Abstract
A robust redox extraction protocol for quantitative and reproducible metabolite isolation and recovery has been developed for simultaneous measurement of nicotin-amide adenine dinucleotide (NAD) and its reduced form, NADH, from Saccharomyces cerevisiae. Following culture in liquid media, yeast cells were harvested by centrifugation and then lysed under nonoxidizing conditions by bead blasting in ice-cold, nitrogen-saturated 50 mM ammonium acetate. To enable protein denaturation, ice cold nitrogen-saturated CH3CN/50 mM ammonium acetate (3:1 v/v) was added to the cell lysates. Chloroform extractions were performed on supernatants to remove organic solvent. Samples were lyophilized and resuspended in 50 mM ammonium acetate. NAD and NADH were separated by HPLC and quantified using UV–Vis absorbance detection. NAD and NADH levels were evaluated in yeast grown under normal (2% glucose) and calorie restricted (0.5% glucose) conditions. Results demonstrate that it is possible to perform a single preparation to reliably and robustly quantitate both NAD and NADH contents in the same sample. Robustness of the protocol suggests it will be (i) applicable to quantification of these metabolites in other cell cultures; and (ii) amenable to isotope labeling strategies to determine the relative contribution of specific metabolic pathways to total NAD and NADH levels in cell cultures.
Keywords: HPLC, Isotope label, NAD, NADH, Yeast
1 Introduction
Nicotinamide adenine dinucleotides (NADs) are ubiquitous biological molecules that participate in many metabolic reactions. Recent studies indicate that these dinucleotides may play important roles in transcriptional regulation [1, 2], calorie restriction (CR)-mediated lifespan extension [3–5], and age-associated diseases [6–8]. However, current methods for the quantitation of NADs can be complicated by the oxidation of reduced species. Most studies have relied on separate extractions for NAD and reduced nicotinamide adenine dinucleotide (NADH) determinations: a basic extraction for the reduced species and a separate acidic extraction for the oxidized species [3, 9–12]. The extraction conditions are specific for the stabilization of either oxidized compounds, which are more stable in acid, or reduced compounds, which are more stable in base. Metabolites in the separate extracts are then commonly quantitated by enzymatic cycling assays [3, 9–12] that amplify a compound of interest through a series of cycling steps involving coupled reactions with final quantitation achieved flourimetrically.
NAD and NADH measurements in yeast (Saccharomyces cerevisiae) indicate that levels of these metabolites may play important roles in lifespan extension [10]. Yeast has frequently been used as a model organism to study CR where lifespan extension can be achieved by reducing the glucose concentration in the growth media from 2 to 0.5% [3–5]. The benefit of CR appears to require Sir2, a sirtuin family protein that exhibits an NAD-dependent deacetylase activity and whose function, in part, includes regulation of chromatin silencing, genomic stability at the ribosomal DNA loci and lifespan [5]. Sir2 mRNA and protein expression have both been shown to increase when mammalian cells are grown with limited glucose and serum [13]. NADH may serve as a regulator of longevity by inhibiting the activity of Sir2 [10].
NAD is synthesized via two major mechanisms in yeast, the de novo and salvage pathways [1–3, 14–18]. In the de novo pathway, NAD is synthesized from tryptophan present within the cell. In the salvage pathway, NAD can be generated from the recycling of degraded NAD products, such as nicotinamide, and through the incorporation of nicotinic acid from the extracellular environment. Other NAD biosynthesis salvage pathways, the nicotinamide riboside salvage pathway [16, 17] and two nicotinic acid riboside pathways [18], have recently been characterized in yeast. Both the de novo and salvage pathways play redundant yet essential roles in cell growth, but the salvage pathway is thought to play a more important role in lifespan regulation [4, 14, 16]. To elucidate this role, it is important to determine the relative contributions of the salvage and de novo pathways to the total amount of cellular NAD and NADH. While enzymatic cycling assays allow for the quantitation of the total amount of pyridine dinucleotides in a cell, they cannot be used to differentiate between NAD and NADH molecules derived from these pathways. Culturing yeast in media containing isotopically labeled nicotinic acid will specifically label NAD and NADH derived from the salvage pathway which can then be quantified using sensitive isotope measuring techniques [19]. In conjunction with measurements of total cellular NAD and NADH, isotopic measurements should afford estimates of NAD and NADH contents from the de novo pathway.
HPLC is well suited for separating and quantitating metabolites and can be used to isolate isotopically labeled molecules in biological samples [20]. A handful of single sample extraction protocols have been developed using HPLC separation to quantitate NADs within the same sample [21–23]. Some of these techniques rely on acid extraction protocols and HPLC peak integration to measure NAD directly and indirectly measure NADH by its acid degraded products [23]. Another procedure involves a rapid postextraction labeling reaction with cyanide in basic solution that leads to the reaction product NAD–CN for NAD [21]. NADH and the derivatized NAD–CN isomers are then quantified via fluorescence detection. Another method has been developed for direct measurement of underivatized NAD and NADH, along with 37 other small molecules, from tissue samples [22]. This method has been used to measure NAD/NADH ratios of approximately 30, 48, and 154 in liver, brain, and heart tissue, respectively. However, application of this analytical method to yeast cells has yielded NAD/NADH ratios of approximately 20, which are significantly higher than values in yeast cells determined by enzymatic assay, which range from 1.5 to 2.5 NAD/NADH [3, 10]. This raises the possibility that oxidation of NADH to NAD may occur when applying the Lazzarino et al. protocol to yeast.
The contribution of tryptophan, quinolinic acid, nicotinic acid, and nicotinamide to the total cellular NAD has been measured using HPLC/MS [24] and isotope labeling has been used in conjunction with HPLC/MALDI/MS and other MS techniques to measure NAD [25–27]. These techniques rely on measured ratios of compound-specific radiolabeled spikes to unlabeled NAD for quantitation and have been successfully used to measure NAD, but have not been established for NADH quantitation. As with all NADH extraction approaches, these isotope labeling methods will have to account for the potential oxidation of NADH to NAD if they are to be applied for NADH quantitation.
