are quiescent vs. differentiating. Thus, when investigating the effects of a mutation or a treatment on the hematopoietic system, it is useful to examine the impact on the progenitors, as well as the HSC and the differentiated progeny, which may reveal fine differences between the long-term self-renewing HSC and their immediate (non-self-renewing) progeny. This initial differentiation step of HSCs that gives rise to these committed progenitors is of particular interest as the regulatory events that are involved in these early stages of HSC commitment are key to homeostasis.
The analysis of short-term HSCs and hematopoietic progenitor cells has been pioneered by the Weissman lab and detailed phenotypes exist to identify common lymphoid progenitors (CLPs) and common myeloid progenitors (CMPs) as well as the next successive progenitors of each population comprised of pro-T cells and pro-B cells, and megakaryocyte-erythrocyte progenitors (MEPs) and granulocyte-macrophage progenitors (GMPs) respectively [15,16]. These various hematopoietic progenitor cell compartments of bone marrow can be identified by flow cytometry (Figure 4). Common lymphoid progenitors can be identified in whole bone marrow using the combination of KLS with Il7rα+; these typically occur at a frequency of approximately 0.02% in mouse bone marrow. The myeloid progenitor population can be identified in the Sca-negative portion of the KSL stain, and are also Il7rα-. This fraction can be further subdivided into CMP (CD34+CD16/32-), MEP (CD34-CD16/32-) and GMP (CD34+CD16/32+). Although these detailed analysis schemes are well documented, careful attention must be applied in instrument setup when using a large number of parameters, particularly compensating for spectral overlap when fluorochromes with close emission wavelengths are used.
Rapid Methods for Peripheral Blood Lineage Analysis
HSC function is generally monitored in terms of functional contribution to blood cell generation. This typically involves transplanting test cells into recipient animals in which their hematopoietic system has been ablated by irradiation. The activity of the transplanted test cells is observed by taking peripheral blood samples from the recipients at various timepoints after the transplant. The level of chimerism and test cell contribution is typically discriminated by having the test and recipient cells carrying distinct markers; frequently different alleles of the CD45 antigen (test cells are typically isolated from CD45.2 mice, as this is the standard C57Bl/6 allele, while recipient mice are normally CD45.1, the congenic strain) which can be readily distinguished using commercially available monocloncal antibodies. The function of test HSCs is described in terms of overall level of contribution to the recipient's peripheral blood (engraftment) and the types of hematopoietic cells generated from the test HSCs (lineage analysis). Typical timepoints for analysis are 4-weeks and 16-weeks after transplantation. After 4-weeks, the peripheral blood cells formed from input test cells can be the progeny of short-term HSCs or long-lived progenitors. However, the only test cells that can self-renew for 16-weeks post-transplant are long-term HSCs; thus, the peripheral blood components generated from test cells after this time period are the progeny of these cells. Therefore, long-term HSCs are typically defined as those than can give rise to all the major hematopoietic lineages at least 4 months after transplantation. In some mutants, there may be classically-defined stem cell activity, but a deficiency in the ability to generate a particular hematopoietic lineage, and thus it is also important to track lineage contribution of the donor cells. We typically use antibodies to track three major peripheral blood cell types in transplanted mice - myeloid cells (Gr-1+, Mac-1+), B cells (B220+) and T cells (CD4+, CD8+). By comparing the distribution of cell types formed from transplanted test cells to wild-type controls, we can determine if the test cells have functional bias for making particular lineages.