Here, we report the development of a single sample extraction and HPLC processing procedure that enables the isolation and quantitation by UV-absorption of total cellular NAD and NADH redox states from pools of yeast. The protocol avoids derivatization or the measurement of degradation products from NAD or NADH, making this approach amenable to isotope labeling strategies as the potential for loss of an isotope label is minimized. Metabolite recovery and stability, as well as reproducibility of the whole procedure are examined. Applicability of this procedure to quantify NAD and NADH levels in yeast is evaluated by culturing yeast under normal (2% glucose) and calorie restricted (0.5% glucose) conditions.
2 Experimental
2.1 Yeast culture
S. cerevisiae (BY4742) were cultured in 25 mL volumes of liquid synthetic complete media at 30°C consisting of 6.7 g/L Bacto yeast nitrogen base without amino acids (Becton-Dickinson, Franklin Lakes, NJ, USA), 1.92 g/L yeast synthetic drop-out media supplement without uracil (Sigma–Aldrich, St. Louis, MO, USA), 0.08 g/L uracil (Sigma–Aldrich) and 20 g/L glucose (anhydrous dextrose) (EMD Chemicals, Gibbstown, NJ, USA) dissolved in distilled H2O. Media was filter sterilized (0.22 μm GP Express Plus Membrane, Millipore, Billerica, MA, USA) before use as previously described [28]. For CR, 5 g/L instead of 20 g/L glucose was used. Approximately 104 yeast were used to inoculate each culture. Aliquots (100 μL) were periodically taken from culture for growth and cell density measurement via hemocytometer. Cultures were maintained until they contained ~7×106cells/mL corresponding to mid-log phase growth.
2.2 Preparation of yeast pellets for metabolite extraction
Approximately 7×107 yeast were harvested by aliquoting 10 mL of the culture into a 15 mL Falcon tube (Becton-Dickinson) followed by centrifugation for 3 min at 4000 rpm at 4°C in a Sorvall RC 5C Plus (DuPont, Newtown, CT, USA). The supernatant was discarded and the pellet resuspended in 1 mL PBS at 4°C, and transferred to a 2 mL polypropylene bead blasting tube (Outpatient Services, Petaluma, CA, USA). The walls of the Falcon tube were washed with 1 mL ice-cold PBS and the rinsate was added to the bead blasting tube. Samples were centrifuged at 4000 rpm for 3 min in a bench top minicentrifuge (National Labnet, Woodbridge NJ, USA) at 4°C and the supernatant was discarded.
2.3 Single sample metabolite extraction for HPLC speciation
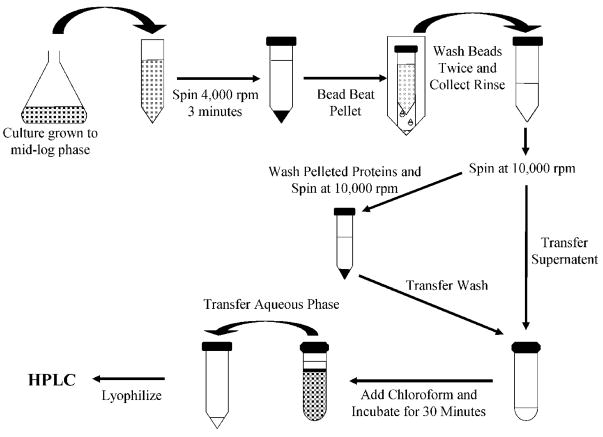
Using the work of Lazzarino et al. [22] as a conceptual basis, an extraction protocol was developed to recover water-soluble metabolites from yeast cells for fractionation by an HPLC (Fig. 1). Ammonium acetate (600 μL of 50 mM; Aldrich, Milwaukee, WI, USA) saturated with N2 gas was added to the pelleted yeast in bead blasting tubes and the tubes filled to the meniscus with 212–300 μm diameter acid-washed glass beads (Sigma–Aldrich). Yeast were bead blasted at maximum speed using a Mini-Bead-beater™ (BioSpec Products, Bartlesville, OK, USA) for 30 s followed by a 2 min incubation on ice and another 30 s of bead blasting. The bead blasting tube was then inverted and its base lanced with a red-hot 0.5 in. long 26 gauge needle (Becton-Dickinson). An open-ended 1.5 mL-microfuge tube (Eppendorf, Westbury NY, USA) was placed over the base of the bead blasting tube and the assembly nested upright in a 50 mL Falcon tube (Becton-Dickinson). The nested tubes were centrifuged at 2000 rpm for 3 min in a Sorvall centrifuge to separate cell lysates from glass beads. Cell lysates were then transferred to a cold 2 mL microfuge tube (Eppendorf) and kept on ice. The glass beads in the bead blasting tube were then washed twice with 600 μL of a N2-saturated solution of ACN (Sigma–Aldrich) and 50 mM ammonium acetate (3:1 v/v), vortexed and centrifuged for 3 min at 2000 rpm at 4°C. After each wash, the rinsate was transferred to the microfuge tube containing the cell lysate.
Figure 1.
Diagrammatic representation of the extraction protocol. Cells are pelleted by centrifugation and then lysed by bead beating. Beads are washed and lysates are collected. The sample is then centrifuged and the supernatant is collected. The remaining protein pellet is washed, centrifuged, and the supernatant is collected. The sample undergoes a chloroform extraction and the aqueous phase is removed, lyophilized, and then separated by HPLC.
Following washing, cell lysates were centrifuged at 10000 rpm for 1 min at 4°C. The supernatant was transferred to a 10 mL glass test tube (Corning, Corning, NY, USA) and stored on ice. The remaining pellet was resuspended in 600 μL of a N2-saturated solution consisting of ACN and 50 mM ammonium acetate (3:1 v/v) and centrifuged at 10 000 rpm for 5 min at 4°C. The resulting supernatant was pooled with the previous supernatant and the remaining pellet was discarded. Lipids were extracted from the liquid volume using 10 mL of ice-cold chloroform (VWR International, West Chester, PA, USA) and 5 s vigorous agitation. Samples were left to separate on ice for 30 min. The upper aqueous phase was collected in a 15 mL Falcon tube and was quickly frozen at −80°C and lyophilized for 15 h in a Christ Alpha 1–2 freeze dryer (Martin Christ, Germany) while the organic phase was discarded. To test whether discarded protein pellets contained NAD or NADH, randomly selected pellets were resuspended in 600 μL of chloroform using vigorous agitation, dried under a stream of nitrogen, resuspended in 600 μL of ACN, and stored in a 4°C refrigerator overnight. The lysate was then vortexed vigorously for 30 s and centrifuged at 10000 rpm for 5 min at 4°C. The supernatant was frozen at −80°C and then lyophilized for at least 15 h.
Lyophilized samples were resuspended in 250 μL ice-cold 50 mM ammonium acetate and vortexed vigorously. The suspension was filtered with a 0.45 μm-polyvinylidene fluoride microspin filter tube (Alltech Associates, Deerfield, IL, USA) using a bench top mini-centrifuge operating at 10000 rpm for 1 min and aliquots analyzed by HPLC.
2.4 HPLC speciation of metabolites
HPLC measurements were performed using a Hydrosphere C18 column (5 μm, 150 mm×4.6 mm id (Waters, Milford, MA, USA)) with a direct connection UNIGUARD guard column (Thermo Electron, Newington, NH, USA) fitted with the appropriate PEEK tip and Hypersil Gold guard cartridge (5 μm, 10 mm×4.0 mm id (Thermo Electron)) on an Agilent 1100 HPLC (Hewlett-Packard Wilmington, DE, USA). The C18 column and guard column were maintained at room temperature (22°C). Up to 100 μL aliquots of metabolites were injected and fractionated by RP chromatography. A 1 mL/min gradient of mobile phase A (50 mM ammonium acetate) and mobile phase B (100% ACN) initially consisting of 100% mobile phase A, with mobile phase B increasing to 5% over 25 min at a rate of 0.2% per minute was utilized. The column was washed after each separation by increasing mobile phase B to 90% for 7 min. UV absorbance was monitored at 260 and 340 nm and pertinent peak areas integrated using area under the curve algorithms. Quantitation of NAD was assessed using absorbance at 260 nm. Since NADH absorbs at 340 nm whereas NAD does not, quantitation of NADH was assessed using absorbance at both 260 and 340 nm.
Peak identification and quantitation of NAD and NADH were assessed using standard solutions of NAD and NADH (Sigma–Aldrich) dissolved in 50 mM ammonium acetate. Standard stock solutions were prepared fresh every week, while working standard solutions were prepared daily by appropriate dilution of the stock solutions. Working standards were always analyzed on the same day as samples of interest to generate standardization curves.
Intracellular metabolite contents were calculated from the measured absorbance, corrected for recovery efficiency and converted to concentrations (mM) using the number of cells in the extraction, the molecular weight of the metabolite of interest, and an assumed volume of 70 μm3 per cell [29].
2.5 MS measurements
Fractions obtained via HPLC analysis of standards or metabolite extracts from yeast were dried in a centrifugal vacuum evaporator (Jouan, Winchester, VA, USA) at ambient temperature and stored at −80°C prior to MS analysis. Samples were resuspended in an 89.9:10:0.1 water/ACN/formic acid (Sigma–Aldrich) solution prior to direct infusion into the MS system by a syringe pump without a column at a flow rate of 10 μL/min. ESI-MS analyses were carried out using a Quattro MicroTM API (Micromass UK, Manchester, UK) mass spectrometer. High-resolution MS (HR/MS) analyses were done in ESI mode on a Micromass Fourier transform MS (Micromass UK). All ESI-MS spectra were obtained in positive ion mode with an electrospray capillary potential of 3.0 kV and a cone potential of 21 V. Standard solutions of NAD and NADH, resuspended in an 89.9:10:0.1 water/ACN/formic acid mixture, were used for NAD and NADH identification in the MS measurements.
2.6 Reproducibility, linearity, and sensitivity provided by the chromatographic method
Reproducibility, linearity, and sensitivity provided by the chromatographic method were studied with solutions containing up to 1 μg/μL NAD or NADH standards with HPLC injection volumes varying between 5 and 100 μL. The quantities of NAD and NADH used in the linearity experiments spanned the working range of metabolite contents obtained from processing pools of yeast. Regression analysis was utilized to determine linear correlation coefficients of standard curves for each standard. LODs were considered to be the quantities that produced a peak whose area was three times that of the square root of the integrated background underneath the peak. Reproducibility of the chromatographic method was evaluated using up to 100 μL injections of standard solutions on the same day (intraday) and on various days (interday) over a three-month period. Fresh standard solutions were prepared weekly for the interday reproducibility measurements.
2.7 Recovery and reproducibility of the extraction protocol
Recovery efficiency of the extraction protocol was determined from cell-free samples containing 2.5 μg of either NAD or NADH standard solutions dissolved in 50 mM ammonium acetate. For standard addition experiments to determine the recovery of standard spikes from cell lysates, yeast cultures were split into two samples prior to bead beating. One sample was processed, as previously described, to determine yeast NAD and NADH contents. Standard solutions containing 2.5 or 10 μg of either NAD or NADH (these amounts span the range of NAD and NADH contents within yeast metabolite extracts) dissolved in 50 mM ammonium acetate were added to bead beaten cell lysates from the other sample for subsequent processing. Total quantities of NADH and NAD in spiked samples were corrected for the contribution of yeast metabolites and recovery of NAD and NADH from the spikes assessed.
2.8 Statistical analysis
Differences in NAD and NADH contents of yeast grown in 20 or 5 g/L glucose were assessed by unpaired two-tailed Student’s t-tests. A significance level of less than 0.02 was considered meaningful. A significance level between 0.02 and 0.10 was considered evidence of a possible trend, while a significance level of greater than 0.10 was considered to indicate no significant difference.