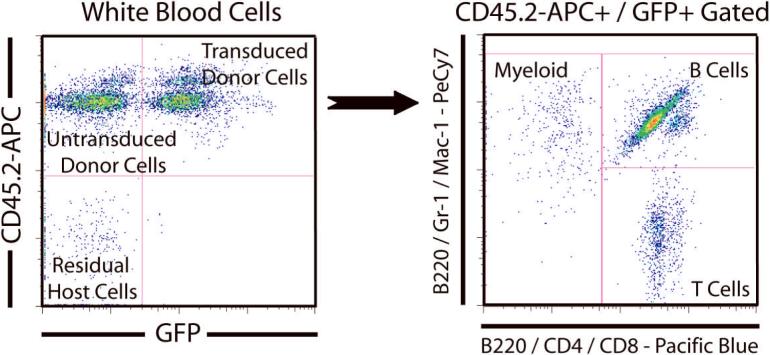
With the development of more fluorochromes for flow cytometry, it is now possible to analyze engraftment of the test cell population and multiple peripheral blood lineages with a single tube. We use a dual labeling strategy to simplify the staining scheme and show the distribution of major peripheral blood lineages (myeloid, B cells, T cells) on a single dot-plot. An example of an experiment in which recipient mice (CD45.1) were transplanted with donor HSCs (CD45.2) that had been transduced with a GFP-tagged retrovirus is presented in Figure 5. At 16-weeks after the transplant, peripheral blood was collected, red blood cells were lysed and remaining nucleated cells were stained for flow cytometric analysis with the following antibodies - CD45.2-APC, Gr-1-PeCy7, Mac-1-PeCy7, B220-PeCy7, B220-Pacific Blue, CD4-Pacific Blue and CD8-Pacific Blue. In the analysis, white blood cells were gated for viability based on propidium iodide (PI) staining and displayed on a FITC (GFP) versus APC (CD45.2) dot-plot. Untransduced donor cells appear in the CD45.2+GFP- quadrant, and transduced donor cells appear in the CD45.2+GFP+ quadrant. Some cells appear in the CD45.2-GFP- quadrant which represents residual recipient cells that survived irradiation or un-lysed red blood cells (which do not express CD45 at all and should be gated out on FSC/SSC). The CD45.2+GFP+ population can then be gated to a PeCy7 versus Pacific Blue dot-plot to analyze lineage distribution of transduced donor cells. With our dual labeling strategy, B cells are stained with both B220-PeCy7 and B220-Pacific Blue, which then allows us to observe the three major lineages on a two-color dot-plot. Myeloid cells (Gr-1+, Mac-1+) appear as PeCy7+Pacific Blue-, T cells (CD4+, CD8+) appear as PeCy7-Pacific Blue+ while the B cells (B220+) are the double positive population. This strategy saves time by reducing the number of tubes required to be processed for each sample and the amount of flow cytometric analysis required. The lineage distribution of the transduced (GFP+) donor cells can then be compared to control virus or wild-type donor cells to ascertain if the gene harbored by the retrovirus has any effect on lineage differentiation of HSCs. We anticipate this dual labeling strategy could be readily adapted to analyze different tissue samples where multiple cell types are required to be analyzed simultaneously and in which a single antigen identifying a specific cell type can be stained with the same antibody carrying different fluorochromes.
Figure 5.
Rapid flow cytometric analysis of peripheral blood of mice transplanted with HSCs transduced with MSCV-GFP retrovirus. For this analysis, red blood cells are depleted or lysed, and the remainder gated out on a FSC/SSC plot. Then viable white blood cells are gated and displayed on a CD45.2-APC versus GFP dot-plot. Progeny of donor HSC (CD45.2) can be discriminated from recipient cells (CD45.1) by CD45 alleles, and the donor HSCs that were successfully transduced to over-express a test gene are GFP+. The CD45.2+ / GFP+ population can then be gated to a PeCy7 versus Pacific Blue dot-plot to analyze distribution of the blood lineages. Myeloid cells are labeled with Gr1 and Mac-1 conjugated to PeCy7. T-cells are labeled with CD4 and CD8 antibodies conjugated to Pacific Blue (Pac-Blue). By labeling B cells with both B220-PeCy7 and B220-Pac-Blue, all major hematopoietic lineages can be displayed simultaneously on the same plot. The B cells are the double positive population, the myeloid cells (Gr-1+, Mac-1+) are the PeCy7+PacBlue- population, while the T cells (CD4+, CD8+) are the PeCy7-PacBlue+ population. If we were analyzing transplanted mice in which the GFP was not used to follow retroviral marking, we typically use CD45.1-FITC to track the recipient cells in addition to the donor cells.