3 Results and discussion
3.1 Reproducibility, linearity, and sensitivity provided by the chromatographic method
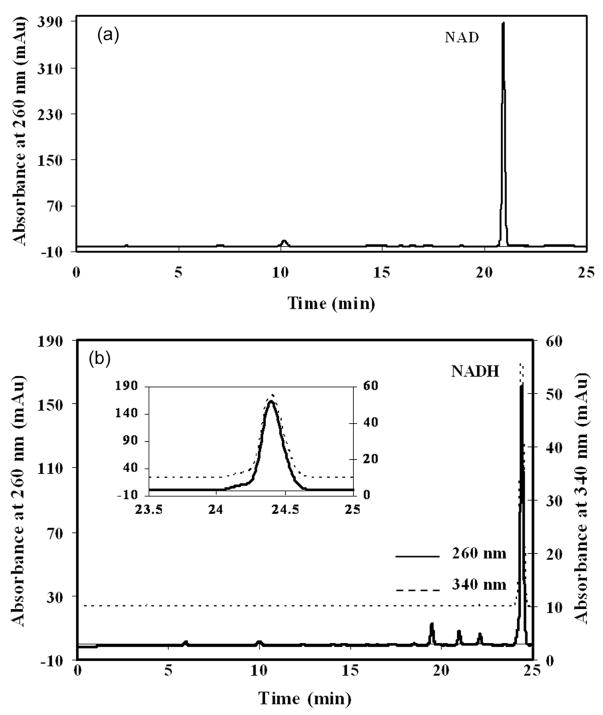
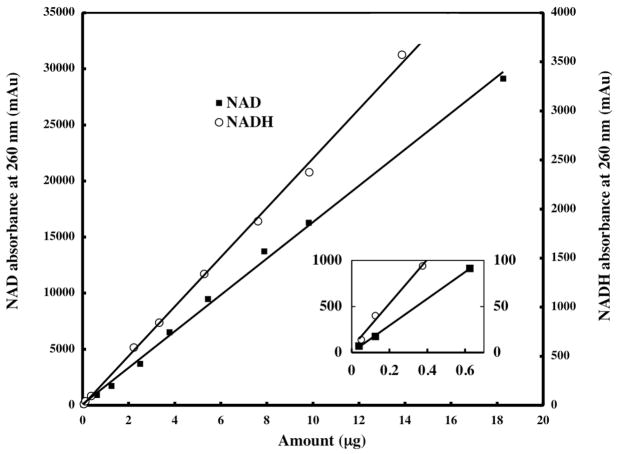
Typical chromatograms of standard solutions containing NAD and NADH are shown in Fig. 2. Intraday variability observed in retention time of each authentic compound was typically less than 2% of the mean retention time. Interday variability observed in retention times of the authentic compounds was typically less than 60 s. Linearity for peak area quantitation at 260 nm absorbance of standard solutions of NAD and NADH is shown in Fig. 3. Linearity for peak area quantitation at 340 nm absorbance of standard solutions of NADH (data not shown) was virtually identical to that observed for NADH in Fig. 3. LODs at 260 nm absorbance were 10 ng for NAD and 25 ng for NADH, while the LOD at 340 nm absorbance was 20 ng for NADH. Linearity and LOD for the chromatographic method were insensitive to the range of injection volumes (5–100 μL) utilized for each standard. The dynamic range of the chromatographic method was linear for 38 ng to 18.3 μg quantities of the authentic NAD standard studied and 68 ng to 13.9 μg quantities of the authentic NADH standard studied. The sensitivity of the chromatographic method for NAD and NADH detection compares favorably with that of other chromatographic methods [21] and suggest that NAD contents from as few as 2×105 yeast cells and NADH contents from as few as 5×105 yeast cells can be quantified. However, such cell numbers are at least as high as those used for enzymatic cycling measurements of NAD and NADH in yeast, which are routinely performed on 105cells [10].
Figure 2.
Typical HPLC chromatograms of 10 μL aliquots of 50 mM ammonium acetate containing (a) 2.50 μg NAD and (b) 2.34 μg NADH (absorbance at 340 nm has been offset vertically by 10 mAu for clarity and an expanded view of the NADH peak is inset). Retention times (tr) of the authentic compounds are: NAD tr = 20.9 min., and NADH tr = 24.4 min.
Figure 3.
Standard curves for the chromatographic quantitation of authentic compounds of NAD and NADH. NAD contents range from 0.038 ng to 18.26 μg. NADH contents range from 0.068 ng to 13.87 μg. An expanded view of the standard curves near the origin is inset. Lines represent linear least squares fits to the data. Coefficients of correlation are 0.9975 for NAD and 0.9984 for NADH.
Table 1 summarizes the intra- and interday precision of the chromatographic method for the detection of NAD and NADH at 260 nm absorbance. The intra- and interday precision of the chromatographic method for the detection of NADH at 340 nm absorbance was virtually identical to that shown for NADH in Table 1 (data not shown). RSDs (SD expressed as a percentage of the corresponding mean value) for detection of the authentic compounds ranged from 1.6 to 2.3% for the intraday reproducibility experiments and 3.3–4.0% for the interday reproducibility experiments. The chromatographic method produces acceptable intra- and interday reproducibility in retention time, speciation, and subsequent quantitation of NAD and NADH. The higher interday variation observed for the quantitation of the authentic compounds plausibly arises as the intraday measurements were performed on the same set of standard solutions while fresh solutions were prepared weekly for the interday measurements. The higher inter- than intraday variation observed for retention time of the authentic compounds plausibly arises as day to day variations in pump and column performance are likely greater than variation within a single day.
Table 1.
Intra- and interday reproducibility for the detection of NAD and NADH in at 260 nm absorbance
| NAD | NADH | |
|---|---|---|
| Amount (μg) | 0.125 | 0.125 |
| Intraday precision (% RSD)a (n = 3) | 2.0 | 2.3 |
| Interday precision (% RSD) (n = 5) | 3.9 | 4.0 |
| Amount (μg) | 0.625 | 0.375 |
| Intraday precision (% RSD) (n = 4) | 1.6 | 1.9 |
| Interday precision (% RSD) (n = 5) | 4.0 | 3.6 |
| Amount (μg) | 5.44 | 3.33 |
| Intraday precision (% RSD) (n = 3) | 1.9 | 2.3 |
| Inter-day precision (% RSD) (n = 5) | 3.4 | 3.3 |
| Amount (μg) | 15.26 | 13.87 |
| Intraday precision (% RSD) (n = 4) | 1.8 | 2.0 |
| Interday precision (% RSD) (n = 5) | 3.5 | 3.5 |
RSD is the SD expressed as a percentage of the corresponding mean value.
3.2 Recovery and reproducibility of the metabolite extraction protocol
Table 2 shows recoveries of NAD and NADH obtained with our extraction and speciation protocol for standard solutions containing 2.5 μg of either NAD or NADH. Differences in measured NADH values via absorbance at 340 and 260 nm in the recovery experiments for standard solutions were typically less than 1%. Recovery of NAD was 95.9 ± 4.9% (mean ± SD) while no detectable conversion or degradation of NAD to other metabolites was observed. Recovery of NADH was 83.4 ± 6.6% and 12.9 ± 6.2% of the NADH oxidized to NAD during sample processing. The replacement of 50 mM ammonium acetate with other buffers such as Tris, PBS, or KH2PO4 at either pH 6.8 or 7.5 yielded lower recovery rates and significantly higher NADH oxidation (data not shown).
Table 2.
Percent recoveries obtained from NAD and NADH standard extractions
| Standard contents | % Recovered (SD)a | % Conversion (SD) | % Total recovered (SD) |
|---|---|---|---|
| NAD (2.5 μg) | 95.9 (4.9) | None detected | 95.9 (4.9) |
| NADH (2.5 μg) | 83.4 (6.6) | 12.9 (6.2) | 96.3 (2.1) |
Values represent the mean and SD of nine separate experiments.
Bead blasting, a standard technique for lysing yeast cell membranes, generates a significant amount of heat and incorporates oxygen into the sample causing oxidation of NADH. Bead blasting for 60 s significantly and inconsistently increased rates of oxidation up to 20% (data not shown). Our studies into the effects of bead blasting on oxidation rates indicate that several brief periods of bead blasting ice-cold samples minimizes oxidation of NADH. Accordingly, yeast were bead blasted for two 30 s periods and left on ice for the intervening period with a single 10 s vortexing during the incubation period to redistribute heated glass beads. Greater consistency in NADH recovery was obtained using this protocol over that from bead blasting for one 60 s period or without ice-incubations and vortexing which generated noticeably warmer samples and higher rates of oxidation.
After lysis, frozen extracts could be kept overnight in a −80°C freezer; however, standard solutions containing NADH frozen for more than 72 h started to display signs of NADH oxidation and so extracts were never stored at −80°C for more than 12 h.
We also found that multiple washings of the glass beads in the bead blasting tubes following bead beating, careful attention to vigorous vortexing and agitation during resuspension or mixing activities, and conservative centrifugation times could improve metabolite recoveries by as much as 20%.
Recovery of NADH was found to be dependent on incubation time in chloroform. Sample incubation on ice during chloroform extraction of less than 25 min resulted in a nearly 60% loss of NADH (data not shown). Amounts of extracted NADH were consistent between 30 and 60 min incubations. Because we observed that lyophilization of standard solutions of NADH in 50 mM ammonium acetate can result in oxidation of NADH to NAD, the longer settling times during lipid extraction possibly allow ammonium acetate in the metabolite extracts to partition into the organic phase.
Table 3 shows recoveries of NAD and NADH obtained from the standard addition experiments performed on yeast cultures grown in 20 g/L glucose after correction for the contribution from yeast to the NAD and NADH contents. Differences in measured NADH values via absorbance at 340 and 260 nm in the standard addition experiments were typically less than 1%. Recoveries from the standard addition experiments are consistent with the metabolite recovery efficiency data shown in Table 2. Recovery of 2.5 μg NAD matrix spikes was 95.6 ± 8.2%, while recovery of 10 μg NAD matrix spikes was 96.5 ± 6.0%. No detectable conversion or degradation of NAD matrix spikes to other metabolites was observed. Recovery of 2.5 μg NADH matrix spikes was 82.6 ± 12.3%, while recovery of 10 μg NADH matrix spikes was 83.2 ± 9.9%; NADH matrix spikes (13.3 ± 11.3%, 2.5 μg) oxidized to NAD during sample processing, while 12.0 ± 8.0% of the 10 μg NADH matrix spikes oxidized to NAD. Standard addition experiments performed on yeast cultures grown in 5 g/L glucose produced similar matrix spike recoveries (data not shown) to those reported in Table 3.
Table 3.
Percent recoveries of authentic standard solutions spiked into yeast cultures
| Spike contents | % Recovereda (SD) | % Conversion (SD) | % Total recovered |
|---|---|---|---|
| NAD (2.5 μg) | 95.6 (8.2) | None detected | 95.6 (8.2) |
| NAD (10 μg) | 96.5 (6.0) | None detected | 96.5 (6.0) |
| NADH (2.5 μg) | 82.6 (12.3) | 13.3 (11.3) | 96.0 (9.2) |
| NADH (10 μg) | 83.2 (9.9) | 12.0 (8.0) | 95.2 (1.9) |
Each value represents the mean and SD of at least three separate experiments performed on different days.
Great care must be exercised at all points in the extraction protocol to prevent artefactual oxidation or degradation of NADH. We observed that oxidation of NADH occurs at a high rate in the presence of Tris, PBS, and KH2PO4. Reliance on KH2PO4 as an extraction buffer could account for the published high NAD/NADH ratios [22]. Precautions were taken at each step in the metabolite extraction protocol to minimize processing time and sample temperature to minimize NADH oxidation while optimizing metabolite recovery. We found that samples must be kept cold during all steps of the extraction. Allowing the sample temperature to supersede room temperature can cause oxidation of NADH to occur by as much as 50%.