Cell Cycle Status and Proliferation Assays for Purified HSCs
HSCs are a primarily quiescent population, with around 1-3% in cycle when isolated HSC are examined using PI staining [1]. It has become clear that this property is important for maintenance of stem cell activity, as stem cells that are unable to return to quiescence, isolated from mutant mice, exhibit defective repopulating capacity [17]. Thus, when studying HSCs, it is helpful to examine their proliferation status. Standard methods are applicable, such as BrdU labeling, Ki-67 staining and Pyronin Y, and each is useful for addressing slightly different questions. Ki-67 protein is present during all active phases of the cell cycle (G1, S, G2/M), but is absent from resting cells (G0). Ki-67 is an excellent marker to determine the growth fraction of a given cell population at any given point in time. Pyronin Y is an RNA stain which has been used to differentiate the different cell cycle states of various populations, including HSCs [18], and is most useful for distinguishing cells in G1 and G0, i.e. those that are in real quiescence. These two measures can show the cell cycle status of a given cell population at any given point in time, like a proliferation snap-shot. The recent proliferative history of a cell population can be assayed using the synthetic nucleoside BrdU (bromodeoxyuridine). BrdU can be incorporated into the newly synthesized DNA of dividing cells (during the S phase of the cell cycle), by substituting for thymidine residues during DNA replication, so BrdU can reveal what proportion of stem cells have entered or completed cell-cycle over the entire labeling period, which can range from minutes / hours (generally in vitro labeling) to several days (usually in vivo); the amount of BrdU incorporation reflects the proliferation history of a cell population over that period.
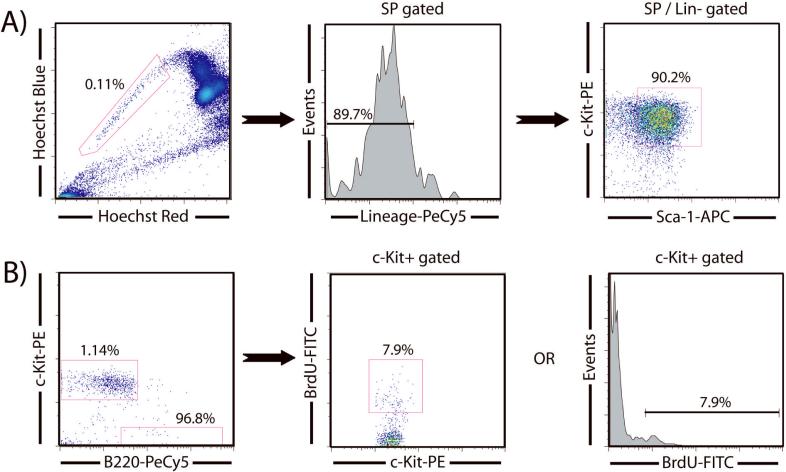
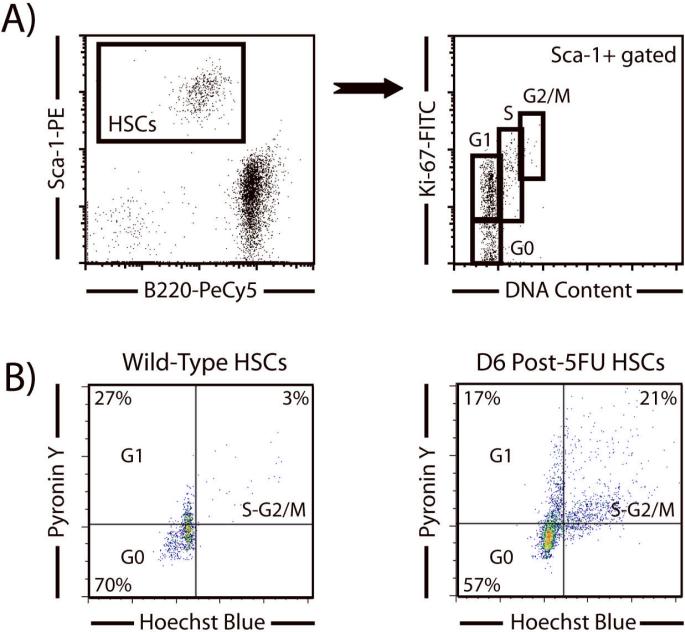
The analysis of cell cycle kinetics in HSCs is challenging because of the limiting numbers of cells available per mouse. In addition, because SP sorting using the dye efflux phenotype requires cells to be viable, any analysis of intracellular properties cannot be performed simultaneously. As BrdU and Ki-67 staining requires fixation and permeabilization, SPKLS cells must be purified first and then subsequently reanalyzed for these cell cycle assays. Due to small cell numbers associated with HSC sorting, and the cells lost in the subsequent reanalysis processing, we have developed a carrier cell method to alleviate cell loss. With this technique, a given number of carrier cells are added to the purified SPKLS cells before the fix and permeabilization procedure to minimize cell loss when dealing with low cell numbers. Notably, one must choose an appropriate fluorochrome to label carrier cells distinctly (typically B cells from spleen). To distinguish HSCs from the carrier cells, one can choose a fluorochrome that HSCs are lacking (for example the flurochrome designated for