3.3 Measurement of NAD and NADH contents in yeast
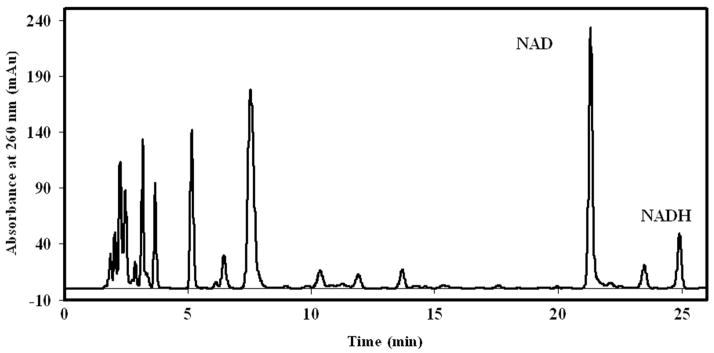
Figure 4 shows a typical HPLC chromatogram of metabolites extracted from approximately 7×107 yeast grown in 20 g/L glucose. HPLC chromatograms of metabolites extracted from yeast grown in 5 g/L glucose were qualitatively similar to that shown in Fig. 4. Fractions of yeast metabolite extracts with retention times corresponding to those of authentic NAD and NADH standards were collected and their identity confirmed with MS. MS data from these metabolite fractions were highly consistent with MS data obtained from corresponding authentic standards of NAD and NADH. Briefly, MS of metabolite fractions with retention times corresponding to that of the authentic NAD standard revealed an m/z of 664.2 ± 0.2 (expected 664.1 for m) and a pattern of fragmentation ions that was similar to that of the authentic standard (data not shown). Likewise, MS of metabolite fractions with retention times corresponding to that of the authentic NADH standard revealed an m/z of 666.1 ± 0.2 (expected 666.1 for m) and a pattern of fragmentation ions of smaller m/z that was similar to that of the authentic standard (data not shown). These data indicate that the NAD and NADH HPLC peaks from yeast extract samples contain only NAD or NADH and are free of co-eluting contaminants.
Figure 4.
Typical HPLC chromatogram of a 75 μL aliquot of yeast metabolite extracts from approximately 7×107 yeast cells grown in 20 g/L glucose. Retention times (tr) of the examined compounds are: NAD tr = 21.3 min and NADH tr = 24.9 min.
Metabolite extracts from yeast cultured in 20 g/L glucose typically contained 4 μg each of NAD and NADH, while those cultured in 5 g/L glucose contained 5 μg of NAD and 2.5 μg NADH. Differences in measured NADH values via absorbance at 340 and 260 nm for the metabolite extracts were typically less than 1%. Quantities of authentic standards of NAD and NADH used in studying the linearity of the chromatographic method correspond to NAD and NADH contents that one could expect to extract from ~2×106to ~2×108yeast cells.
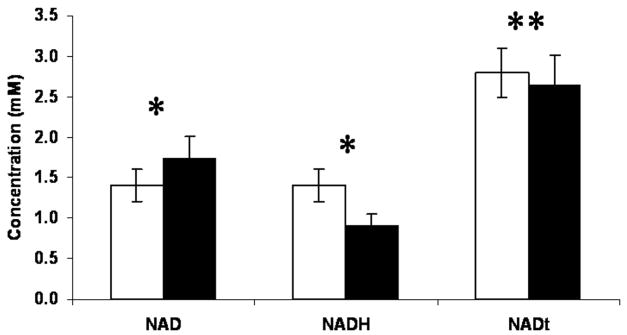
Figure 5 and the top row in Table 4 show concentrations (mM) of NAD and NADH in yeast grown in synthetic complete media containing 20 or 5 g/L glucose. HPLC analysis of extracts from the remaining protein pellet revealed no detectable NAD or NADH (data not shown). For growth in 20 g/L glucose (non-CR), intracellular NAD is present in concentrations of 1.39 ± 0.18 mM. This value increases significantly (p < 0.02, n ≥ 7 for each condition) to 1.74 ± 0.27 mM when cells are cultured in 5 g/L glucose (CR). NADH concentrations, however, significantly decrease (p < 0.02, n ≥ 7 for each condition) from 1.42 ± 0.19 mM under noncalorie restricted conditions to 0.91 ± 0.14 mM with CR. The total NAD and NADH concentration is not significantly different for both growth conditions at approximately 2.73 mM. The ratio of NAD to NADH increases significantly (p < 0.02, n ≥ 7 for each condition) from 1.0 for yeast cultured in 20 g/L glucose to 1.9 for yeast cultured in 5 g/L glucose.
Figure 5.
Metabolite concentrations (mM) of yeast grown in synthetic complete media containing 20 g/L glucose (normal) or 5 g/L glucose (CR). Data are presented as mean and SD of seven separate experiments for each culture condition. Student’s t-test assessments were used to determine whether NAD, NADH, and NAD + NADH are similar for the 20 and 5 g/L glucose growth conditions (*p < 0.02, **p < 0.10).
Table 4.
Comparison of intracellular metabolite concentrations (mM) in yeast
| Normal |
CR |
|||||||
|---|---|---|---|---|---|---|---|---|
| NAD | NADH | NAD/NADH ratio | NAD + NADH | NAD | NADH | NAD/NADH ratio | NAD + NADH | |
| Present studya | 1.39 (0.18) | 1.42 (0.19) | 1.00 (0.10) | 2.81 (0.32) | 1.74 (0.27) | 0.91 (0.14) | 1.94 (0.30) | 2.65 (0.37) |
| Lin et al. [10] (2004) | 1.26 (0.06) | 0.85 (0.13) | 1.48 | 2.11 | 1.19 (0.08) | 0.39 (0.11) | 3.05 | 1.58 |
| Anderson et al. [3] (2002)b | 2 | 0.78 | 2.56 | 2.78 | – | – | – | – |
| Lin et al. [11] (2001) | 2–3 | – | – | – | 2–3 | – | – | – |
| Ashrafi et al. [9] (2000)b | 1.14 | – | – | – | – | – | – | – |
| Smith et al. [12] (2000)b | 2.14 | – | – | – | – | – | – | – |
Data are presented as mean and SD of replicates (n ≥ 7).
Reported values were converted to concentrations (mM) as reported [10].
Intracellular NAD concentrations for normal growth conditions (i. e., 20 g/L glucose) have been reported between 1.14 and 2.14 mM [3, 9–12], which is consistent with our findings of 1.39 ± 0.18 mM. Although intracellular concentrations of NAD have been shown to decrease with increase in cell density [16], NAD and NADH measurements in this study were made on cell cultures grown to the same density (approximately 7×106 cells/mL); so the measured differences in intracellular concentrations of NAD and NADH are not attributable to changes in cell density. There are few published reports of measured NADH values in yeast presumably because of the difficulty in making measurements on such readily oxidizable molecules. NADH values obtained from the current method are nearly 40% higher than those obtained through enzymatic cycling assays [3, 10]. Cells grown under CR (i. e., 5 g/L glucose) have been reported to contain a similar range of NAD values as cells grown without CR [3, 10]. We measured calorie-restricted NAD concentrations of 1.74 ± 0.27 mM using the current method, which falls well within the published range. NADH concentrations obtained from yeast grown under CR have been reported at 0.39 ± 0.11 mM [10]. Using the current method for metabolite extraction, we measure 0.91 ± 0.14 mM NADH from calorie-restricted cells. However, similar to the study of Lin et al. [10], we found that under CR conditions, NADH contents are significantly reduced and that the NAD/NADH ratio significantly increases compared to yeast cultured in 20 g/L glucose.
Enzymatic cycling reactions for indirectly measuring both NAD and NADH typically rely on a heating step under either acidic or alkali conditions for cell lysis and preservation of either NAD (acidic) or NADH (alkali) [3, 9–12] during which redox reactions may continue thus altering the redox state. Enzymatic assays are also subject to a large loss (more than half) of the metabolite of interest during the harsh extraction conditions. Interestingly, during development of our assay, we observed that the choice of buffer the yeast are maintained in, both prior to and during bead beating, had a profound influence on intracellular NAD and NADH contents. Compared to bead beating in 50 mM ammonium acetate, bead beating yeast in an ACN/10 mM KH2PO4 buffer (3:1 v/v) resulted in oxidation of cellular NADH levels by as much as 90%, while bead beating yeast in a 50 mM KH2PO4 buffer resulted in reduction of cellular NADH levels by as much as 20%. Stored cellular NADH has been shown in various animal models to be rapidly utilized by cells under stressful conditions including oxidative stress [30] and high osmolarity [31]. Additionally, NADH has been shown to degrade more rapidly in KH2PO4 buffers than other buffers such as Pipes [32]. It is plausible that the lower NADH levels observed in the enzymatic assays and by bead beating in ACN are a result of a cellular stress response to the buffer. These observations suggest that it is crucial that exposure of intact cells to chemical or environmental stressors prior to, and during, cell lysis be minimized.
3.4 General observations
The work of Lazzarino et al. [22] served as a conceptual basis for the development of the metabolite extraction protocols reported here. Their work was developed for recovering a broad range of metabolites representative of both the redox and energy states of cells in animal tissue and utilized ACN and KH2PO4-containing buffers during both cell lysis and metabolite extraction [22]. To enable robust, reliable measurement of NAD and NADH redox states in yeast, it was necessary to develop significantly different cell lysis and metabolite extraction methods. ACN-containing buffers were avoided during cell lysis and KH2PO4-containing buffers were avoided throughout the entire protocol. Owing to their robust cell wall, bead-beating procedures were used to rapidly and efficiently lyse yeast. We also found that chloroform extraction performed on protein pellets, as proposed by Lazzarino et al. [22], did not increase the yields of NAD and NADH recovered from yeast. Elimination of this step reduced total sample processing time by at least 12 h. The extraction method reported here relies on rapid and efficient cell lysis and relatively benign chemical conditions prior to lysis and during metabolite extraction. These careful considerations likely minimize cellular use and degradation of NADH resulting in recovery of higher amounts of NADH.
The protocol reported here measures the total amount (i. e., free + bound) of NAD and NADH in yeast. Ratios of total NAD to total NADH in yeast seem to be within a reasonable range for NAD regulation [33]. However, baseline ratios of the free pool of NAD/NADH have been debated [19, 34–38]. The free pool NAD/NADH ratio has been estimated from concentrations of the intracellular metabolites, pyruvate and lactate, for various tissues in different species animal species, to range from 0.1 to 10 [34–38] or at least 20 for studies conducted using 13C NMR spectroscopy [19]. During development of the assay reported here, we found that yeast cell lysates that were passed through a 0.45 μm glass fiber filter, immediately after bead beating, possessed NADH levels that were around one sixth of those obtained from extractions of unfiltered lysates (data not shown). NAD levels, however, were not altered resulting in NAD/NADH ratios of approximately 7. This observation is consistent with the hypothesis that the majority of cellular NADH is likely membrane or protein bound, while the majority of NAD is not protein bound. In our protocol, bound NADH is likely effectively released from proteins after bead beating by the use of ACN, which is well known for its ability to denature proteins [39].
4 Concluding remarks
Minimization of artefactual oxidation of reduced metabolite species is challenging to achieve for any metabolite extraction protocol. Here, we have improved on previous approaches and developed a method for measuring NAD and NADH in the same sample of yeast using a simple extraction protocol that provides a steady-state view of redox states within yeast by minimizing and accounting for oxidation of the reduced species. RP-HPLC provided excellent speciation and resolution of NAD and NADH, as confirmed by MS, from other metabolites within a 25 min run time.
Results of this study and that of others [10] reveal that yeast NADH contents are significantly reduced under calorie restricted conditions while NAD/NADH ratios increase. It has been suggested that this shift may lead to an activation of Sir2, a sirtuin protein family member [4]. NADH has been shown to function as an inhibitor of Sir2 [10], so lower levels of NADH found in yeast cells grown under CR could result in less inhibition of Sir2 and therefore an increased lifespan in yeast [10]. By culturing yeast in 14C-labeled growth media, we are investigating the relative contributions of the de novo and salvage pathways to total NAD and NADH levels in yeast strains as a function of nutrient status [40]. The protocol developed here avoids derivitization or the measurement of degradation products from NAD or NADH so that the potential for loss of the isotope label is minimized. Initial radiolabeling of yeast cells and subsequent accelerator MS (AMS) measurement for 14C quantitation indicate that: (i) yeast cells are amenable to such labeling and quantification [40]; and (ii) the single sample extraction and HPLC speciation protocol developed here are amenable to AMS quantitation of 14C in isolated fractions of the HPLC trace [40] enabling us to quantify total NAD and NADH (by UV–Vis absorption) and the contribution of NAD and NADH from a specific pathway (by AMS) from the same sample.