the lineage markers when sorting SPKLS since this is sorted against) or can be a color that has been spared during the HSC sorting for later intracellular analysis. In the reanalysis of intracellular staining, the cells are then displayed under a two dimensional dot-plot with the carrier flurochrome (positive for carriers, negative for HSCs) and the Sca-1 or c-Kit fluorochrome (negative for carriers, positive for HSCs) subsequently allowing one to distinguish HSCs clearly from the carrier population (Figure 6). A typical strategy for this would be to pre-sort B220-PeCy5+ cells from spleen into a collection tube, then sort SP+ / Sca-1-APC+ / c-Kit-PE+ / Lineage-PeCy5- HSCs into the same tube. The cells are then fixed overnight (or this can be done the same day, but due to the long preparation time for HSC it is convenient to leave these overnight) and the next day permeabilized and stained for anti-BrdU-FITC and reanalyzed. This carrier cell technique has been adapted to also analyze intracellular staining of Ki-67, and Pyronin Y to reveal the proliferation properties of HSCs (Figure 7). It is also worth mentioning that the carrier cells, if chosen wisely, can serve as an internal control for the intracellular staining experiment.
Figure 6.
Analysis of HSCs turnover by BrdU labeling using flow cytometry. (A) HSCs from mice injected with BrdU are purified from Sca-1-enriched bone marrow (increasing the proportion of SP cells 10-fold) and then fixed and permeabilized overnight. (B) Reanalysis of the sorted cells following intracellular staining for BrdU. A PE versus PeCy5 dot-plot allows for discrimination between sorted HSCs and carrier B cells (the majority of carrier cells are lined up against the x-axis). The HSCs can then be gated to either a stem cells marker versus BrdU dot-plot or to a histogram to determine BrdU incorporation.
Figure 7.
Cell cycle reanalysis of purified SPKLS cells by Ki-67 and Pyronin Y. (A) Using the carrier cell technique, SPKLS cells were purified then fixed and permeabilized for Ki-67 staining. On the reanalysis, HSCs (Sca-1+B220-) were easily identified from carrier cells (Sca-1-B220+) and gated to show a distribution of Ki-67 versus DNA content using propidium iodide. This plot allows discrimination of the various stages of cell cycle of the HSC population. (B) Analysis of HSC cell cycle status by Pyronin Y staining. As above, SPKLS were sorted into carrier cells and then both populations were stained for Pyronin Y analysis. On reanalysis, HSCs are gated away from carrier cells to a Hoechst versus Pyronin Y plot which shows the different stages of cell cycle. The use of this assay is clearly demonstrated when comparing normal HSCs to those which have been stimulated with the chemotherapeutic agent 5-flurouracil (5-FU) which brings them out of quiescence and into cell activation programs. In normal HSCs, the vast majority are resting in the G0 stage, while six days after 5-FU stimulation a much higher proportion are actively engaged in the cell cycle in S/G2-M.
CONCLUSIONS
The hematopoietic system has served as the paradigm for understanding much of adult stem cell biology. In this article we have reviewed some of the common methods for HSC purification by flow cytometry, discussed analysis of hematopoietic progenitors and peripheral blood samples of transplanted mice, and provided novel methods for working with limiting cell numbers when dealing with HSCs. While we have suggested methods for HSC identification and analysis that work well in our laboratory, ultimately each investigator must validate the techniques in their own hands before confidently setting forth on large-scale experimental programs. One caveat to the assays discussed here is that while phenotype can be informative, HSCs are ultimately defined by their functional capacity to repopulate the bone marrow and generate all the major blood lineages in a stem cell-ablated host. The phenotype of stem cells has been well documented to change developmentally as well as when regeneration is stimulated by agents such as 5-flurouracil. Thus, phenotype alone cannot be relied upon to definitively identify stem cells.
METHODS AND MATERIALS
Hematopoietic Stem Cell Identification and Isolation
All animal procedures were conducted in accordance with the Baylor College of Medicine (Houston, Texas, USA) institutional guidelines. Whole bone marrow was isolated from femurs and tibias of mice and SP cell staining was performed with the vital dye Hoechst 33342 (Sigma-Aldrich, St Louis, MO) as previously described [1]. Briefly, whole bone marrow was resuspended in staining media at 106 cells/mL and incubated with 5 mg/mL Hoechst 33342 for 90 minutes at 37°C. For Sca-1 enrichment of whole bone marrow prior to sorting, cells were resuspended at 108 cells/mL, stained on ice with anti-mouse Sca-1-biotin (eBioscience, San Diego, CA) for 15 minutes, resuspended at 1.25 × 108 cells/mL, incubated with 200 mL/mL of magnetic antibiotin microbeads (Miltenyi Biotech, Auburn, CA) for 10 minutes at 4°C, rinsed with staining buffer, resuspended at 2 × 108 cells/mL, and magnetically enriched on an AutoMACS instrument (Miltenyi). For antibody staining, cells were suspended at a concentration of 108 cells/mL and incubated on ice for 20 minutes with various combinations of the following antibodies (all 1:100 dilution); PeCy5-conjugated Mac-1, Gr-1, CD4, CD8, B220 and Ter119 (eBioscience); FITC-conjugated Mac-1, Gr-1, CD4, CD8, B220 and Ter119 (BD Pharmingen, Franklin Lakes, NJ); Sca-1-APC (eBioscience) -FITC (BD Pharmingen) - PE (BD Pharmingen) -PeCy7 (eBioscience); c-Kit-APC (eBioscience) -FITC (BD Pharmingen) -PE (BD Pharmingen) -AF750 (eBioscience); CD150-PE (BioLegend, San Diego, CA); EPCR-FITC (StemCell Technologies, Vancouver, BC, Canada). Cell sorting and analysis were performed on a MoFlo cell sorter (Dako North America, Carpinteria, CA) and a FACSAria Cell-Sorting System (BD Biosciences) and additional analysis was accomplished with an LSRII (BD Biosciences).