Acknowledgments
Authors wish to thank Dr. Magnus Palmblad for performing some of the initial sample extraction and HPLC speciation development, Dr. Sang Soo Hah for performing the ESI MS measurements, and Dr. Giuseppe Lazzarino for useful discussions pertaining to metabolite analysis. This work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344 and was supported by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41 RR013461).
Abbreviations
- AMS
accelerator MS
- CR
calorie restriction
- NAD
nicotinamide adenine dinucleotide
- NADH
reduced nicotinamide adenine dinucleotide
Footnotes
The authors declared no conflict of interest.
References
- 1.Sandmeier JJ, Celic I, Boeke JD, Smith JS. Genetics. 2002;160:877–889. doi: 10.1093/genetics/160.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JS, Boeke JD. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 3.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 4.Lin SJ, Defossez PA, Guarente L. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 5.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 6.Garcia Soriano F, Virag L, Jagtap P, Szabo E, Mabley JG, Liaudet L, Marton A, Hoyt DG, Murthy KG, Salzman AL, Southan GJ, Szabo C. Nat Med. 2001;7:108–113. doi: 10.1038/83241. [DOI] [PubMed] [Google Scholar]
- 7.Greenamyre JT, Sherer TB, Betarbet R, Panov AV. IUBMB Life. 2001;52:135–141. doi: 10.1080/15216540152845939. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, Piston DW, Goodman RH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 9.Ashrafi K, Lin SS, Manchester JK, Gordon JI. Genes Dev. 2000;14:1872–1885. [PMC free article] [PubMed] [Google Scholar]
- 10.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin SS, Manchester JK, Gordon JI. J Biol Chem. 2001;276:36000–36007. doi: 10.1074/jbc.M103509200. [DOI] [PubMed] [Google Scholar]
- 12.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nemoto S, Fergusson MM, Finkel T. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 14.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubmeyer CT, Gross JW, Rajavel M. Meth Enzymol. 1999;308:28–48. doi: 10.1016/s0076-6879(99)08004-0. [DOI] [PubMed] [Google Scholar]
- 16.Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C. Cell. 2007;129:473–484. doi: 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 17.Bieganowski P, Brenner C. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- 18.Tempel W, Rabeh WM, Bogan KL, Belenky P, Wojcik M, Seidle HF, Nedyalkova L, Yang T, Sauve AA, Park HW, Brenner C. PLoS Biol. 2007;5:263. doi: 10.1371/journal.pbio.0050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson RM, Latorre-Esteves M, Neves AR, Lavu S, Medvedik O, Taylor C, Howitz KT, Santos H, Sinclair DA. Science. 2003;302:2124–2126. doi: 10.1126/science.1088697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turteltaub KW, Vogel JS. Curr Pharm Des. 2000;6:991–1007. doi: 10.2174/1381612003400047. [DOI] [PubMed] [Google Scholar]
- 21.Klaidman LK, Leung AC, Adams JD., Jr Anal Biochem. 1995;228:312–317. doi: 10.1006/abio.1995.1356. [DOI] [PubMed] [Google Scholar]
- 22.Lazzarino G, Amorini AM, Fazzina G, Vagnozzi R, Signoretti S, Donzelli S, Di Stasio E, Giardina B, Tavazzi B. Anal Biochem. 2003;322:51–59. doi: 10.1016/j.ab.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Pogolotti AL, Jr, Santi DV. Anal Biochem. 1982;126:335–345. doi: 10.1016/0003-2697(82)90524-3. [DOI] [PubMed] [Google Scholar]
- 24.Evans J, Wang TC, Heyes MP, Markey SP. Anal Biochem. 2002;306:197–203. doi: 10.1006/abio.2002.5715. [DOI] [PubMed] [Google Scholar]
- 25.Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, Zhao W, Thiyagarajan M, MacGrogan D, Rodgers JT, Puigserver P, Sadoshima J, Deng H, Pedrini S, Gandy S, Sauve AA, Pasinetti GM. J Biol Chem. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- 26.Sauve AA, Moir RD, Schramm VL, Willis IM. Mol Cell. 2005;17:595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 27.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman F, Fink GR, Lawrence CW. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1978. [Google Scholar]
- 29.Sherman F. Meth Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 30.Navas P, Villalba JM, de Cabo R. Mitochondrion. 2007;7:S34–S40. doi: 10.1016/j.mito.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Kaeberlein M, Andalis AA, Fink GR, Guarente L. Mol Cell Biol. 2002;22:8056–8066. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rover Junior L, Fernandes JC, de Oliveira Neto G, Kubota LT, Katekawa E, Serrano SH. Anal Biochem. 1998;260:50–55. doi: 10.1006/abio.1998.2656. [DOI] [PubMed] [Google Scholar]
- 33.Lin SJ, Guarente L. Curr Opin Cell Biol. 2003;15:241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 34.Gaikwad A, Long DJ, II, Stringer JL, Jaiswal AK. J Biol Chem. 2001;276:22559–22564. doi: 10.1074/jbc.M101053200. [DOI] [PubMed] [Google Scholar]
- 35.MacDonald MJ, Marshall LK. Arch Biochem Biophys. 2000;384:143–153. doi: 10.1006/abbi.2000.2107. [DOI] [PubMed] [Google Scholar]
- 36.Mongan PD, Capacchione J, West S, Karaian J, Dubois D, Keneally R, Sharma P. Am J Physiol Heart Circ Physiol. 2002;283:H1634–H1644. doi: 10.1152/ajpheart.01073.2001. [DOI] [PubMed] [Google Scholar]
- 37.Ramasamy R, Trueblood N, Schaefer S. Am J Physiol. 1998;275:H195–H203. doi: 10.1152/ajpheart.1998.275.1.H195. [DOI] [PubMed] [Google Scholar]
- 38.Sanni LA, Rae C, Maitland A, Stocker R, Hunt NH. Am J Pathol. 2001;159:1105–1112. doi: 10.1016/S0002-9440(10)61786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang JY. J Biol Chem. 1999;274:123–128. doi: 10.1074/jbc.274.1.123. [DOI] [PubMed] [Google Scholar]
- 40.Vogel JS, Palmblad NM, Ognibene T, Kabir MM, Buchholz BA, Bench G. Nucl Instrum Methods Phys Res, Sect B. 2007;259:745–751. [Google Scholar]