Hematopoietic Progenitor Staining
Whole bone marrow was isolated and stained on ice with various antibody cocktails to identify each progenitor compartment (all antibodies were obtained from BD Pharmingen and used at a concentration of 1:100 unless otherwise indicated). LT-HSC, ST-HSC and MPP were first stained with the vital dye Hoechst 33342 (Sigma-Aldrich). They were then stained with FITC-conjugated lineage markers (Mac-1, Gr-1, CD4, CD8, B220 and Ter119), Sca-1-PeCy7 (eBioscience), c-Kit-APC-AlexaFluor-750 (eBioscience), Flk-2-PE (at 1:50), and CD34-AlexaFluor-647 (at 1:50; eBioscience) for 20 minutes. CLPs were stained with biotinylated lineage markers (Mac-1, Gr-1, CD4, CD8, B220, CD3 and Ter119), IL7ra-PeCy7 (eBioscience), Sca-1-FITC, and c-Kit-PE for 20 minutes. Cells were then spun down, resuspended, and stained with strepavidin-Pacific Blue (1:50) for 20 minutes. CMP, GMP, and MEP were stained with biotinylated lineage markers (Gr-1, Ter119, CD4, CD8, CD3, B220, CD19), IL7ra-PeCy7 (eBioscience), Sca-1-FITC, c-Kit-PE, CD34-AlexaFluor-647(at 1:50; eBioscience), and CD16/32-AlexaFlour-700 (at 1:50; eBioscience) for 20 minutes. Cells were then spun down, resuspended, and stained with strepavidin-Pacific Blue (1:50) for 20 minutes. Finally, cells were spun down resuspended in a propidium iodide solution, and analysis was accomplished on live cells with an LSRII (Becton Dickinson).
Peripheral Blood Analysis
Blood was collected from mice by retro-orbital bleeding and samples were subject to red blood cell lysis. Samples were incubated with the following antibodies (all 1:100 dilution; eBioscience) on ice for 20 minutes - CD4-Pacific Blue, CD8-Pacific Blue, B220-Pacific Blue, B220-PeCy7, Mac1-PeCy7, and Gr-1-PeCy7 for lineage and CD45.1-FITC plus CD45.2-APC (for competitive transplants) or CD45.2-APC alone (for MSCVGFP over-expression transplants with the FITC channel spared to monitor GFP expression). Cells were then spun down, resuspended in a propidium iodide solution, and analysis was accomplished on live cells with an LSRII (Becton Dickinson).
BrdU Staining
Mice received an initial intraperitoneal injection of BrdU (Sigma-Aldrich; 1 mg/6 g mouse weight) 12 hours prior to sacrifice. Mice were then killed, and SPKLS cells (sparing the FITC channel) were sorted into a previously sorted carrier cell population of 400,000 B220-PeCy5+ splenocytes. Samples were prepared for analysis of BrdU incorporation using the FITC BrdU Flow Kit (BD Pharmingen), and samples were reanalyzed by flow cytometry. On reanalysis, HSCs (c-Kit-PE+ / B220-PeCy5-) were readily distinguishable from carrier cells (c-Kit-PE- / B220-PeCy5+) and were gated for analysis of BrdU incorporation.
Pyronin Y Staining
200,000 B220-FITC+ splenocytes were pre-sorted into collection tubes. HSCs (more than 1000) were then sorted into the same tube using the gating scheme SP+ / Sca-1-APC+ / Lineage-FITC-. The sorted cells were then incubated for 45 minutes with 20 μg/mL Hoechst 33342 and 50 μg/mL Verapamil (Sigma-Aldrich) in phosphate buffered saline supplemented with 3% fetal bovine serum. Pyronin Y (Sigma-Aldrich) was then added at 1 μg/ml, and the cells were incubated for another 15 min at 37°C, washed, and immediately analyzed on a BD LSRII. During flow analysis, both Hoechst 33342 and Pyronin Y signal were displayed under linear mode. The carrier cells, B cells, were then a control to define the G0/G1 DNA content (2N).
ACKNOWLEDGEMENTS
We thank Chris Threeton, Joel Sederstrom, Candice Sellers and Xuejun (Tony) Zhang for help with flow cytometric sorting and analysis, and Megan Tierney for assistance with Pyronin Y staining. G.A.C. was supported by an Australian National Health and Medical Research Council (NHMRC) CJ Martin Fellowship. K.K.L. was partially supported by T32 DK064717 from the Departments of Immunology and Cell And Gene Therapy. This work was also supported by NIH grants HL081007, EB005173, and DK58192, the Ellison foundation (AGSS178706), and the American Heart Association (0740020N).
REFERENCES
- 1.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, Sasaki Y, Jacobsen SE. Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 3.Kiel MJ, Radice GL, Morrison SJ. Lack of evidence that hematopoietic stem cells depend on N-cadherin-mediated adhesion to osteoblasts for their maintenance. Cell Stem Cell. 2007;1:204–217. doi: 10.1016/j.stem.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Dykstra B, Ramunas J, Ken D, McCaffrey L, Szumsky E, Kelly L, Farn K, Blaylock A, Eaves C, Jervis E. High-resolution video monitoring of hematopoietic stem cells cultured in single-cell arrays identifies new features of self-renewal. Proc Natl Acad Sci USA. 2006;103:8185–8190. doi: 10.1073/pnas.0602548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balazs AB, Fabian AJ, Esmon CT, Mulligan RC. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood. 2006;107:2317–2321. doi: 10.1182/blood-2005-06-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camargo FD, Chambers SM, Drew E, McNagny KM, Goodell MA. Hematopoietic stem cells do not engraft with absolute efficiencies. Blood. 2006;107:501–507. doi: 10.1182/blood-2005-02-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weksberg DC, Chambers SM, Boles NC, Goodell MA. CD150- side population cells represent a functionally distinct population of long-term hematopoietic stem cells. Blood. 2008;111:2444–2451. doi: 10.1182/blood-2007-09-115006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Goodell MA, Rosenzweig M, Kim H, Marks DF, DeMaria M, Paradis G, Grupp SA, Sieff CA, Mulligan RC, Johnson RP. Dye efflux studies suggest that hematopoietic stem cells expressing low or undetectable levels of CD34 antigen exist in multiple species. Nat Med. 1997;3:1337–1345. doi: 10.1038/nm1297-1337. [DOI] [PubMed] [Google Scholar]
- 10.Pearce DJ, Bonnet D. The combined use of Hoechst efflux ability and aldehyde dehydrogenase activity to identify murine and human hematopoietic stem cells. Exp Hematol. 2007;35:1437–1446. doi: 10.1016/j.exphem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Novelli EM, Ramirez M, Civin CI. Biology of CD34+CD38- cells in lymphohematopoiesis. Leuk Lymphoma. 1998;31:285–293. doi: 10.3109/10428199809059221. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, Cabana R, Shariatmadar S, Krishan A. Cellular volume and marker expression in human peripheral blood apheresis stem cells. Cytometry A. 2008;73:160–167. doi: 10.1002/cyto.a.20524. [DOI] [PubMed] [Google Scholar]
- 13.Keeney M, Gratama JW, Sutherland DR. Critical role of flow cytometry in evaluating peripheral blood hematopoietic stem cell graphs. Cytometry A. 2004;58:72–75. doi: 10.1002/cyto.a.10103. [DOI] [PubMed] [Google Scholar]
- 14.Brown M, Wittwer C. Flow Cytometry: Principles and Clinical Applications in Hematology. Clin Chem. 2000;46:1221–1229. [PubMed] [Google Scholar]
- 15.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 16.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 17.Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada S, Nakauchi H, Nagayoshi K, Nishikawa S, Miura Y, Suda T. In vivo and in vitro stem cell function of c-kit- and Sca-1-positive murine hematopoietic cells. Blood. 1992;80:3044–3050. [PubMed] [Google Scholar]
- 20.Yang L, Bryder D, Adolfsson J, Nygren J, Mansson R, Sigvardsson M, Jacobsen SE. Identification of Lin(-)Sca1(+)kit(+)CD34(+)Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]